Metabolic Stability of Eight Airborne OrganoPhosphate Flame Retardants (OPFRs) in Human Liver, Skin Microsomes and Human Hepatocytes
Abstract
:1. Introduction
2. Materials and Methods
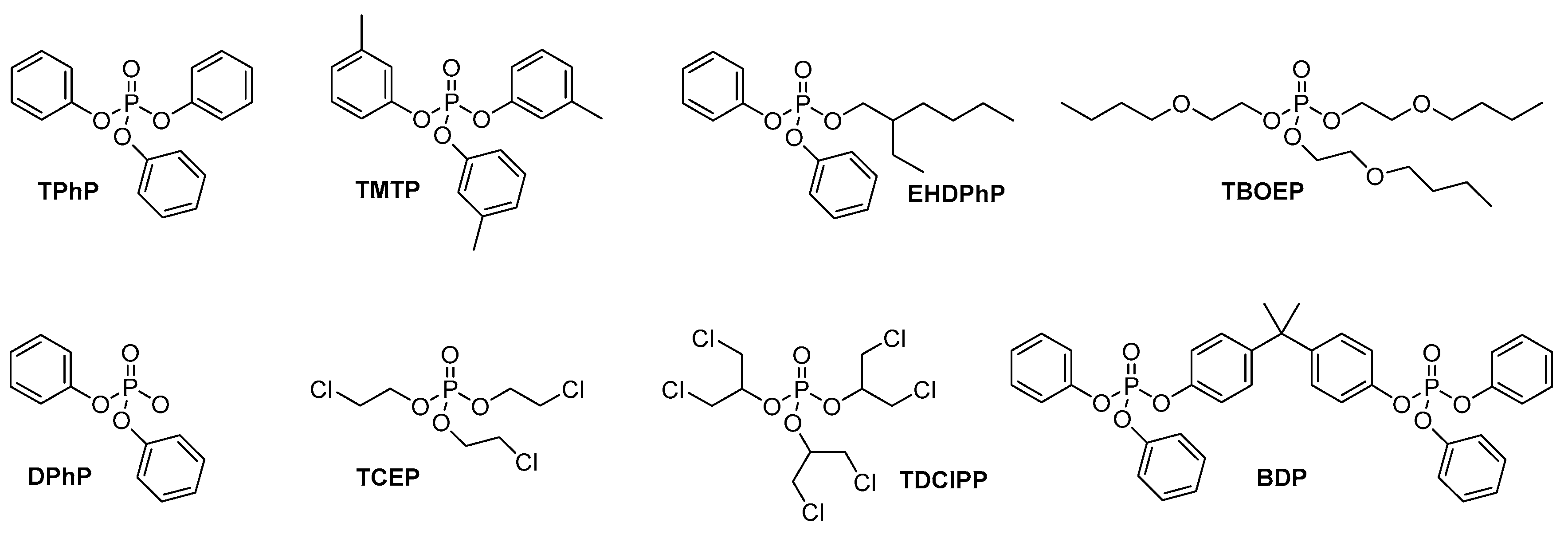
2.1. Chemicals and Reagents
2.2. In Vitro Incubations
2.3. LC-QTOF Method
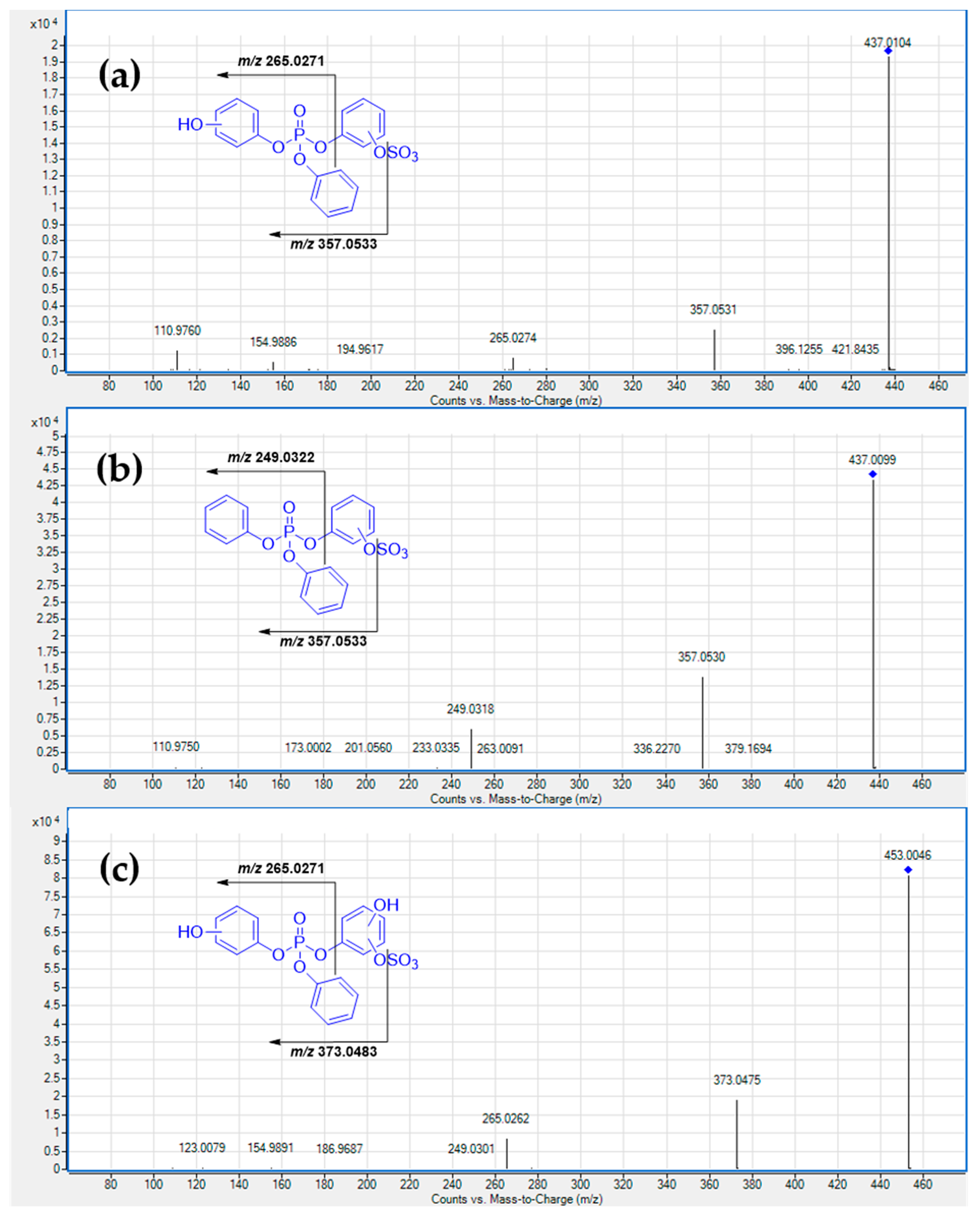
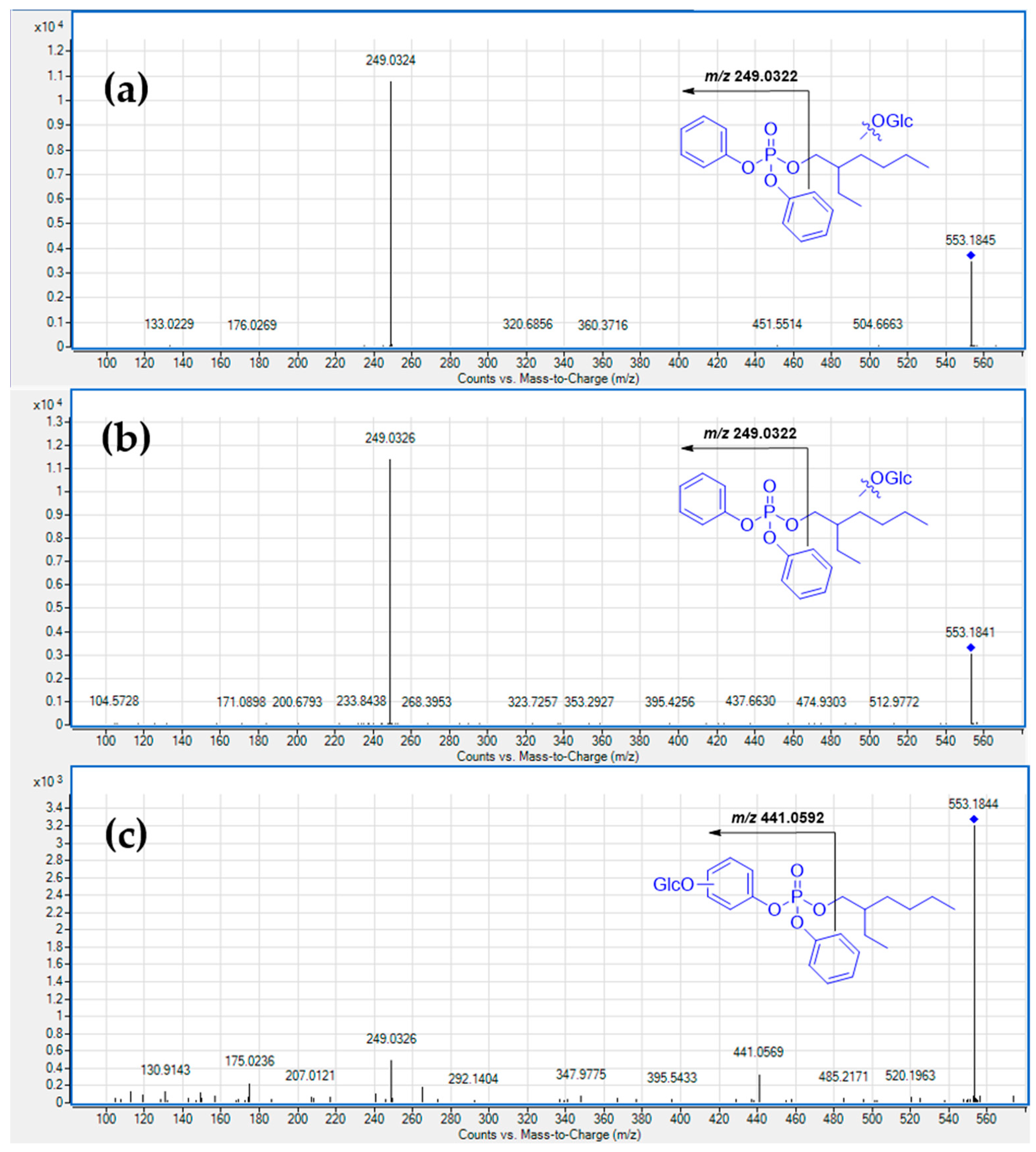
3. Results and Discussion
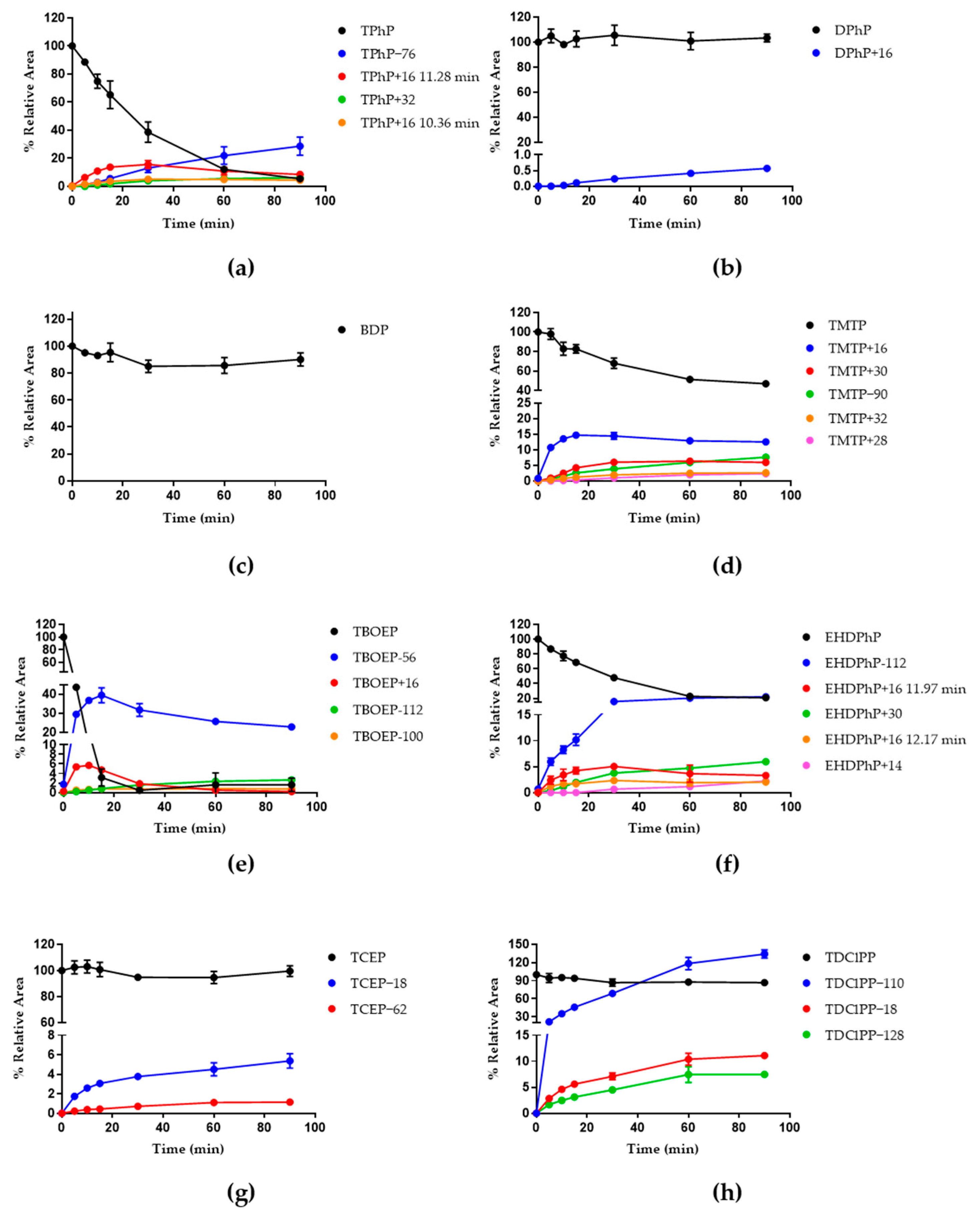
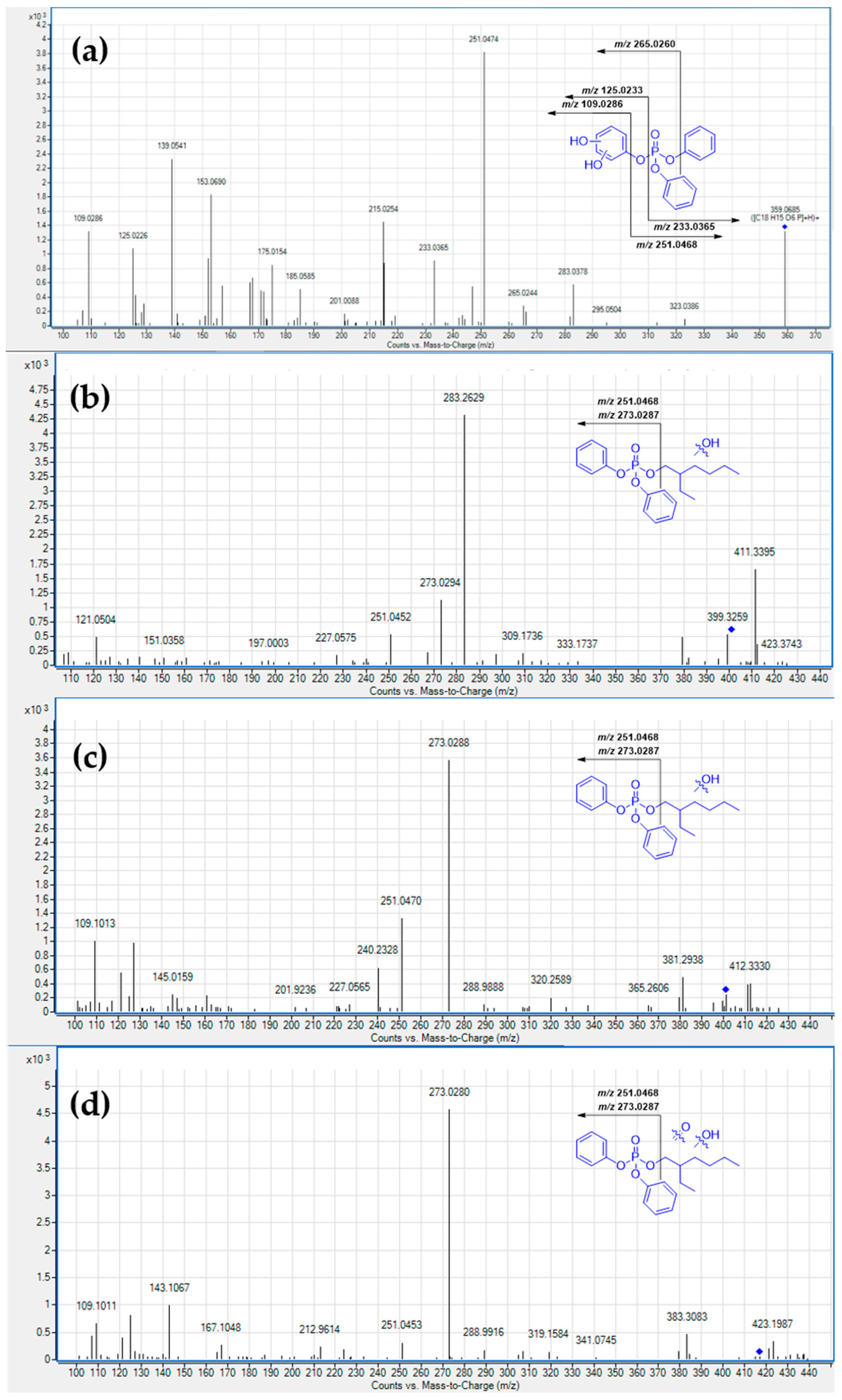
3.1. Metabolism in HLM
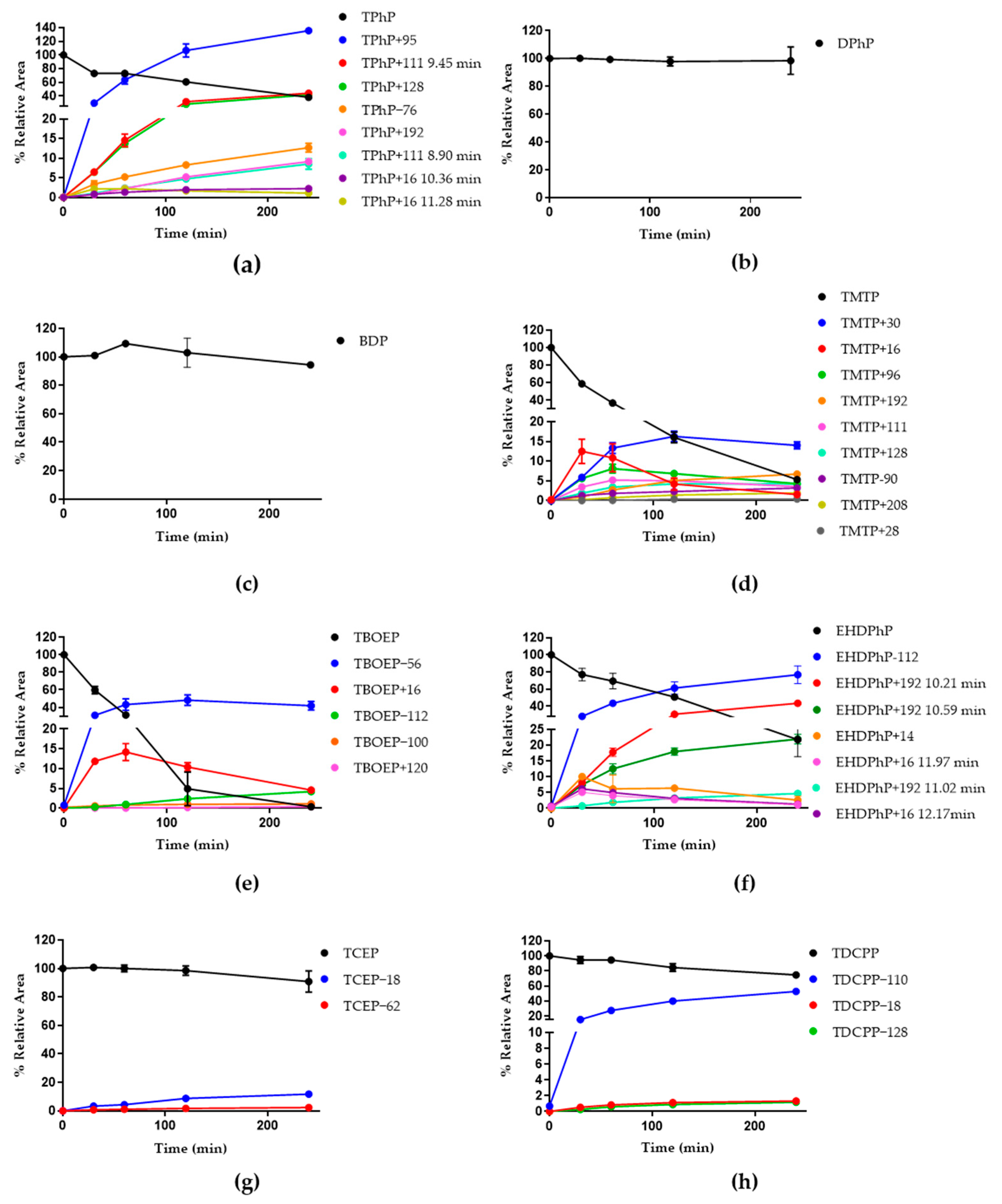
3.2. HuHep Incubation
3.3. HSM Incubation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Blum, A.; Behl, M.; Birnbaum, L.; Diamond, M.L.; Phillips, A.; Sipes, N.S.; Stapleton, H.M.; Venier, M. Organophosphate Ester Flame Retardants: Are They a Regrettable Substitution for Polybrominated Diphenyl Ethers? Environ. Sci. Technol. Lett. 2019, 6, 638–649. [Google Scholar] [CrossRef]
- Sharkey, M.; Harrad, S.; Abdallah, M.A.; Drage, D.S.; Berresheim, H. Phasing-out of legacy brominated flame retardants: The UNEP Stockholm Convention and other legislative action worldwide. Environ. Int. 2020, 144, 106041. [Google Scholar] [CrossRef]
- Wang, J.; Guo, Y.; Zhao, S.; Huang, R. A novel intumescent flame retardant imparts high flame retardancy to epoxy resin. Polym. Adv. Technol. 2019, 31, 932–940. [Google Scholar] [CrossRef]
- Van Der Veen, I.; De Boer, J. Phosphorus flame retardants: Properties, production, environmental occurrence, toxicity and analysis. Chemosphere 2012, 88, 1119–1153. [Google Scholar] [CrossRef]
