Abstract
Industrial wastewater usually contains a large amount of organic and inorganic pollutants, and many microorganisms. However, the types of microorganism present in industrial wastewater are still unclear. The aim of this study was to analyze the physicochemical properties and drug resistance of Pseudomonas aeruginosa isolated from industrial wastewater containing high concentrations of sulfate compounds. Pseudomonas aeruginosa was isolated from industrial wastewater from industrial produce with high concentrations of sulfate and phosphate, and mass spectrometry identification, gene identification, biochemical analysis and genomic and proteomic property identification were carried out. According to the results of matrix-assisted flight mass spectrometry and 16S rDNA sequencing, the isolated bacterium was identified as Pseudomonas aeruginosa, and was positive for reactions of ONPG, ACE, GLU, MNE, etc. Through growth experiments, it can be seen that Pseudomonas aeruginosa had a significant growth rate in the LB medium. Antibiotic sensitivity tests showed that Pseudomonas aeruginosa was susceptible to most antibiotics and moderately resistant to Polymyxin B and Polymyxin E. The drug resistance gene experiment showed that Pseudomonas aeruginosa had the gyrB gene related to antibiotic resistance. Proteomic analysis revealed that six proteins were involved in antibiotic resistance. This experiment isolated Pseudomonas aeruginosa from industrial produce wastewater containing high concentrations of sulfate and phosphate ions, providing a new perspective for further research on the characteristics and drug resistance of microorganisms in industrial wastewater and their potential functions when using them to deal with environmental pollution.
1. Introduction
Increasing urbanization and industrialization have brought domestic sewage and industrial wastewater discharge into local water sources and groundwater. Water pollution not only aggravates the deterioration of the ecological environment but also has a certain impact on human health [1]. The increase in industrial wastewater containing organic pollutants further increases toxicity of ecosystems. Removing certain hazardous substances from industrial wastewater is crucial for ensuring environmental and human safety. Biodegradation is a treatment method based on microorganisms naturally present in wastewater or activated sludge, and is relatively inexpensive and environmentally friendly [2]. Research has shown that some selected microorganisms can effectively remove certain hazardous substances from industrial wastewater [3,4]. Therefore, the study of adaptive microorganisms in industrial wastewater with different chemical compositions is of great significance in exploring new treatment methods for industrial wastewater and in mitigating the polluting effects of industrial wastewater.
Pseudomonas aeruginosa, a kind of Pseudomonas, is a human pathogen that can cause many body infections and even cause very serious diseases. Pseudomonas aeruginosa has wide genetic diversity and is able to survive in a variety of environmental conditions, including in soil and many aquatic environments [5]. Meanwhile, some studies have shown that Pseudomonas aeruginosa has strong resistance to antibiotics [6]. The carbapenem-resistant characteristics of Pseudomonas aeruginosa can be life-threatening as well as producing healthcare-associated infections, and resistance to other antibiotics such as Ceftazidime still demonstrates multigenic and complex effects [7]. The resistance nodulation-division (RND) family plays a very important role related to multidrug efflux pumps with Gram-negative bacteria, as their function increases the susceptibility of bacteria to antimicrobial agents [8]. Several studies have confirmed that multidrug resistance in Pseudomonas aeruginosa is caused by RND efflux pumps [9,10]. In addition, a study has shown that Pseudomonas aeruginosa can inhibit the biofilm formed by exposure to a combination of chlorinated polyfluorinated ether sulfonate and chromium [11,12]. In recent years, it has been found that Pseudomonas aeruginosa can reduce nitrate and chromium in wastewater [13], but it has not been studied in industrial wastewater containing high concentrations of sulfate compounds. Therefore, in this study, Pseudomonas aeruginosa was isolated from industrial wastewater, and biochemical identification and drug resistance gene and proteomic analysis were performed. These works provide a theoretical basis for the further study of the application of Pseudomonas aeruginosa in industrial wastewater management.
2. Materials and Methods
2.1. Isolation and Cultivation of Bacteria from Industrial Wastewater
First, 500 mL of industrial wastewater was collected from Henan Baili New Energy Materials Co., Ltd. (Jiaozuo, China) The main components of the industrial wastewater were sulfate ions (SO2-, 50,000 ppm), ammonia nitrogen (NH3-N, 11,000 ppm), sodium (Na, 5500 ppm), magnesium (Mg, 2100 ppm), phosphorus (P, 1200 ppm), manganese (Mn, 220 ppm), iron (Fe, 120 ppm) and calcium (Ca, 60 ppm). Bacteria were isolated from the industrial wastewater using the inoculation ring and incubated on a nutrient agar medium (Hopebiol, Qingdao, China) in an incubator at 37 °C. Then, a single colony was selected and inoculated in fresh nutritional broth medium (Hopebiol, Qingdao, China) and incubated for 14 h at 240 rpm.
