Abstract
Rolling circle amplification (RCA) and loop mediated isothermal amplification (LAMP) were combined to establish the rolling circle and loop mediated isothermal amplification (RC-LAMP) method for miRNA detection. With the participation of Bst 2.0 DNA Polymerase, the method enabled RCA and LAMP amplification to occur simultaneously without thermal cycling. The limit of detection of RC-LAMP was 500 amol/L, which is comparable to previously reported amplification strategies. Moreover, its upper limit of quantitation was higher and showed a stronger resistance to matrix interference. Therefore, it is possible to detect low concentrations of miRNA in samples by increasing the total RNA added. Owing to its facile detection mode and simple operation, this method has great potential in clinical sample detection.
1. Introduction
MicroRNAs (miRNAs) are small, non-coding RNAs with 19–24 nucleotides, which regulate two thirds of the protein-coding genes in humans [1]. MiRNAs have been confirmed to be involved in a variety of cellular functions and biological and pathological processes, including the expansion, differentiation, metabolism and apoptosis of cancer cells [2]. As a regulator of many cancer-related genes, miRNAs are expected to become biomarkers for the diagnosis and prognosis of cancer [3]. A miRNA has the functions of binding to functional proteins, regulating mitochondria-related gene mRNAs, directly activating gene transcription and targeting other non-coding RNAs [4]. Based on the above biological functions, miRNAs become a promising therapeutic drug in a variety of pathological processes. Thus, it is imperative to establish sensitive and selective miRNA detection strategies for the sake of medical applications or to understand their functions. However, miRNAs are still challenging analytical targets on account of their small size, sequence homology among family members and low abundance [5].
In recent years, some new technologies, such as assays based on isothermal nucleic acid amplification, have been applied to detect miRNAs [6,7,8,9,10,11,12,13,14]. Among them, rolling circle amplification (RCA) has become more popular, owing to its specificity and simplicity [7,13,14,15,16,17,18,19,20,21,22,23,24]. The miRNA always serves as a ligation template, which is hybridized with the padlock probe. Then, this DNA complex will be ligated by an enzyme to form a circular single-stranded DNA (ssDNA). An external primer or miRNA itself works as primer to extend on the circular ssDNA, and eventually displace the linked miRNA, then long cascaded products, which are the repeated sequences from circular ssDNA, are produced [14,15,17,18,19,22,23,24,25,26,27]. However, the detecting of RCA products is exactly laborious and time-consuming, although several kinds of methods based on RCA were designed for simple and specific miRNA expression analysis including MNAzyme-mediated [13], nicking-mediated [17,27,28] and dumbbell probe-mediated RCA [29]. However, the sensitivity of these RCA-based assays is still generally unsatisfactory, with low levels of miRNA detection, which limits its biomedical applications. Recently, a miRNA quantification method named rolling circle-quantitative PCR (RC-qPCR) was developed by our group, where the Vent (exo-) DNA Polymerase was firstly utilized to combine RCA and qPCR in one step, due to its high thermostability, which brought high sensitivity and specificity [30]. Unfortunately, this technique requires variable temperature amplification and special laboratory skills, which restricts its wide applications.
Loop mediated isothermal amplification (LAMP) is another novel technique that is suitable for genetic diagnosis with high efficiency. In regular protocol of LAMP, four different primers are used to identify six different sequence areas, which brings an excellent selectivity, while the sensitivity is improved by the exponential amplification feature. In most LAMP-based assays, the target miRNAs serve as triggers to activate the reaction, only in the presence of which can the primers be extended by the strand displacement DNA synthesis function of DNA polymerase [31]. The target-triggered LAMP (TT-LAMP) with a simple probe design remarkably improved detection sensitivity [32]. In ligation-based LAMP (ligation-LAMP), the high fidelity of ligase in discriminating single-base mismatches can be applied to improve the specificity [33]. Real-Time LAMP (RT-LAMP) applying a new electrochemical DNA chip enables a multiplex LAMP reaction in a simple device that is easy to operate [34]. These technologies have the characteristics of high specificity and sensitivity, and have been widely used in the diagnosis of clinical diseases and the qualitative and quantitative detection of epidemic bacteria of viruses. In LAMP-based miRNA detection, as the LAMP template contains 4–6 pre-determined stem-loop sequences, the design of the probes can be quite complicated [25]. Another shortcoming of LAMP is the use of indirect signal detection methods, such as precipitation and dye, which are ineffective to discriminate target products from nonspecific products.
In this paper, we designed a facile and ultrasensitive strategy for miR-200a detection by combining the benefits of RCA with high specificity and LAMP with high sensitivity (refers to RC-LAMP strategy). Periodontal disease is one of the most prevalent infectious disease in humans and it is the main cause of tooth loss. Increasing evidence implicates that periodontitis may link to multiple systemic diseases. In miRNA profiling studies of periodontal disease tissues, the miR-200 family has been consistently identified as a potential candidate miRNA biomarker in periodontitis-related processes [35,36]. In this RC-LAMP assay, the target miR-200a worked as templates for the ligation of padlock probe. Then, the RCA reaction was initiated by the backward inner primer (BIP) from LAMP, bringing tandem ssDNA which was long and repeated. The cascade extension and strand displacement reactions along the RCA product (similar to LAMP process) was carried out with the forward inner primer (FIP) and forward outer primer (F3). As the reaction went on, many double hairpin structures with different stem lengths would be produced, which could independently initiate subsequent LAMP process. Thus, compared with RCA or the LAMP method, the RC-LAMP method greatly improves detection sensitivity. By utilizing the proposed RC-LAMP, even miRNA targets at 1 fM can be definitely detected. Bst 2.0 DNA Polymerase with strand displacement activity is used in this assay to enable the RCA and LAMP to be performed simultaneously without the need for temperature changing equipment, which simplifies the experimental operation and avoids the possibility of contamination caused by repeated opening of the tube.
2. Materials and Methods
2.1. Apparatus
The real-time fluorescence measurements of the RC-LAMP were performed on an Applied Biosystem StepOnePlus real-time PCR instrument (Life Technologies, Inc., Carlsbad, CA, USA).
2.2. Materials and Reagents
Bst 2.0 DNA Polymerase (New England Biolabs, Ipswich, MA, USA) was purchased from New England Biolabs. Betaine solution (Sigma-Aldrich, St. Louis, MO, USA) and EvaGreen (Biotium, Fremont, CA, USA) were supplied by Merck. dNTP Set 100mM Solutions (Thermo Scientific, Waltham, MA, USA) were bought from Thermo Fisher Scientific. Diethyl pyrocarbonate (DEPC) treated deionized water was purified using an Elga “Purelab Option” Water System. A miRNeasy Mini Kit and miRNeasy Serum/Plasma Advanced Kit (QIAGEN, Hilden, Germany) were purchased from QIAGEN. The miRNAs were acquired from Shanghai GenePharma Co., Ltd. (Shanghai, China). Oligonucleotides including padlock probe, FIP, BIP and F3 were synthesized and purified by Sangon Biotech (Shanghai, China) and had the following sequences (Table 1).

Table 1.
miRNA and DNA sequences used in this work.
2.3. Preparation of Solution
LAMP primer mix solution: 5 μL F3 (100 μM), 20 μL FIP (100 μM), 20 μL BIP (100 μM) and 55 μL water were mixed together to get the final concentration of F3 to 5 μM and FIP and BIP to 20 μM. This solution was stored at −20 °C.
2.4. RC-LAMP Assay Procedures
The ligation of the padlock probe is firstly introduced. Briefly, 5 μL target the miRNA sample and 1 μL 20 pM padlock probe in T4 buffer (66 mM Tris-HCl, 6.6 mM MgCl2, 10 mM DTT and 100 μM ATP, pH 7.6) were mixed and preheated for 3 min at 65 °C, then cooled down to room temperature. T4 DNA Ligase (70 U) was added (with the solution up to 10 μL) and incubated at 37 °C for 45 min to accomplish the ligation reaction and then heated at 65 °C for 10 min to deactivate the enzyme (Table S1).