- Azizi, S.; Hadi, M.; Naddafi, K.; Nabizadeh, R. Occurrence of organophosphorus esters in outdoor air fine particulate matter and comprehensive assessment of human exposure: A global systematic review. Environ. Pollut. 2023, 318, 120895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, Y.; Zhang, C.; Yao, Y.; Wang, L.; Sun, H. A review of organophosphate esters in soil: Implications for the potential source, transfer, and transformation mechanism. Environ. Res. 2022, 204, 112122. [Google Scholar] [CrossRef]
- Chupeau, Z.; Bonvallot, N.; Mercier, F.; Bot, B.L.; Chevrier, C.; Glorennec, P. Organophosphorus Flame Retardants: A Global Review of Indoor Contamination and Human Exposure in Europe and Epidemiological Evidence. Int. J. Environ. Res. Public Health 2020, 17, 6713. [Google Scholar] [CrossRef] [PubMed]
- Brits, M.; Brandsma, S.H.; Rohwer, E.R.; De Vos, J.; Weiss, J.M.; De Boer, J. Brominated and organophosphorus flame retardants in South African indoor dust and cat hair. Environ. Pollut. 2019, 253, 120–129. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Kannan, K. Occurrence, distribution and human exposure to 20 organophosphate esters in air, soil, pine needles, river water, and dust samples collected around an airport in New York state, United States. Environ. Int. 2019, 131, 105054. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, P.; Wang, X.; Castro-Jiménez, J.; Kallenborn, R.; Liao, C.; Mi, W.; Lohmann, R.; Vila-Costa, M.; Dachs, J. Organophosphate ester pollution in the oceans. Nat. Rev. Earth Environ. 2022, 3, 309–322. [Google Scholar] [CrossRef]