2.2. Plasmid and Genome DNA Purification
The DNA and plasmids were extracted and purified using the plasmid and genome DNA Extraction Kit (Tiangen, Shanghai, China). The universal primers 27F (5′- AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′- GGTTACCTTGTTACGATT-3′) were used to execute 16S rDNA gene PCR amplification. The conditions for PCR amplification were 98 °C for 5 min, 98 °C for 15 s, 55 °C for 25 s, 72 °C for 20 s, the above program executed 40 cycles and then 72 °C for 5 min. The PCR product was observed using 1% agarose gel with Gelred dye, and the size of the DNA was confirmed by electrophoresis.
2.3. Establishment of a Phylogenetic Tree
The neighbor-joining method was used to produce the phylogenetic tree with a Max Seq difference of 0.5 through the NCBI Website. B21 was seen as the strain Pseudomonas aeruginosa 16S DNA sequence in the study.
2.4. Matrix-Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry Was Used for Rapid Identification
The matrix-assisted laser desorption ionization time of flight mass spectrometry (M-Discover 100 Excellence, MEIHUA, Zhuhai, China) and the sample pretreatment reagents of mass spectrometry detector (2 × 50 model) were used to implement bacterial identification. First, the target sample plate of the mass spectrometry was used to hold an appropriate amount of bacteria and the bacterial body was smeared to form a thin layer of bacteria. Absorbed lysis solution M1 (1 μL) was injected onto the thin layer of bacteria, and the film was dried at room temperature. After that, the matrix liquid (1 μL) was absorbed and it was covered with the previously mentioned layer of bacteria. After drying at the environment temperature again, bacteria could be detected on the machine.
2.5. Biochemical Experiments on Microorganisms
The biochemical experiments on the isolated strains were carried with the instruments for microbial identification and antibiotic resistance analysis (MA120 MEIHUA, Zhuhai, China). The biochemical experiment was implemented as follows: Firstly, a single colony was selected for pure culture, and then the bacterial suspension (0.5 McFarland standard) was produced by diluting the bacterial solution into sterile physiological saline. Then, the above bacterial solution (100 μL) was added to the wells of the biochemical identification plate. For some biochemical identification wells, it was necessary to add sterile paraffin oil. The adhesive sticker was torn off and attached to the biochemical identification plate. After incubation at 37 °C for 24 h, the plate was put into the machine for interpretation.
2.6. Experiments of Antimicrobial Resistance
The antibiotic resistance experiments used the instruments for microbial identification and antibiotic resistance analysis (MA120, MEIHUA, Zhuhai, China). The antibiotic resistance analysis was implemented as follows: First, the bacterial suspension (0.5 McFarland standard) was produced, and 50 μL was added to the M-H broth culture medium. Then, a drop of drug sensitivity chromogenic solution was added and mixed evenly. Then, 100 μL of the bacterial solution that was prepared as described above was added to each well of the drug sensitivity test plate. The adhesive sticker was torn off and attached to the antibiotic resistance identification plate. After incubation at 37 °C for 24 h, the plate was put into the machine for interpretation. The antibiotics were polymyxin B (PB), polymyxin E (CT), ceftazidime (CAZ), tetracycline (TET), piperacillin/tazobactam (P/T), cefoperazone/sulbactam (CPS), gentamicin (GEN), piperacillin (PIP), tobramycin (TOB), minocycline (MIN), imipenem (IPM), ceftriaxone (CRO), ciprofloxacin (CIP), ticarcillin/clavulanic acid (TIM), cefepime (FEP), doxycycline (DOX), aztreonam (ATM), ampicillin/sulbactam (AMS), meropene M (MRP), chloramphenicol (CHL), compound xinnuomin (SXT), cefotaxime (CTX), levofloxacin (LEV) and amikacin (AMK).
2.7. Observation of the Growth Pattern of This Bacterium
First, the 200 μL of bacterial culture medium that included the novel isolated strain was absorbed and injected into the column of 96 well plates. We set the OD600 absorbance value to determine bacterial density on the microplate reader (Tecan (Shanghai) Trading Co., Ltd., Shanghai, China). We adjusted the concentration of bacteria to 1 OD to observe the growth efficiency of different concentrations of bacteria. We set up one control group and seven experimental groups. Group 1 was a blank medium as the control. Group 2 consisted of 1 μL of bacterial fluid + 999 μL of sterile LB culture medium. Group 3 consisted of 10 μL of bacterial fluid + 990 μL of sterile LB culture medium. Group 4 consisted of 20 μL of bacterial fluid + 980 μL of sterile LB culture medium. Group 5 consisted of 40 μL of bacterial fluid + 960 μL of sterile LB culture medium. Group 6 consisted of 60 μL of bacterial fluid + 940 μL of sterile LB culture medium. Group 7 consisted of 80 μL of bacterial fluid + 920 μL of sterile LB culture medium. Group 8 consisted of 100 μL of bacterial fluid + 900 μL of sterile LB culture medium. A total of 1 mL of culture medium was included in the above dilution concentrations of bacteria to the 48-well plate, with 1 mL per well. The microbial high throughput growth detector (MicroScreen HT, JIELING, Tianjin, China) was used to execute the growth detection experiment. The experimental conditions of this device were set to 37 °C and 600 rpm, and OD values were collected per hour. The collected OD values were used to create the growth curve of the bacteria after 87 h of growth.