Then, the ligation products are amplified by RCA using BIP as a primer. Exactly 1 μL of the ligation products were pipetted into a pre-degeneration mixture containing 1 μL LAMP primer mix, 0.4 mM dNTPs, 1 M betaine and 2 mM MgSO4 into a final volume of 20 μL, then the mixture was pre-degenerated at 95 °C for 5 min and kept on ice for another 2 min. A 5 μL solution containing Bst 2.0 DNA Polymerase (6 U) and 1.25 μL 20× EvaGreen was added into pre-degeneration solution, then the 25 μL mixture was put into the StepOnePlus real-time PCR instrument to conduct the RC-LAMP at 62 °C. The fluorescence intensity was real-time monitored at intervals of 3 min (Table S2).
2.5. Cell Lysis and RNA Preparation
Human colon cancer cell HT-29 was purchased from the Cell Bank of Chinese Academy of Sciences under catalog number TCHu 103. HT-29 was firstly centrifuged at 3000 rpm for 5 min in culture medium, then washed with PBS buffer once and spun down with centrifugation for 5 min (3000 rpm). A miRNeasy Mini Kit was used for total RNA extraction from cells following the manufacturer’s quick-start protocol. The serum sample was obtained using a miRNeasy Serum/Plasma Advanced Kit to extract miRNA. The miR-200a sample was kept at −80 °C.
2.6. Detection of miRNA in Cell and Serum
The cells chosen were human colorectal cancer cell HT-29 (with high expression of miR-200a). The total RNA from the cells were extracted and added at a concentration not exceeding 1000 ng. The serum samples were collected from the patients who have severe periodontal disease in the dental clinic of Sir Run Run Shaw hospital, Zhejiang University. The related clinical data were accessed with appropriate ethical approvals. The POI value (the point of inflection value) referred to the time when the amplification curve reaches the maximum slope, which was used for the quantitative calculation of the LAMP reaction. The concentration of miRNA was calculated by substituting POI value into the accompanying standard curve.
3. Results and Discussion
3.1. Design Principle of RC-LAMP Assay
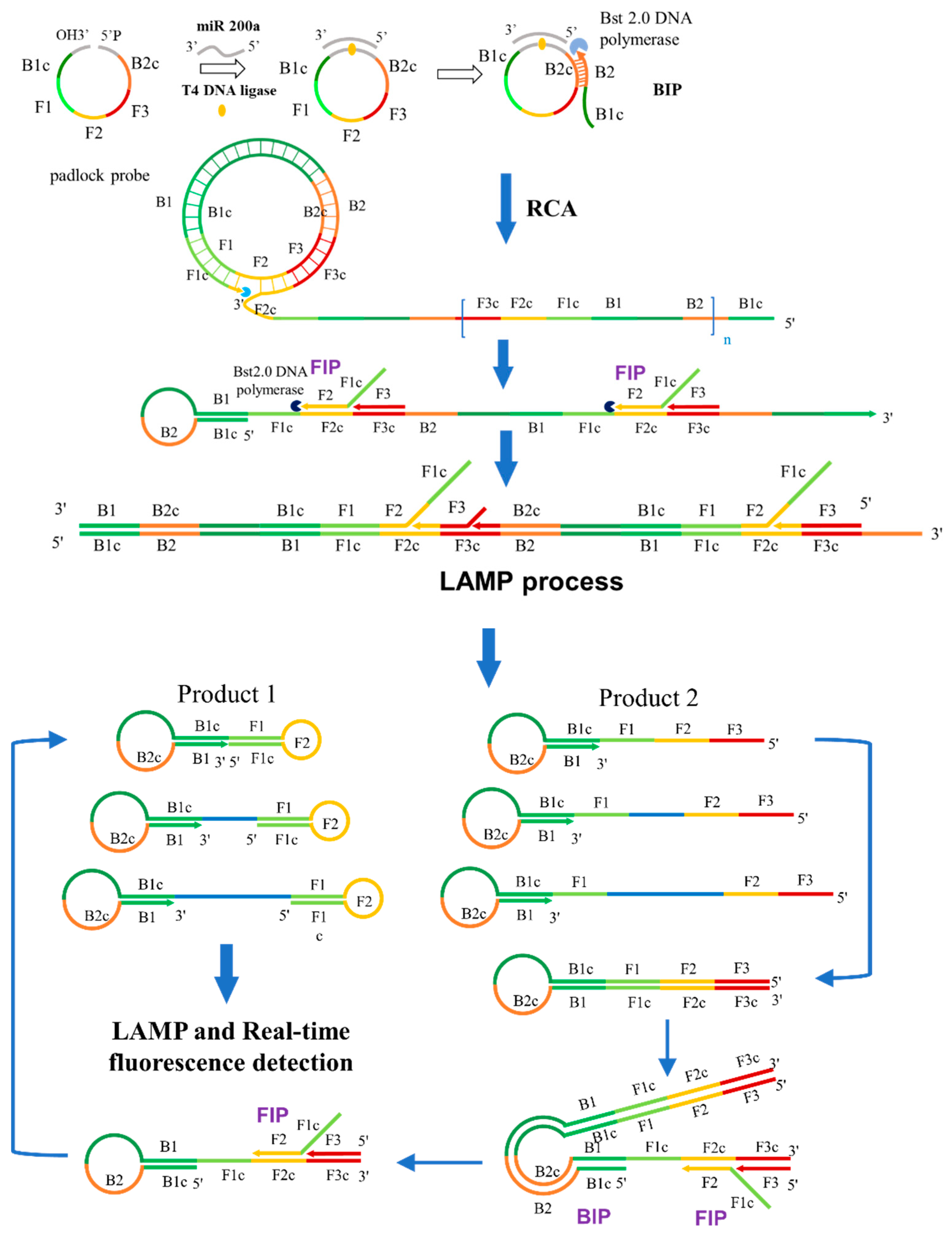
The schematic representation of miRNA detection by RC-LAMP method is shown in Scheme 1. MiR-200a will hybridize to 5′ and 3′ ends of padlock probe (containing B1c, F1, F2, F3 and B2c sequence area from LAMP), then this DNA complex would be ligated by T4 DNA Ligase to form a circular ssDNA, a substrate for RCA. The RC-LAMP reaction is carried out in one step with the participation of circular ssDNA, LAMP primer (FIP, BIP and F3) and Bst 2.0 DNA Polymerase. Firstly, the BIP which contains B2 and B1c (B1 complementary) sequences bind to the B2c (B2 complementary) sequence on the circular ssDNA to initiate the RCA reaction under the action of Bst 2.0 DNA Polymerase with strand displacement activity, creating a single-stranded repeat sequence. Then, the internal primer (FIP) and the external primer (F3) are hybridized with this single stranded product, forming amplicons of various stem-loop structures by amplification and strand displacement, which is further amplified by following LAMP process. The final amplification products bind to the double-stranded fluorescent dye EvaGreen to produce a fluorescent signal, which can be detected to quantify the target miRNA.
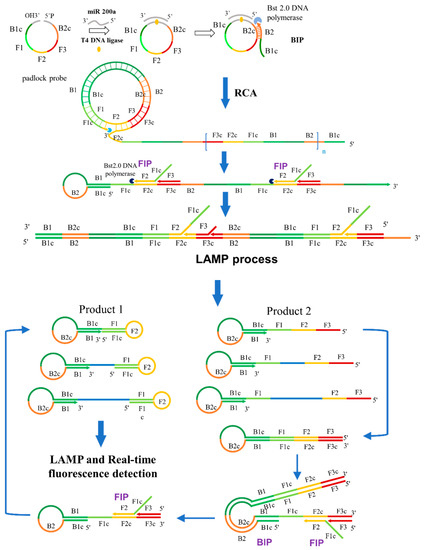
Scheme 1.