- Liang, C.; Mo, X.; Xie, J.; Wei, G.; Liu, L. Organophosphate tri-esters and di-esters in drinking water and surface water from the Pearl River Delta, South China: Implications for human exposure. Environ. Pollut. 2022, 313, 120150. [Google Scholar] [CrossRef] [PubMed]
- Kim, U.; Kannan, K. Occurrence and Distribution of Organophosphate Flame Retardants/Plasticizers in Surface Waters, Tap Water, and Rainwater: Implications for Human Exposure. Environ. Sci. Technol. 2018, 52, 5625–5633. [Google Scholar] [CrossRef]
- Bajard, L.; Melymuk, L.; Blaha, L. Prioritization of hazards of novel flame retardants using the mechanistic toxicology information from ToxCast and Adverse Outcome Pathways. Environ. Sci. Eur. 2019, 31, 14. [Google Scholar] [CrossRef]
- Behl, M.; Hsieh, J.-H.; Shafer, T.J.; Mundy, W.R.; Rice, J.R.; Boyd, W.A.; Freedman, J.H.; Hunter, E.S.; Jarema, K.A.; Padilla, S.; et al. Neurotoxicology and Teratology Use of alternative assays to identify and prioritize organophosphorus flame retardants for potential developmental and neurotoxicity. Neurotoxicol. Teratol. 2015, 52, 181–193. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, Z.; Ren, X.; Chen, X.; Zhou, X.; Zhou, X.; Zhang, T.; Liu, Y.; Wang, S.; Qin, C. Ecotoxicology and Environmental Safety Genetic comprehension of organophosphate flame retardants, an emerging threat to prostate cancer. Ecotoxicol. Environ. Saf. 2021, 223, 112589. [Google Scholar] [CrossRef]