2.8. Drug Resistance Genes Testing
We synthesized primers for gyrA, gyrB, AAC (3)—II, cmlA, CTX-M-l, qnrA, qnrB, qnrC, qnrD, qnrS, blaKPC, oqxB, OXA, NDM-1, Sul2, oqxA, parC and qepA genes (14–17) using Tsingke Biotechnology Co., Ltd. (Beijing, China). These antibiotic resistant genes were amplified using PCR (C1000 Touch, BIORAD, Hercules, CA, USA), nucleic acid electrophoresis (Pow-erpac Basic, BIORAD, Hercules, CA, USA) was used to observe the nucleic acid migration, and the molecular weight of the gene was identified by gel imaging (GelGel Go, BI-ORAD, USA). We sequenced DNA fragments of expected molecular weight and size and compared them with the NCBI database.
2.9. Proteomic Analysis
The pretreatment of protein samples included protein extraction, denaturation, reductive alkylation, enzymatic hydrolysis and peptide desalting. In addition, in order to improve sample quality, the iST Sample Preparation kit (PreOmics, Planegg, Germany) was used to preprocess proteins. The specific method was as follows: An appropriate amount of protein was added to 50 µL lyse buffer and heated at 95 °C for 10 min at 1000 rpm with agitation. After the sample was cooled to room temperature, trypsin digestion buffer was added and incubated at 37 °C for 2 h. A termination buffer was added to stop the digestion process. Peptides were desalted, eluted and then lyophilized by SpeedVac and further analyzed by the Q-Exactive Plus coupled to an EASY nanoLC 1200 system (Thermo Fisher Scientific, Waltham, MA, USA).
Processed peptide segments were re-dissolved in 0.1% formic acid in water. A total of 3 μL of peptide solution was injected onto a 25 cm analytical column (Dr Maisch C18). The separation process was as follows: Starting at 2% buffer (80% ACN with 0.1% FA) transfer to a stepwise increase to 30% in 47 min, 100% in 1 min and maintain for 12 min. The column flow rate was controlled at 300 nL/min and the column temperature was 40 °C. The data-dependent acquisition (DDA) mode was manipulated when the mass spectrometer was run, and MS and MS/MS modes were automatically switched during the operation of this machine. Orbitrap was set to 70,000 resolution as a standard mode for the survey of full scan MS spectra (m/z 350–1800). Other settings included an automatic gain control (AGC) target of 3e6 and a maximum injection time of 50 ms.
Tandem mass spectra were processed using PEAKS Studio version 10.6 (Bioinformat-ics Solutions Inc., Waterloo, ON, Canada). The database was Pseudomonas aeruginosa (strain ATCC 15692/DSM 22644/CIP 104116/JCM 14847/LMG 12228/1C/PRS 101/PAO1) (version2023, 5564 entries), which was download from UniProt.
3. Results
3.1. The Rapid Identification of Pseudomonas aeruginosa by Matrix-Assisted Flight Mass Spectrometry
Matrix-assisted flight mass spectrometry and its software were used for the analysis, and the ion mass–charge ratio (m/z) was taken as the horizontal axis, and the detected ion peak (intensity) was taken as the vertical axis. The baseline of the bacterial protein fingerprint was stable, the peak of the main protein was obvious, and the results of mass spectrometry were good. The main ion peaks were around 5211 and 5738, respectively. The isolated bacterial protein fingerprint is shown in Figure 1. Compared with its database, the bacterium we isolated was identified as Pseudomonas aeruginosa.
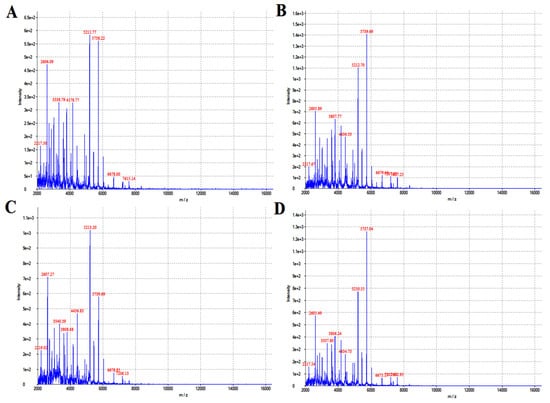
Figure 1.