Schematic representation of the RC-LAMP method for miRNA detection.
3.2. Optimization of Reaction Parameters
Several reaction parameters including the concentration of padlock probe, FIP/BIP and amplification temperature were systematically optimized to get the optimal experimental conditions for RC-LAMP. We evaluate each experimental parameter by ∆POI value (POI sample—POI negative control), and take ∆POI maximum value as the optimal experimental condition.
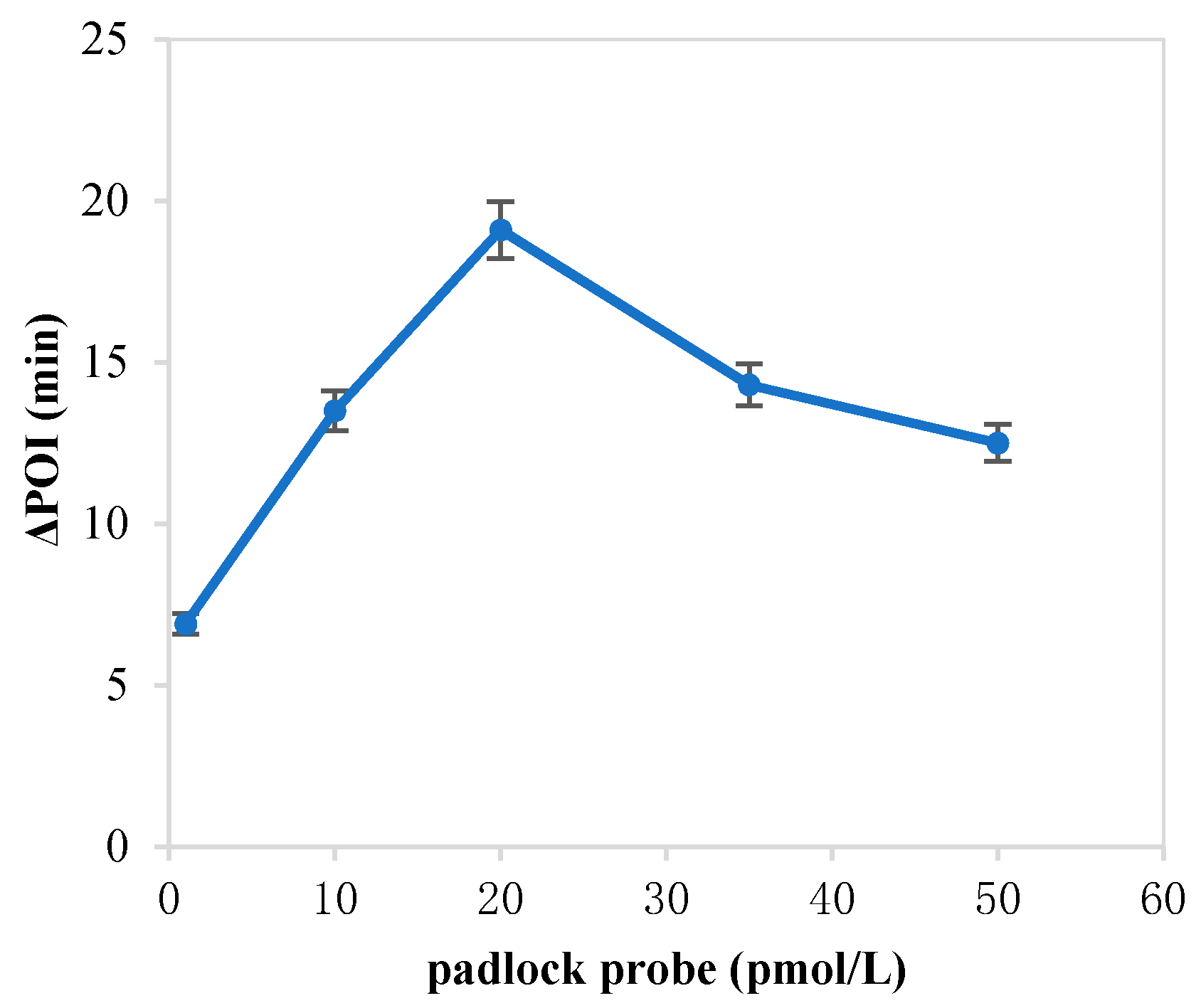
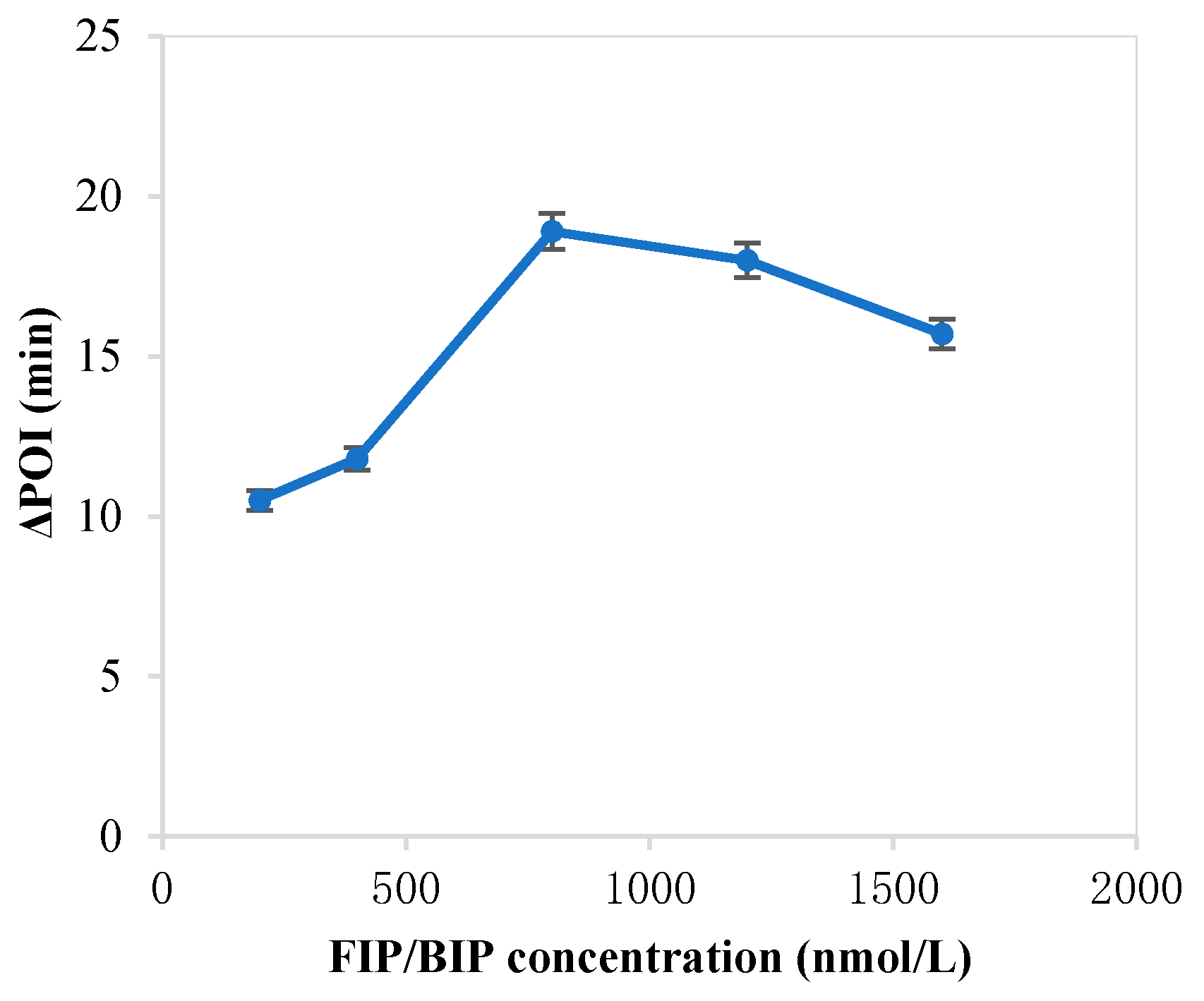
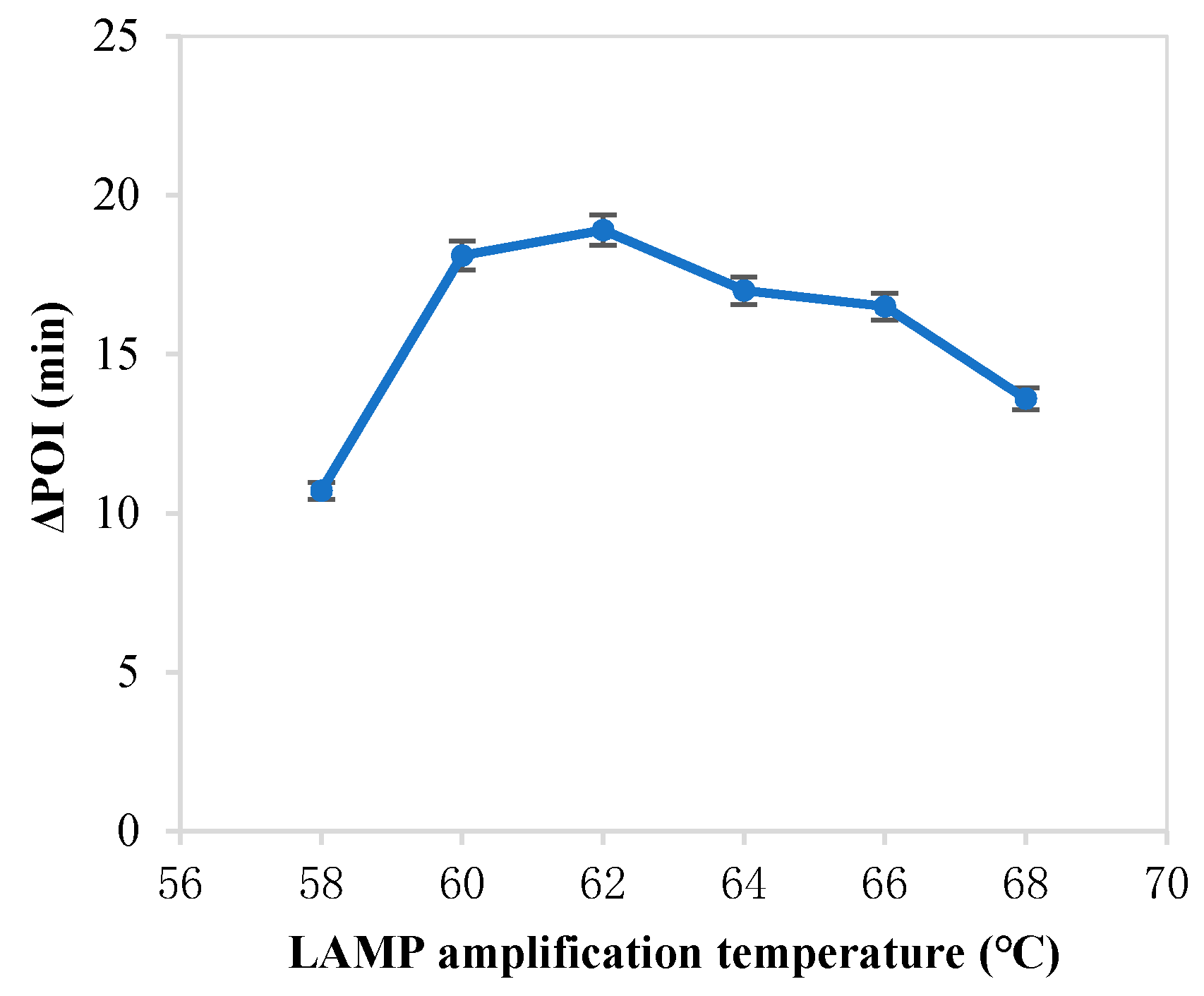
The forming of circular ssDNA needed the help of padlock probe; BIP and FIP also played an important role in our RC-LAMP method. Thus, we investigated various concentrations of padlock probe, BIP and FIP to screen the optimal concentration. As shown in Figure 1, the maximum ∆POI value was obtained using the 20 pM padlock