- Li, Y.; Fu, Y.; Hu, K.; Zhang, Y.; Chen, J.; Zhang, S.; Zhang, B.; Liu, Y. Positive correlation between human exposure to organophosphate esters and gastrointestinal cancer in patients from Wuhan, China. Ecotoxicol. Environ. Saf. 2020, 196, 110548. [Google Scholar] [CrossRef] [PubMed]
- Böckers, M.; Paul, N.W.; Thomas, E. Organophosphate ester tri-o-cresyl phosphate interacts with estrogen receptor α in MCF-7 breast cancer cells promoting cancer growth. Toxicol. Appl. Pharmacol. 2020, 395, 114977. [Google Scholar] [CrossRef]
- Liu, M.; Li, A.; Meng, L.; Zhang, G.; Guan, X.; Zhu, J.; Li, Y.; Zhang, Q.; Jiang, G. Exposure to Novel Brominated Flame Retardants and Organophosphate Esters and Associations with Thyroid Cancer Risk: A Case—Control Study in Eastern China. Environ. Sci. Technol. 2022, 56, 17825–17835. [Google Scholar] [CrossRef]
- Hu, L.; Zhou, B.; Li, Y.; Song, L.; Wang, J.; Yu, M.; Li, X.; Liu, L.; Kou, J.; Wang, Y.; et al. Independent and combined effects of exposure to organophosphate esters on thyroid hormones in children and adolescents. Environ. Geochem. Health 2023, 45, 3833–3846. [Google Scholar] [CrossRef]