The isolated strains were identified with flight mass spectrometry. (A–D) refers to the identification result of flight mass spectrometry after 12, 16, 20 and 24 h of cultivation.
3.2. Molecular Biological Identification of Pseudomonas aeruginosa
The expected size of the 16S rDNA gene amplification product was approximately 1500 bp. Only one specific DNA fragment appeared in the amplified product, which had good specificity. Then, the results of 16S rDNA sequencing were compared with the NCBI database to further confirm that the isolated bacterium was Pseudomonas aeruginosa. Meanwhile, we conducted a phylogenetic analysis of Pseudomonas aeruginosa isolated from the industrial wastewater and found that it had a high degree of homology with Pseudomonas aeruginosa in the gene database, as shown in Figure 2.
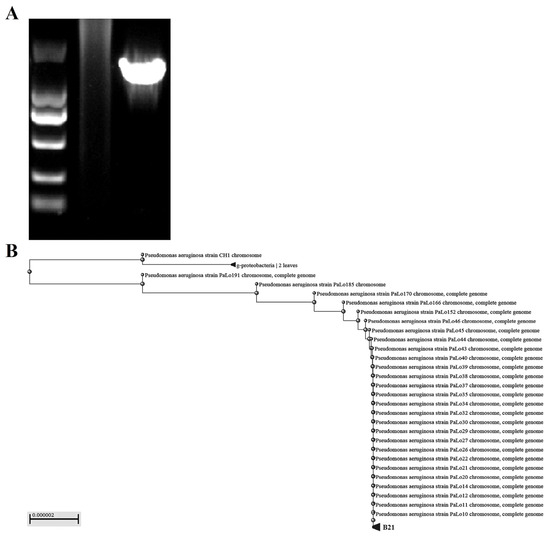
Figure 2.
Molecular biological identification and phylogenetic tree analysis of Pseudomonas aeru-ginosa isolated from industrial wastewater. (A) The 16S rDNA gene amplification products were displayed using gel electrophoresis. (B) Phylogenetic tree analysis of Pseudomonas aeruginosa based on 16S rDNA sequences.
3.3. Biochemical Identification of Pseudomonas aeruginosa
To further identify the metabolic effects of the newly isolated Pseudomonas aeruginosa on different chemical components, we conducted analysis using efficient and rapid microbial biochemical identification instruments. Our biochemical identification experiments showed that ONPG, ACE, GLU, MNE, XYL, ADH, GEL, NIT, MTE, CIT and MLT were positive reactions. GLUf, ESC, IND, MAN, H2S, ODC, SAC, LAC, MAL, LDC, C, URE and FRU were negative reactions (see Table 1).

Table 1.
Biochemical identification of Pseudomonas aeruginosa.
3.4. Analysis of Antibiotic Resistance in Pseudomonas aeruginosa
To further analyze the antibiotic resistance of the newly isolated Pseudomonas aeruginosa, we conducted a minimum inhibitory test (MIC) for drug resistance evaluation. Antibiotic sensitivity tests showed that the newly isolated Pseudomonas aeruginosa from industrial wastewater was sensitive to most of the antibiotics in the experiment, but its resistance to PB and CT was intermediate. These results indicate that most antibiotics in Pseudomonas aeruginosa still have good sensitivity (see Table 2).

Table 2.
Antibiotic sensitivity experiment on Pseudomonas aeruginosa.
3.5. Growth Experiment on Pseudomonas aeruginosa
We evaluated the growth of Pseudomonas aeruginosa with different dilutions after 87 h of growth. From Figure 3, we can see that it reached a peak at 12–13 h after cultivation, dropped to a low point around 40 h, and then gradually decreased. However, the peak times of Pseudomonas aeruginosa were different. The bacteria with low dilution took longer to peak than the bacteria with high dilution. However, the peak OD value of the bacteria with low dilution was higher than that of the bacteria with high dilution. The higher the OD value, the more bacteria there were. As shown in Table 3, it can be seen that the OD value obtained by 10 μL of diluted bacterial solution was the highest, and the OD value obtained by 100 μL of diluted bacterial solution was the lowest.