probes. Figure 2 showed that suitable concentration of FIP/BIP were 800 nM. Then, different amplification temperatures for the RC-LAMP were tested, and 62 °C was chosen as the reaction temperature (Figure 3).
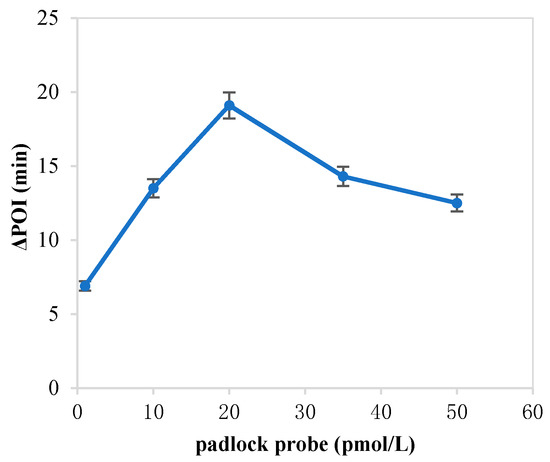
Figure 1.
The optimization of the concentration of padlock probe on the RC-LAMP assay. The graph shows the ΔPOI value vs. the concentration of padlock probe. Experimental conditions: miR-200a 1 pmol/L, T4 DNA ligase 70 U, FIP/BIP 800 nmol/L and amplification temperature 62 °C, respectively.
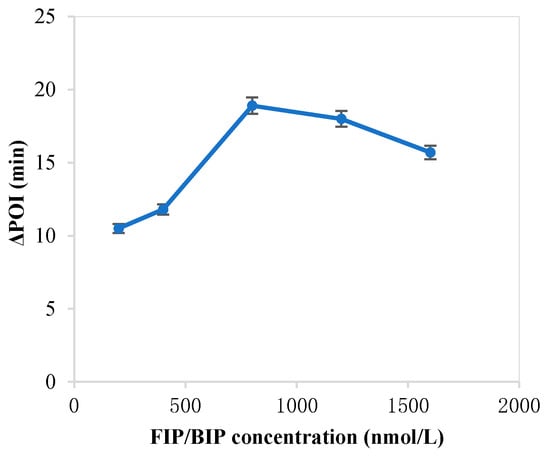
Figure 2.
The optimization of the concentration of FIP/BIP on the RC-LAMP assay. The graph shows the ΔPOI value vs. the concentration of FIP/BIP. Experimental conditions: padlock probe 20 pmol/L, miR-200a 1 pmol/L, T4 DNA ligase 70 U and amplification temperature 62 °C, respectively.
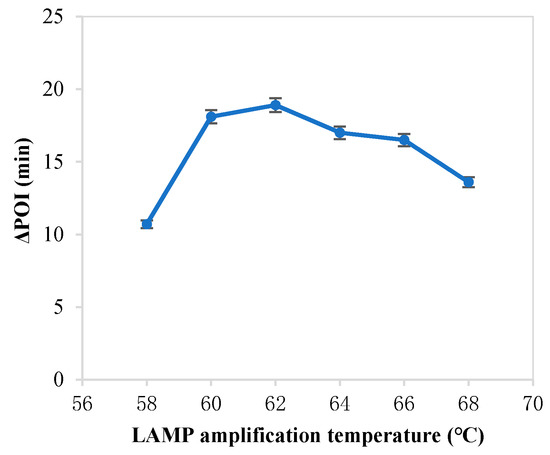
Figure 3.