- Zhang, Q.; Ji, C.; Yin, X.; Yan, L.; Lu, M.; Zhao, M. Thyroid hormone-disrupting activity and ecological risk assessment of phosphorus-containing flame retardants by in vitro, in vivo and in silico approaches. Environ. Pollut. 2016, 210, 27–33. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, M.; Dong, X.; Wang, C.; Zhang, C.; Liu, W.; Zhao, M. Potential Estrogenic Effects of Phosphorus-Containing Flame Retardants. Environ. Sci. Technol. 2014, 48, 6995–7001. [Google Scholar] [CrossRef]
- Cui, D.; Bi, J.; Zhang, Z.-N.; Li, M.-Y.; Qin, Y.-S.; Xiang, P.; Ma, L.Q. Organophosphorus flame retardant TDCPP-induced cytotoxicity and associated mechanisms in normal human skin keratinocytes. Sci. Total Environ. 2020, 726, 138526. [Google Scholar] [CrossRef]
- Johnson, C.H.; Patterson, A.D.; Idle, J.R.; Gonzalez, F.J.; Sciences, B. Xenobiotic Metabolomics: Major Impact on the Metabolome. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 37–56. [Google Scholar] [CrossRef]
- Lipscomb, J.C.; Poet, T.S. In vitro measurements of metabolism for application in pharmacokinetic modeling. Pharmacol. Ther. 2008, 118, 82–103. [Google Scholar] [CrossRef] [PubMed]
- Cederbaum, A.I. Molecular mechanisms of the microsomal mixed function oxidases and biological and pathological implications. Redox Biol. 2015, 4, 60–73. [Google Scholar] [CrossRef]
- Pazzi, G.; Buiarelli, F.; Di Filippo, P.; Pomata, D.; Riccardi, C.; Lucarelli, F.; Giardi, F.; Sonego, E.; Galarini, R.; Lorenzetti, S.; et al. Metals and organic species associated with fine and coarse aerosol particles in an electronic waste recycling plant. Air Qual. Atmos. Health 2023, 16, 841–856. [Google Scholar] [CrossRef]