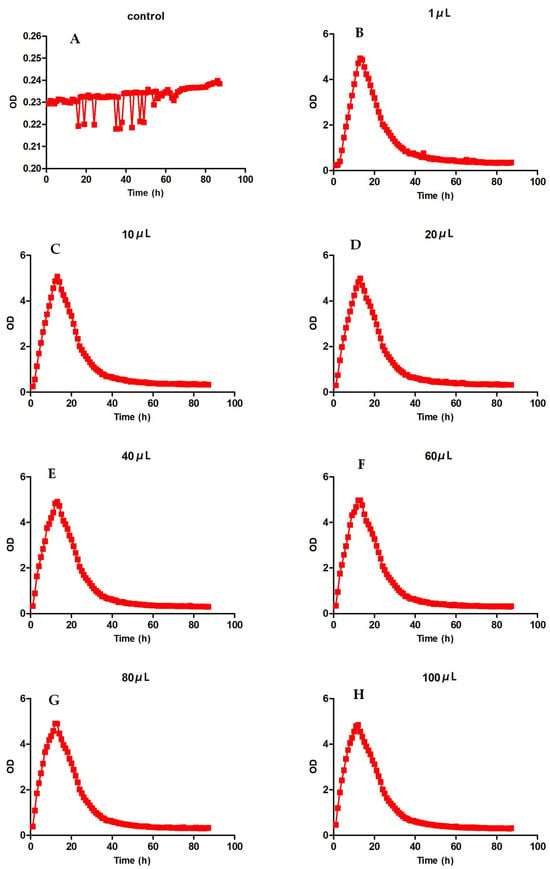
Figure 3.
Growth curves of Pseudomonas aeruginosa. (A) refers to the blank control without dilution, and (B–H) refer to the growth level of Pseudomonas aeruginosa with different dilution degrees.

Table 3.
The growth peak point of Pseudomonas aeruginosa with different concentrations.
3.6. Drug Resistance Gene Detection Test
Considering the uncertainty of the genetic distribution of drug resistance genes, we conducted genetic material amplification of different samples. Bacteria, genomes and plasmid were used as amplification templates to amplify drug resistance genes, and the amplification products were sequenced. The results showed the presence of gyrB resistance genes in the bacteria (see Figure 4 and Figure 5).
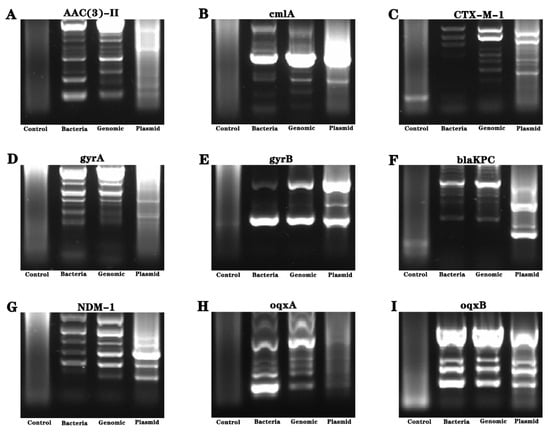
Figure 4.
Analysis of drug resistance genes of Pseudomonas aeruginosa isolated from industrial wastewater. (A–I) refer to the nucleic acid electrophoresis images of the resistance gene (AAC (3)—II, cmlA, CTX-M-l, gyrA, gyrB, blaKPC, NDM-1, oqxA and oqxB genes).
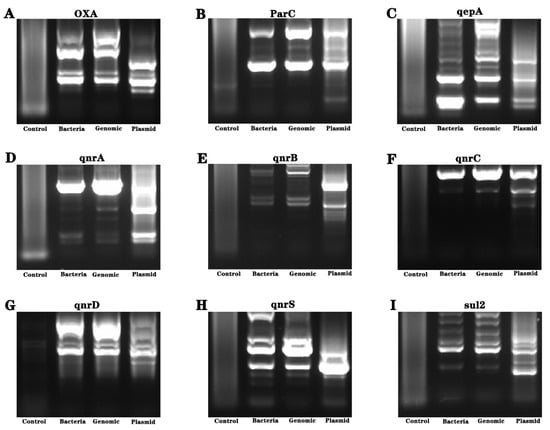
Figure 5.
Analysis of drug resistance genes of Pseudomonas aeruginosa isolated from industrial wastewater. (A–I) are the nucleic acid electrophoresis images of the resistance gene (AAC (3)—II, cmlA, CTX-M-l, gyrA, gyrB, blaKPC, NDM-1, oqxA, oqxB, OXA, parC, qepA, qnrA, qnrB, qnrC, qnrD, qnrS and Sul2 genes).
3.7. Proteomics Analysis
To further screen for possible drug resistance components, we found more than 2500 proteins of Pseudomonas aeruginosa through proteomics. After screening, we found six proteins related to drug resistance, as shown in Table 4.

Table 4.
Proteomics screens out proteins related to drug resistance.
4. Discussion
In recent decades, with the development of global industry, the pollution of the environment with heavy metals has become increasingly serious, and now poses a serious threat to human life and health. With the increasing demand for industrial wastewater treatment, the screening of suitable strains for the biological treatment of specific chemical pollutants will become more important for pollutant treatment and environmental protection.