The effect of the LAMP amplification temperature on the RC-LAMP assay. The graph shows the ΔPOI value vs. the LAMP amplification temperature. Experimental conditions: padlock probe 20 pmol/L, miR-200a 1 pmol/L, T4 DNA ligase 70 U and FIP/BIP 800 nmol/L, respectively.
3.3. Selectivity
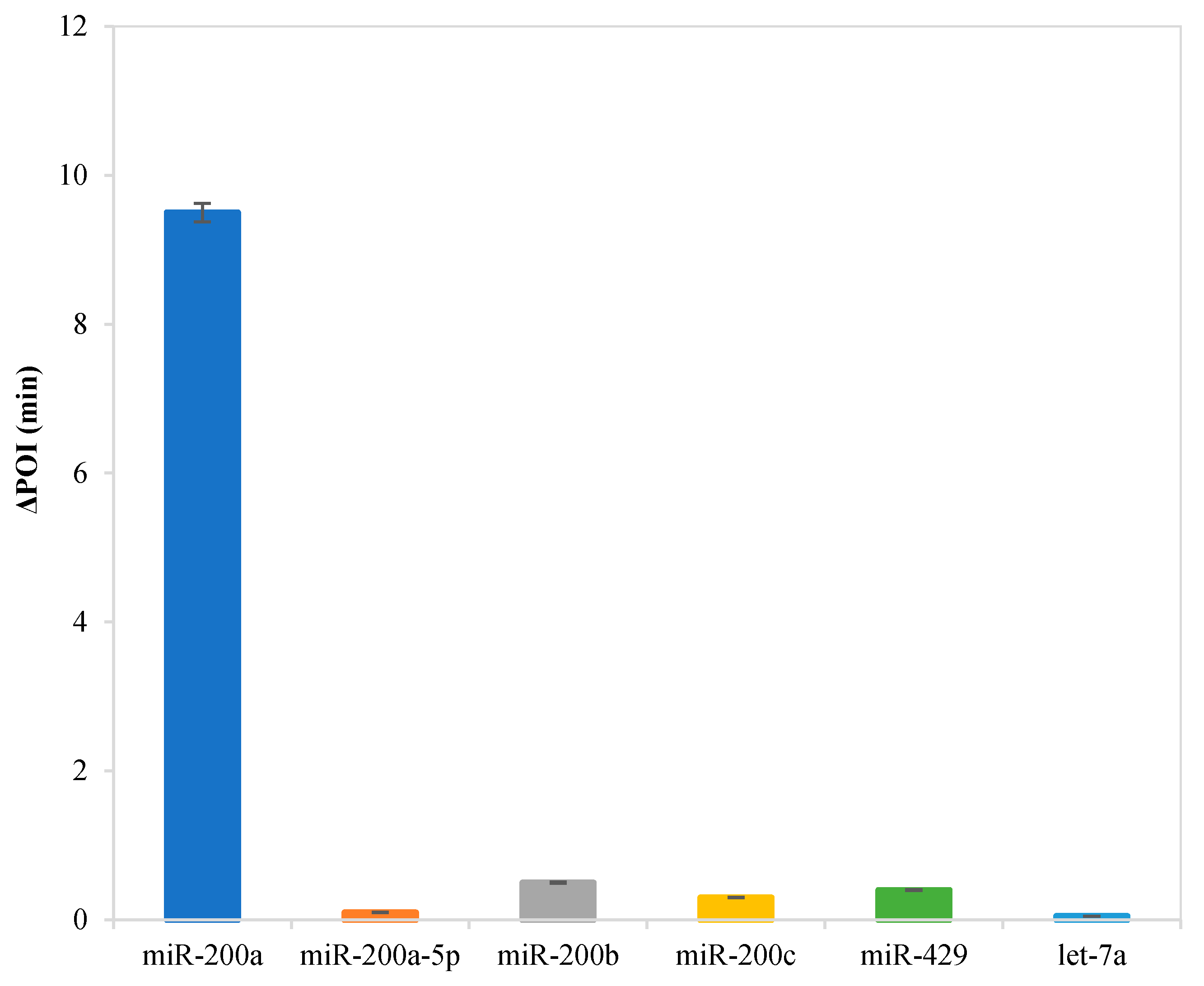
In order to evaluate the selectivity of this RC-LAMP method for miR-200a analysis, some other miRNAs that have high sequence homology with miR-200a, such as miR-200a-5p, miR-200b, miR-200c, miR-429 and let-7a, were selected to perform interference assays under identical conditions. The target and interfering miRNAs are detected by the RC-LAMP strategy using miR-200a specific padlock probe, and all miRNAs are at the same concentration (10 fM). The result was examined by detecting the ∆POI value, which was the POI value difference of blank and target. The higher the ∆POI value, the better the response of the miRNA to detection method. As shown in Figure 4, the ∆POI value of other miRNAs is much lower than target miR-200a, meaning interferences of other miRNAs are almost negligible. Our RC-LAMP method has excellent selectivity for miRNA detection.
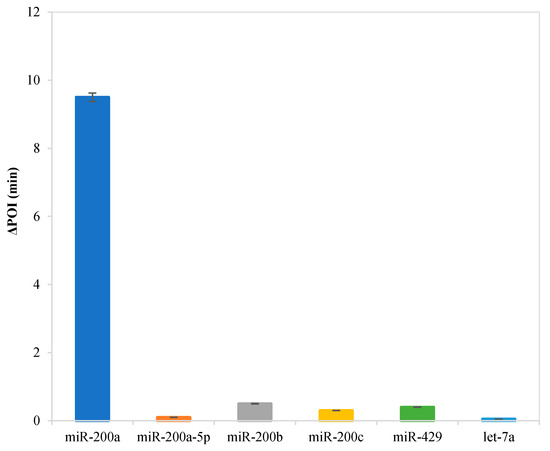
Figure 4.
Selectivity evaluation of the RC-LAMP assay for miRNA detection. Experimental conditions: padlock probe 20 pmol/L, miR-200a / mismatch miRNA 10 fmol/L, T4 DNA ligase 70 U, FIP/BIP 800 nmol/L and amplification temperature 62 °C, respectively.