- Pomata, D.; Di Filippo, P.; Riccardi, C.; Buiarelli, F.; Marini, F.; Romani, L.; Lucarelli, F.; Pazzi, G.; Galarini, R.; Simonetti, G. Concentrations and co-occurrence of 101 emerging and legacy organic pollutants in the ultrafine, fine and coarse fractions of airborne particulates associated with treatment of waste from electrical and electronic equipment. Chemosphere 2023, 338, 139443. [Google Scholar] [CrossRef] [PubMed]
- Pyo, S.M.; Maibach, H.I. Skin Metabolism: Relevance of Skin Enzymes for Rational Drug Design. Skin Pharmacol. Physiol. 2019, 32, 283–293. [Google Scholar] [CrossRef]
- Van Den Eede, N.; Maho, W.; Erratico, C.; Neels, H.; Covaci, A. First insights in the metabolism of phosphate flame retardants and plasticizers using human liver fractions. Toxicol. Lett. 2013, 223, 9–15. [Google Scholar] [CrossRef]
- Selmi-Ruby, S.; Marín-Sáez, J.; Fildier, A.; Buleté, A.; Abdallah, M.; Garcia, J.; Deverchère, J.; Spinner, L.; Giroud, B.; Ibanez, S.; et al. In vivo characterisation of the toxicological properties of DPhP, one of the main degradation products of aryl phosphate esters. Environ. Health Perspect. 2020, 128, 127006. [Google Scholar] [CrossRef]
- Alves, A.; Erratico, C.; Lucattini, L.; Cuykx, M.; Leonards, P.E.G.; Voorspoels, S.; Covaci, A. Mass spectrometric identi fi cation of in vitro-generated metabolites of two emerging organophosphate flame retardants: V6 and BDP. Chemosphere 2018, 212, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Van Den Eede, N.; Erratico, C.; Exarchou, V.; Maho, W.; Neels, H.; Covaci, A. In vitro biotransformation of tris (2-butoxyethyl) phosphate (TBOEP) in human liver and serum. Toxicol. Appl. Pharmacol. 