Pseudomonas aeruginosa, a common Gram-negative opportunistic pathogen in clinical practice, is resistant to a variety of antibiotics and is one of the main pathogens of nosocomial infection that can cause healthcare-associated infections in humans and animals [14,15] Pseudomonas aeruginosa is more likely to grow in environments with water sources [16]. Pseudomonas aeruginosa can not only cause diseases, but can also develop complex drug resistance [17]. Pseudomonas aeruginosa and some of its isolates can also participate in the degradation of specific pollutants [18,19]. Phenolic substances are highly soluble in water; they can pollute freshwater, groundwater, oceans and soil, which has a negative impact on plant germination and growth, and animal and human health. In order to detoxify and bioremediate phenol in wastewater, previous studies have shown that Pseudomonas aeruginosa can effectively biodegrade phenol and restore the planting function of soil [20]. In current research, Pseudomonas aeruginosa has been found to be resistant to high cadmium and nickel which was isolated from the Industries Factory in Sari, Mazandaran, Iran [21]. In recent years, it has been found that Pseudomonas aeruginosa can reduce nitrate and chromium in wastewater [13]. Other studies have found that Pseudomonas aeruginosa SD-1 has a certain value in the treatment of livestock wastewater [22]. In addition, adding Pseudomonas aeruginosa S5 to coking wastewater can effectively promote the biodegradation of high-weight molecular polycyclic aromatic hydrocarbons in the sludge phase [23,24]. However, it has not been studied in high-salt wastewater. Therefore, a Pseudomonas aeruginosa strain was successfully isolated from industrial wastewater in this study, which was confirmed by matrix-assisted flight mass spectrometry and 16S rDNA sequencing results. Further biochemical analysis showed that Pseudomonas aeruginosa was positive for ONPG, ACE, GLU, MNE, etc. Taken together, the strain isolated from industrial wastewater in this study was identified as Pseudomonas aeruginosa.
Pseudomonas aeruginosa develops strong resistance to antibiotics commonly used in clinical settings. Pseudomonas aeruginosa isolated from hospital wastewater is mainly resistant to clinically relevant anti-pseudomonas drugs [25]. Recent reports have shown that the frequency of Pseudomonas aeruginosa, antibiotic resistance, antibiotic resistance coding genes, virulence factor coding genes and multidrug resistance isolated from marine samples were higher [26]. This study found that Pseudomonas aeruginosa from industrial wastewater was sensitive to most of the antibiotics (CAZ, P/T, CPS, GEN, etc.) in the experiment. Pseudomonas aeruginosa has been found in various environments, and various resistance genes have also been found. The gyrB gene is widely found in various bacteria; its sequence is conserved and variable, and it can be used as a target molecule in the classification and identification of bacteria based on nucleotide sequencing [27,28]. There is evidence that gyrB is a potential target for breaking resistance in Staphylococcus aureus [29]. Current studies have shown that gyrB mutation plays an important role in in vitro screening for resistance to fluoroquinolones in Pseudomonas aeruginosa [30]. Cbrera et al. [31] confirmed that Pseudomonas aeruginosa isolated from patients with bronchiectasis had a high level of resistance to ciprofloxacin, and gyrB gene mutation was detected in this strain. In this study, gyrB was expressed in the genome, plasmid and fluid of Pseudomonas aeruginosa. In addition, growth tests showed that the Pseudomonas aeruginosa grew at a moderate rate in the LB medium. The above results indicate that Pseudomonas aeruginosa can grow in culture medium and has good sensitivity to most antibiotics.
Many countries around the world have reported the discovery of Pseudomonas aeruginosa with different resistance mechanisms [32]. The overexpression of efflux pumps is one of the most important mechanisms of intrinsic and acquired resistance of Pseudomonas aeruginosa to various antibiotics. Current studies have shown that mexAB-oprM and mexXY-oprA efflux operons play an important role in antibiotic resistance in Pseudomonas aeruginosa isolates from Tabriz, Iran [33]. Pseudomonas aeruginosa was isolated from clinical specimens from a tertiary care hospital in Nepal, and MexB was detected in the multi-drug-resistant Pseudomonas aeruginosa [34]. Recent studies have shown that Pseudomonas aeruginosa isolates were multidrug-resistant with a high level of efflux pump expression of the MexA gene [35]. The tripartite multi-drug efflux system MexAB-OprM is the main factor in the resistance of Pseudomonas aeruginosa through the export of multiple antimicrobial compounds, and the antibiotic output of MexB multidrug efflux transporter is controlled by the allosteric MexA-OprM chaperone-like complex [36]. In recent years, it has been found that the expression of the RND efflux pump is significantly correlated with the resistance of most anti-pseudomonas antibiotics in Pseudomonas aeruginosa [9]. In this study, the proteomic analysis showed that Pseudomonas aeruginosa contained six drug-resistance-related proteins, including MexA, MexB, Multidrug resistance operon repressor, Multidrug ABC transporter ATPase, RND multidrug efflux membrane fusion protein and Multidrug transporter. This suggests that the resistance of Pseudomonas aeruginosa is closely related to MexA, MexB and RND multidrug efflux membrane fusion protein.