3.4. Quantification of miRNA
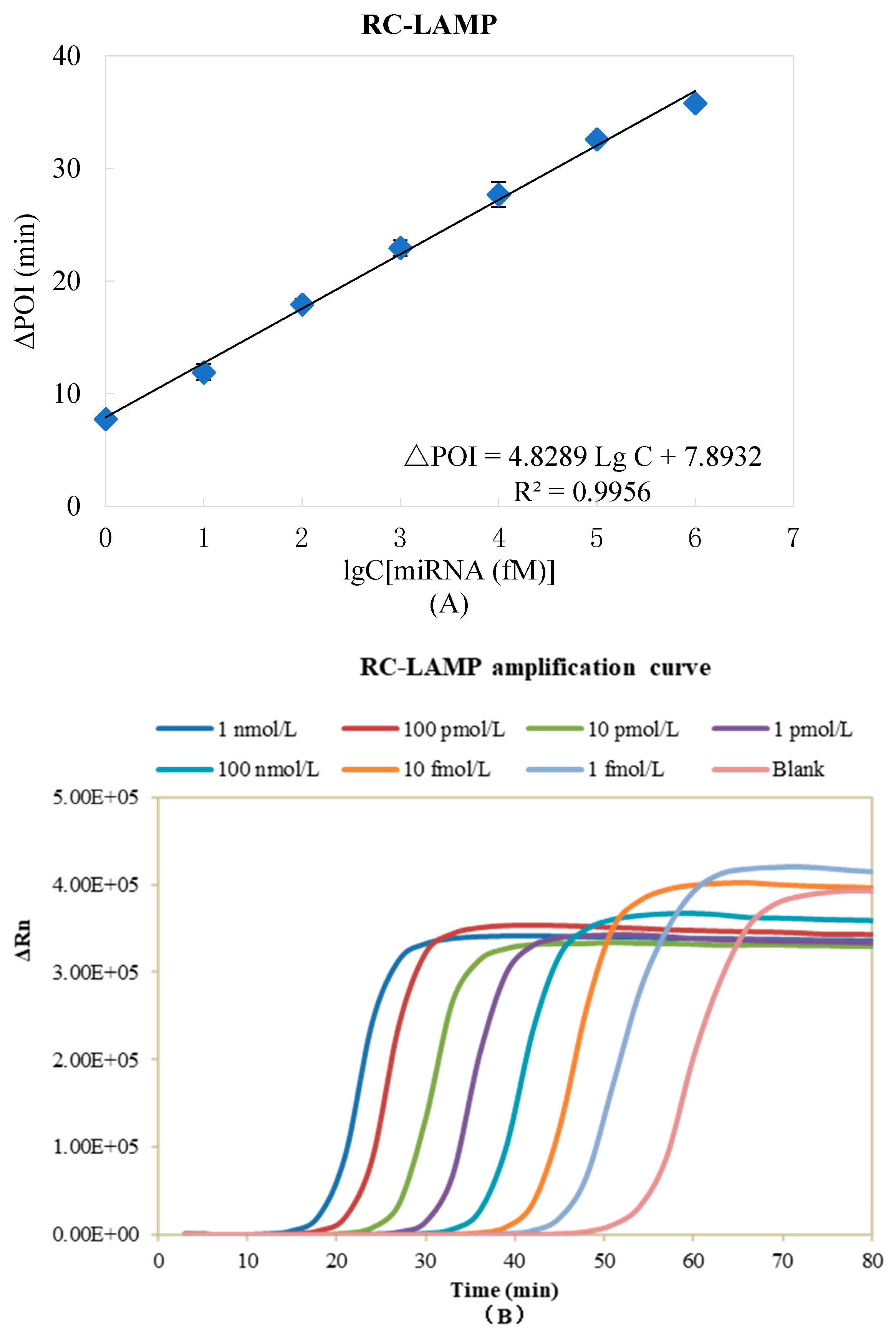
The analytical performance of the RC-LAMP method is estimated under the optimized reaction parameters. The ∆POI value was recorded and used to quantitatively determine the miR-200a level. As shown in Figure 5A, a linear relationship was observed with the logarithm (lg) of the miR-200a concentrations in the range of 1 fM to 1 nM to the ∆POI value. The correlation equation was ∆POI = 4.8289 Lg C + 7.8932, (R2 = 0.9956, C was the concentration of miR-200a). The real-time fluorescence signal curves were shown in Figure 5B, and the signal aroused by 1 fM miR-200a can be clearly discriminated from that of the blank control. Table S3 displays the limit of detection of this method (500 aM), which was similar to our RC-qPCR for miRNA detection reported earlier [30]. The sensitivity of the proposed method is comparable to previously reported amplification strategies for miRNA detection, as summarized in Table S4.
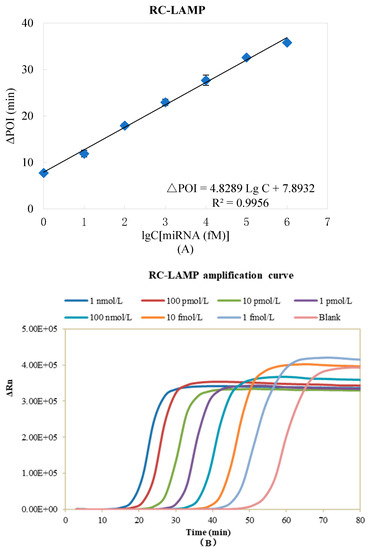
Figure 5.
Standard curve of miR-200a detected by RC-LAMP method. (A) ΔPOI values vs. logarithm (lg) of miR-200a concentrations. (B) Real-time fluorescence curves vs. miR-200a concentrations. Experimental conditions: Padlock probe 20 pmol/L, T4 DNA ligase 70 U, FIP/BIP 800 nmol/L and amplification temperature 62 °C, respectively.
The analytical sensitivity, precision and accuracy of this assay investigated referring to the requirements in the MIQE (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) guidelines. For the sensitivity investigation, miR-200a was diluted to a concentration of 1 fM, 750 aM, 500 aM, 250 aM and 100 aM, respectively. The samples without miRNA were used as negative control, and 10 samples were detected in parallel for each concentration. As shown in Table S3, the sensitivity of RC-LAMP is 500 aM. In order to investigate the precision and accuracy, the miR-200a concentration was set to 1 fM (the lower limit of quantification), 50 fM (low concentration), 5 pM (medium concentration) and 500 pM (high concentration). The samples without miRNA were used as negative control to measure the ΔPOI value of each concentration of miR-200a, and each concentration was tested in parallel five times. The accuracy was expressed in (measured value/true value) × 100% while the precision was expressed in relative standard deviation. The precision of RC-LAMP was 3.47%, 18.2%, 6.24% and 1.81% at the lower limit of quantification and low, medium and high concentration, respectively, while the accuracy was 108%, 89.9%, 113% and 99.4%, respectively. The experimental results indicated that the RC-LAMP method had better precision and accuracy.
3.5. Real Sample Detection
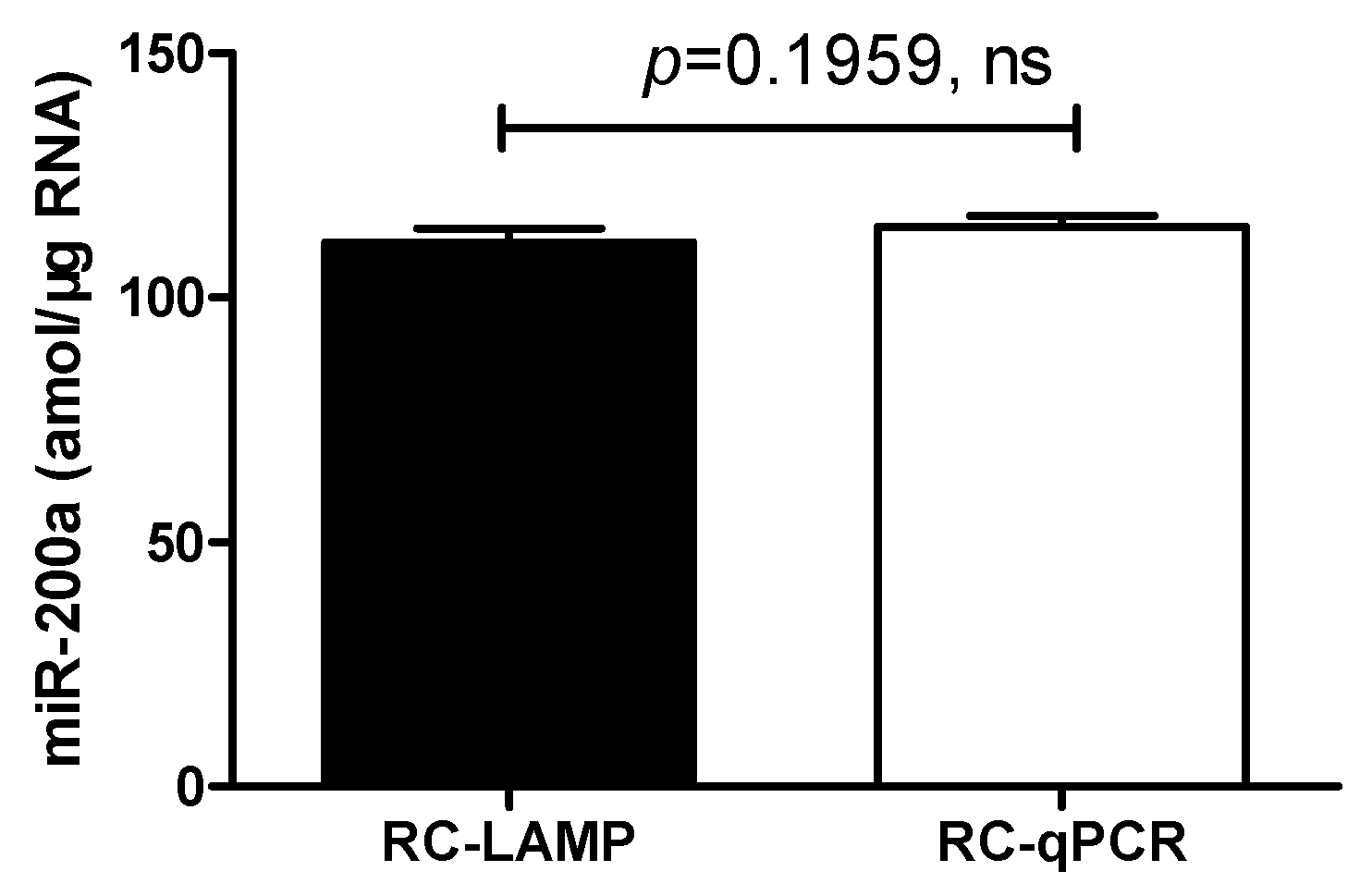
To evaluate the potential of the RC-LAMP method in real sample analysis, the quantification of miR-200a in total RNA, which was extracted from HT-29, was further applied. The content of miR-200a was 111 ± 2.78 amol/μg, a result that was of no significant difference (p = 0.1959) from either the RC-LAMP assay or the RC-qPCR (115 ± 2.21 amol/μg) method reported earlier [2,9] (Figure 6).