2015, 284, 246–253. [Google Scholar] [CrossRef]
- Zamora, I.; Fontaine, F.; Serra, B.; Plasencia, G. High-throughput, computer assisted, specific MetID. A revolution for drug discovery. Drug Discov. Today Technol. 2013, 10, e199–e205. [Google Scholar] [CrossRef]
- Radchenko, T.; Zamora, I.; Fontaine, F.; Morettoni, L.; Design, L.M.; Cugat, S. WebMetabase: Cleavage sites analysis tool for natural and unnatural substrates from diverse data source. Bioinformatics 2018, 35, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Goracci, L.; Desantis, J.; Valeri, A.; Castellani, B.; Eleuteri, M.; Cruciani, G. Understanding the Metabolism of Proteolysis Targeting Chimeras (PROTACs): The Next Step toward Pharmaceutical Applications. J. Med. Chem. 2020, 63, 11615–11638. [Google Scholar] [CrossRef] [PubMed]
- Rolsted, K.; Kissmeyer, A.; Rist, M.G.; Hansen, S.H. Evaluation of cytochrome P450 activity in vitro, using dermal and hepatic microsomes from four species and two keratinocyte cell lines in culture. Arch. Dermatol. Res. 2008, 300, 11–18. [Google Scholar] [CrossRef]