5. Conclusions
Pseudomonas aeruginosa was successfully isolated from industrial wastewater with a high concentration of sulfate compounds and was found to have good sensitivity to most antibiotics. Additionally, Pseudomonas aeruginosa had the gyrB gene and six proteins related to antibiotic resistance. These provide an important basis for subsequent research on the adaptability of the strain and functions in industrial wastewater. However, the potential roles of Pseudomonas aeruginosa in the degradation of pollutants and the management of industrial wastewater need to be further explored.
Author Contributions
Methodology, Z.W., W.T. and S.S.; editing, X.C.; conceptualization and supervision, Z.W. and H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Key Scientific Research Project of Higher Education of Henan Province (23A610015), the Henan Province Science and Technology Research and Development Plan Joint Funds (232103810037), the Science and Technology Development Plan of Kaifeng in 2020 (No. 2003048) and the Education and Teaching Reform and Research Project of the National Food Industry Vocational Education and Teaching Guidance Committee in 2023 (SHK2023027).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, H.; Sun, C.; Chen, X.; Yan, K.; He, H. Isolation of Pseudomonas oleovorans Carrying Multidrug Resistance Proteins MdtA and MdtB from Wastewater. Molecules 2023, 28, 5403. [Google Scholar] [CrossRef] [PubMed]
- Koul, Y.; Devda, V.; Varjani, S.; Guo, W.; Ngo, H.H.; Taherzadeh, M.J.; Chang, J.S.; Wong, J.W.C.; Bilal, M.; Kim, S.H.; et al. Microbial electrolysis: A promising approach for treatment and resource recovery from industrial wastewater. Bioengineered 2022, 13, 8115–8134. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, A.; Kumar, P.S.; Duc, P.A.; Rangasamy, G. Strategies for microbial bioremediation of environmental pollutants from industrial wastewater: A sustainable approach. Chemosphere 2023, 313, 137323. [Google Scholar] [CrossRef] [PubMed]
- Brunner, S.; Klessing, T.; Dotsch, A.; Sturm-Richter, K.; Gescher, J. Efficient Bioelectrochemical Conversion of Industrial Wastewater by Specific Strain Isolation and Community Adaptation. Front. Bioeng. Biotechnol. 2019, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Sommer, L.M.; Johansen, H.K.; Molin, S. Antibiotic resistance in Pseudomonas aeruginosa and adaptation to complex dynamic environments. Microb. Genom. 2020, 6, e000370. [Google Scholar] [CrossRef] [PubMed]
- Roulova, N.; Mot’kova, P.; Brozkova, I.; Pejchalova, M. Antibiotic resistance of Pseudomonas aeruginosa isolated from hospital wastewater in the Czech Republic. J. Water Health 2022, 20, 692–701. [Google Scholar] [CrossRef] [PubMed]
- Papanikolopoulou, A.; Gargalianos-Kakolyris, P.; Stoupis, A.; Moussas, N.; Pangalis, A.; Theodoridou, K.; Chronopoulou, G.; Pantazis, N.; Kantzanou, M.; Maltezou, H.C.; et al. Carbapenem-Resistant Pseudomonas aeruginosa Bacteremia, through a Six-Year Infection Control Program in a Hospital. Microorganisms 2023, 11, 1315. [Google Scholar] [CrossRef] [PubMed]
- Gil-Gil, T.; Laborda, P.; Ochoa-Sanchez, L.E.; Martinez, J.L.; Hernando-Amado, S. Efflux in Gram-negative bacteria: What are the latest opportunities for drug discovery? Expert Opin. Drug Discov. 2023, 18, 671–686. [Google Scholar] [CrossRef]
- Zahedi Bialvaei, A.; Rahbar, M.; Hamidi-Farahani, R.; Asgari, A.; Esmailkhani, A.; Mardani Dashti, Y.; Soleiman-Meigooni, S. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb. Pathog. 2021, 153, 104789. [Google Scholar] [CrossRef]
- Puzari, M.; Chetia, P. RND efflux pump mediated antibiotic resistance in Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa: A major issue worldwide. World J. Microbiol. Biotechnol. 2017, 33, 24. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, Y.; Zhang, M.; Qiao, W. Repairing of rutin to the toxicity of combined F-53B and chromium pollution on the biofilm formed by Pseudomonas aeruginosa. J. Environ. Sci. 2023, 127, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Teerapo, K.; Roytrakul, S.; Sistayanarain, A.; Kunthalert, D. A scorpion venom peptide derivative BmKn—22 with potent antibiofilm activity against Pseudomonas aeruginosa. PLoS ONE 2019, 14, e0218479. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Deng, S.; Xu, J.; Nan, H.; Li, Z.; Song, J.L. Simultaneous reduction of nitrate and Cr(VI) by Pseudomonas aeruginosa strain G12 in wastewater. Ecotoxicol. Environ. Saf. 2020, 191, 110001. [Google Scholar] [CrossRef] [PubMed]