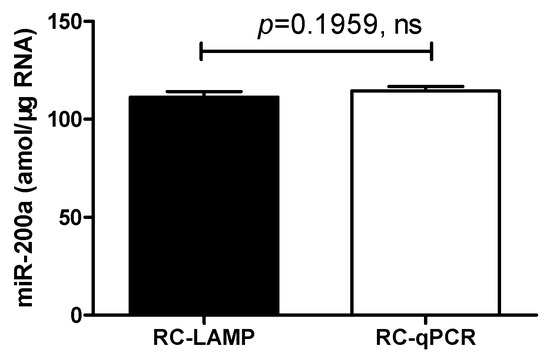
Figure 6.
Comparison with RC-LAMP method and RC-qPCR method applied to miR-200a detection in HT-29 cells. There was no significant difference between RC-LAMP and RC-qPCR method. Experimental conditions (RC-LAMP): padlock probe 20 pmol/L, T4 DNA ligase 70 U, FIP/BIP 800 nmol/L and amplification temperature 62 °C. Experimental conditions (RC-qPCR): padlock probe 20 pmol/L, T4 DNA ligase and Vent (exo-) DNA Polymerase 70 U and 0.2 U, forward and reverse primer 100 nmol/L and annealing temperature 56 °C, respectively.
Moreover, a spike/recovery experiment was carried out by adding certain amount of target RNA (final concentration was 1 pM) to the diluted serum, which was regarded as the sample detected by RC-LAMP method. The recoveries of miR-200a in 250 ng, 500 ng, 1000 ng, 1500 ng and 2000 ng total RNA were 97.3 ± 1.05%, 97.4 ± 1.45%, 98.6 ± 2.97%, 81.3 ± 4.53% and 62.0 ± 3.53%, respectively (Table S5). It is consistent with the recovery of biological sample testing standards when the total RNA concentration is less than 1000 ng.
4. Conclusions
In summary, an ultrasensitive RC-LAMP strategy for miRNA detection was established, coupled with RCA for high specificity and LAMP for efficient amplification. It displays superior detection sensitivity with a detection limit of 500 aM. The sensitivity of the RC-LAMP assay is comparable to the traditional RCA or LAMP-based miRNA assays, showing a wider range of quantification. At the same time, the RC-LAMP assay is more resistant to matrix interference than our RC-qPCR method reported before. Therefore, it is possible to detect lower concentrations of miRNA in samples by increasing the total RNA added. In addition, this method used Bst 2.0 with strand displacement activity as a DNA polymerase, enabling RCA and LAMP to be performed simultaneously without the need for precise temperature changing equipment, simplifying experimental operations and avoiding the possibility of contamination caused by repeated tube opening. In our study, the target miR-200a works as a linker for RCA process. When this method is used to detect other miRNAs, only the complementary sequence for target in padlock probe needs to be modified, which provides a new idea for miRNA detection. Owing to its facile detection mode and simple operation, this method has great potential in clinical sample detection.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/separations8100166/s1, Table S1: Pre-ligation system, Table S2: LAMP system, Table S3: The analytical sensitivity of the RC-LAMP method (n = 10), Table S4: Comparison of sensitivity for different miRNA assay methods, Table S5: The validation of the matrix effect (n = 3).
Author Contributions
Conceptualization, Z.C. and X.J.; methodology, G.X. and Z.C.; validation, Z.C., X.J. and G.X.; writing—original draft preparation, Z.C. and M.X.; writing—review and editing, H.L. and S.C.; supervision, S.C.; funding acquisition, S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant 81673399), the Zhejiang Provincial Natural Science Foundation of China (LY18H300003), the Fundamental Research Funds for the Central Universities (2019FZA7017).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Sir Run Run Shaw Hospital (protocol code 20210504-38, 2021.5.4).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farooqi, A.A.; Qureshi, M.Z.; Coskunpinar, E.; Naqvi, S.K.-U.-H.; Yaylim, I.; Ismail, M. miR-421, miR-155 and miR-650: Emerging trends of regulation of cancer and apoptosis. Asian Pac. J. Cancer Prev. 2014, 15, 1909–1912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, M.A.; Arora, S.; Prakasam, G.; Calin, G.A.; Syed, M.A. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol. Asp. Med. 2019, 70, 3–20. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Huang, Y.; Zou, Q.; Zou, Q.; Wang, S.P.; Wang, S.P.; Tang, S.M.; Tang, S.M.; Zhang, G.Z.; Zhang, G.Z.; et al. The discovery approaches and detection methods of microRNAs. Mol. Biol. Rep. 2011, 38, 4125–4135. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Cao, Z.J.; Lau, C.W.; Lu, J.Z. Label-free technology for the amplified detection of microRNA based on the allosteric hairpin DNA switch and hybridization chain reaction. Analyst 2014, 139, 6022–6027. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.J.; Tang, L.H.; Tian, Q.Q.; Wang, Y.; Lin, L.; Li, J.H. Toehold-initiated Rolling Circle Amplification for Visualizing Individual MicroRNAs In Situ in Single Cells. Angew. Chem. 2014, 53, 2389–2393. [Google Scholar] [CrossRef]
- Huang, J.F.; Zhao, N.; Xu, H.Q.; Xia, H.; Wei, K.; Fu, W.L.; Huang, Q. Sensitive and specific detection of miRNA using an isothermal exponential amplification method using fluorescence-labeled LNA/DNA chimera primers. Anal. Bioanal. Chem. 2016, 408, 7437–7446. [Google Scholar] [CrossRef]
- Jia, H.; Bu, Y.; Zou, B.; Wang, J.; Kumar, S.; Pitman, J.L.; Zhou, G.; Song, Q. Signal amplification of microRNAs with modified strand displacement-based cycling probe technology. Analyst 2016, 141, 6297–6302. [Google Scholar] [CrossRef]
- Miao, J.; Wang, J.S.; Guo, J.Y.; Gao, H.G.; Han, K.; Jiang, C.M.; Miao, P. A plasmonic colorimetric strategy for visual miRNA detection based on hybridization chain reaction. Sci. Rep. 2016, 6, 32219. [Google Scholar] [CrossRef]
- Schwarzkopf, M.; Pierce, N.A. Multiplexed miRNA northern blots via hybridization chain reaction. Nucleic Acids Res. 2016, 44, e129. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.J.; Li, D.D.; Cheng, W.; Hu, R.; Sang, Y.; Yin, Y.B.; Ding, S.J.; Ju, H.X. Chemiluminescence imaging for microRNA detection based on cascade exponential isothermal amplification machinery. Anal. Chim. Acta 2016, 936, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.R.; Tang, M.; Diao, W.; Cheng, W.B.; Zhang, Y.; Yan, Y.R. Electrochemical strategy for ultrasensitive detection of microRNA based on MNAzyme-mediated rolling circle amplification on a gold electrode. Microchim. Acta 2016, 183, 3061–3067. [Google Scholar] [CrossRef]