- Herczegh, S.M.; Chu, S.; Letcher, R.J. Biotransformation of bisphenol-A bis(diphenyl phosphate): In vitro, in silico, and (non-) target analysis for metabolites in rat and bird liver microsomal models. Chemosphere 2023, 310, 136796. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Bona, S.; Artino, E.; Buiarelli, F.; Di Filippo, P.; Galarini, R.; Lorenzetti, S.; Lucarelli, F.; Cruciani, G.; Goracci, L. Metabolic Stability of Eight Airborne OrganoPhosphate Flame Retardants (OPFRs) in Human Liver, Skin Microsomes and Human Hepatocytes. Separations 2023, 10, 548. https://doi.org/10.3390/separations10110548
Di Bona S, Artino E, Buiarelli F, Di Filippo P, Galarini R, Lorenzetti S, Lucarelli F, Cruciani G, Goracci L. Metabolic Stability of Eight Airborne OrganoPhosphate Flame Retardants (OPFRs) in Human Liver, Skin Microsomes and Human Hepatocytes. Separations. 2023; 10(11):548. https://doi.org/10.3390/separations10110548
Chicago/Turabian StyleDi Bona, Stefano, Emanuele Artino, Francesca Buiarelli, Patrizia Di Filippo, Roberta Galarini, Stefano Lorenzetti, Franco Lucarelli, Gabriele Cruciani, and Laura Goracci. 2023. "Metabolic Stability of Eight Airborne OrganoPhosphate Flame Retardants (OPFRs) in Human Liver, Skin Microsomes and Human Hepatocytes" Separations 10, no. 11: 548. https://doi.org/10.3390/separations10110548
APA StyleDi Bona, S., Artino, E., Buiarelli, F., Di Filippo, P., Galarini, R., Lorenzetti, S., Lucarelli, F., Cruciani, G., & Goracci, L. (2023). Metabolic Stability of Eight Airborne OrganoPhosphate Flame Retardants (OPFRs) in Human Liver, Skin Microsomes and Human Hepatocytes. Separations, 10(11), 548. https://doi.org/10.3390/separations10110548