- Hong, C.; Baek, A.; Hah, S.S.; Jung, W.; Kim, D.E. Fluorometric Detection of MicroRNA Using Isothermal Gene Amplification and Graphene Oxide. Anal. Chem. 2016, 88, 2999–3003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, J.; Zhang, L.L.; Liu, S.J.; Yu, R.Q.; Chu, X. A highly sensitive target-primed rolling circle amplification (TPRCA) method for fluorescent in situ hybridization detection of microRNA in tumor cells. Anal. Chem. 2014, 86, 1808–1815. [Google Scholar] [CrossRef]
- Cui, L.; Zhu, Z.; Lin, N.; Zhang, H.; Guan, Z.; Yang, C.J. A T7 exonuclease-assisted cyclic enzymatic amplification method coupled with rolling circle amplification: A dual-amplification strategy for sensitive and selective microRNA detection. Chem. Commun. 2014, 50, 1576–1578. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Wu, C.; Lv, S.; Wang, C.; Zhang, N.; Xiao, S.; Han, Y.; Xu, H.; Zhang, Y.; Li, F.; et al. Nicking-enhanced rolling circle amplification for sensitive fluorescent detection of cancer-related microRNAs. Anal. Bioanal. Chem. 2018, 410, 6819–6826. [Google Scholar] [CrossRef]
- Liu, S.; Fang, H.; Sun, C.; Wang, N.; Li, J. Highly sensitive and multiplexed miRNA analysis based on digitally encoded silica microparticles coupled with RCA-based cascade amplification. Analyst 2018, 143, 5137–5144. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, X.; Chen, X.; Qiu, X.; Qing, G.; Zhang, H.; Zhang, L.; Hu, X.; He, Z.; Zhong, D.; et al. Rolling Circular Amplification (RCA)-Assisted CRISPR/Cas9 Cleavage (RACE) for Highly Specific Detection of Multiple Extracellular Vesicle MicroRNAs. Anal. Chem. 2020, 92, 2176–2185. [Google Scholar] [CrossRef]
- Zhang, J.; He, M.; Nie, C.; He, M.; Pan, Q.; Liu, C.; Hu, Y.; Yi, J.; Chen, T.; Chu, X. Biomineralized Metal-Organic Framework Nanoparticles Enable Enzymatic Rolling Circle Amplification in Living Cells for Ultrasensitive MicroRNA Imaging. Anal. Chem. 2019, 91, 9049–9057. [Google Scholar] [CrossRef]
- Zhang, C.; Li, D.; Li, D.; Wen, K.; Yang, X.; Zhu, Y. Rolling circle amplification-mediated in situ synthesis of palladium nanoparticles for the ultrasensitive electrochemical detection of microRNA. Analyst 2019, 144, 3817–3825. [Google Scholar] [CrossRef]
- Zhao, B.; Song, J.; Guan, Y. Discriminative identification of miRNA let-7 family members with high specificity and sensitivity using rolling circle amplification. Acta Biochim. Biophys. Sin. 2015, 47, 130–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Yang, X.; Yuan, R.; Chai, Y. “Off” to “On” Surface-Enhanced Raman Spectroscopy Platform with Padlock Probe-Based Exponential Rolling Circle Amplification for Ultrasensitive Detection of MicroRNA 155. Anal. Chem. 2017, 89, 2866–2872. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, Y.; Wang, P.; Lu, J. A dual discrimination mode for improved specificity towards let-7a detection via a single-base mutated padlock probe-based exponential rolling circle amplification. Lumin. J. Biol. Chem. Lumin. 2017, 32, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Li, P.; He, W.; Liu, C.; Li, Z. Rolling circle extension-actuated loop-mediated isothermal amplification (RCA-LAMP) for ultrasensitive detection of microRNAs. Biosens. Bioelectron. 2018, 128, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wu, D.; Zhang, Y.F.; Shi, H.M.; Ouyang, C.H.; Li, F.; Jia, L.; Yu, S.H.; Wu, Z.S. RCA-enhanced multifunctional molecule beacon-based strand-displacement amplification for sensitive microRNA detection. Sens. Actuators B 2018, 258, 470–477. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, Y.; Zhang, S.; Sun, M.; Li, W.; Jiang, Y.; Wu, Z.S. Ultrasensitive assay based on a combined cascade amplification by nicking-mediated rolling circle amplification and symmetric strand-displacement amplification. Anal. Chim. Acta 2019, 1047, 172–178. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Li, F.; Zhang, Y.; Zhao, H.; Xu, H.; Wu, Z.-S.; Lyu, J.-X.; Shen, Z.-F. Sensitive detection of cancer gene based on a nicking-mediated RCA of circular DNA nanomachine. Sens. Actuators B 2017, 251, 692–698. [Google Scholar] [CrossRef]
- Zhou, Y.T.; Huang, Q.; Gao, J.M.; Lu, J.X.; Shen, X.Z.; Fan, C.H. A dumbbell probe-mediated rolling circle amplification strategy for highly sensitive microRNA detection. Nucleic Acids Res. 2010, 38, e156. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Ye, J.; Yang, D.; Al-Maskri, A.A.A.; Hu, H.; Jung, C.; Cai, S.; Zeng, S. Ultrasensitive detection of miRNA via one-step rolling circle-quantitative PCR (RC-qPCR). Anal. Chim. Acta 2019, 1077, 208–215. [Google Scholar] [CrossRef]
- Li, C.; Li, Z.; Jia, H.; Yan, J. One-step ultrasensitive detection of microRNAs with loop-mediated isothermal amplification (LAMP). Chem. Commun. 2011, 47, 2595–2597. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tian, H.; Liu, C.; Sun, Y.; Li, Z. One-step detection of microRNA with high sensitivity and specificity via target-triggered loop-mediated isothermal amplification (TT-LAMP). Chem. Commun. 2017, 53, 11040–11043. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Lv, M.; Li, J.; Yu, R.; Jiang, J. A ligation-based loop-mediated isothermal amplification (ligation-LAMP) strategy for highly selective microRNA detection. Chem. Commun. 2016, 52, 12721–12724. [Google Scholar] [CrossRef]
- Hashimoto, K.; Inada, M.; Ito, K. Multiplex Real-Time Loop-Mediated Isothermal Amplification Using an Electrochemical DNA Chip Consisting of a Single Liquid-Flow Channel. Anal. Chem. 2019, 91, 3227–3232. [Google Scholar] [CrossRef] [PubMed]
- Kalea, A.Z.; Hoteit, R.; Suvan, J.; Lovering, R.C.; Palmen, J.; Cooper, J.A.; Khodiyar, V.K.; Harrington, Z.; Humphries, S.E.; D’Aiuto, F. Upregulation of Gingival Tissue miR-200b in Obese Periodontitis Subjects. J. Dent. Res. 2015, 94, 59s–69s. [Google Scholar] [CrossRef] [Green Version]
- Kawakubo-Yasukochi, T.; Morioka, M.; Hazekawa, M.; Yasukochi, A.; Nishinakagawa, T.; Ono, K.; Kawano, S.; Nakamura, S.; Nakashima, M. miR-200c-3p spreads invasive capacity in human oral squamous cell carcinoma microenvironment. Mol. Carcinog. 2018, 57, 295–302. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).