- Veetilvalappil, V.V.; Manuel, A.; Aranjani, J.M.; Tawale, R.; Koteshwara, A. Pathogenic arsenal of Pseudomonas aeruginosa: An update on virulence factors. Future Microbiol. 2022, 17, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Bisht, K.; Baishya, J.; Wakeman, C.A. Pseudomonas aeruginosa polymicrobial interactions during lung infection. Curr. Opin. Microbiol. 2020, 53, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Taudien, S.; Leszczynski, W.; Mayer, T.; Loderstadt, U.; Bader, O.; Kaase, M.; Scheithauer, S. Misidentification as Pseudomonas aeruginosa in hospital water supply samples. J. Hosp. Infect. 2023, 133, 23–27. [Google Scholar] [CrossRef]
- Subedi, D.; Vijay, A.K.; Willcox, M. Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: An ocular perspective. Clin. Exp. Optom. 2018, 101, 162–171. [Google Scholar] [CrossRef]
- Chug, R.; Mathur, S.; Kothari, S.L.; Harish; Gour, V.S. Maximizing EPS production from Pseudomonas aeruginosa and its application in Cr and Ni sequestration. Biochem. Biophys. Rep. 2021, 26, 100972. [Google Scholar] [CrossRef]
- Sutar, V.P.; Mali, G.V.; Upadhye, V.; Singh, V.K.; Sinha, R.P. Purification of Lipase from Pseudomonas aeruginosa VSJK R-9 and Its Application in Combination with the Lipolytic Consortium for Bioremediation of Restaurant Wastewater. Appl. Biochem. Biotechnol. 2023, 195, 1888–1903. [Google Scholar] [CrossRef]
- Mahgoub, S.A.; Qattan, S.Y.A.; Salem, S.S.; Abdelbasit, H.M.; Raafat, M.; Ashkan, M.F.; Al-Quwaie, D.A.; Motwali, E.A.; Alqahtani, F.S.; Abd El-Fattah, H.I. Characterization and Biodegradation of Phenol by Pseudomonas aeruginosa and Klebsiella variicola Strains Isolated from Sewage Sludge and Their Effect on Soybean Seeds Germination. Molecules 2023, 28, 1203. [Google Scholar] [CrossRef]
- Hosseini Zabet, A.; Ahmady-Asbchin, S. Investigation of cadmium and nickel biosorption by Pseudomonas sp. via response surface methodology. World J. Microbiol. Biotechnol. 2023, 39, 135. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Zhang, Y.; Hu, J.; Li, Y.; Li, N.; Wang, M. Mixture of different Pseudomonas aeruginosa SD-1 strains in the efficient bioaugmentation for synthetic livestock wastewater treatment. Chemosphere 2019, 237, 124455. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, Y.; Zang, T.; Wei, J.; Wu, H.; Wei, C.; Qiu, G.; Li, F. A biosurfactant-producing Pseudomonas aeruginosa S5 isolated from coking wastewater and its application for bioremediation of polycyclic aromatic hydrocarbons. Bioresour. Technol. 2019, 281, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Zang, T.; Wu, H.; Zhang, Y.; Wei, C. The response of polycyclic aromatic hydrocarbon degradation in coking wastewater treatment after bioaugmentation with biosurfactant-producing bacteria Pseudomonas aeruginosa S5. Water Sci. Technol. 2021, 83, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Divyashree, M.; Mani, M.K.; Karunasagar, I. Association of exopolysaccharide genes in biofilm developing antibiotic-resistant Pseudomonas aeruginosa from hospital wastewater. J. Water Health 2022, 20, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Dehkordi, S.M.H.; Anvar, S.A.; Rahimi, E.; Ahari, H.; Ataee, M. Molecular investigation of prevalence, phenotypic and genotypic diversity, antibiotic resistance, frequency of virulence genes and genome sequencing in Pseudomonas aeruginosa strains isolated from lobster. Int. J. Food Microbiol. 2022, 382, 109901. [Google Scholar] [CrossRef] [PubMed]
- Collin, F.; Karkare, S.; Maxwell, A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl. Microbiol. Biotechnol. 2011, 92, 479–497. [Google Scholar] [CrossRef]
- Zidar, N.; Tomasic, T.; Macut, H.; Sirc, A.; Brvar, M.; Montalvao, S.; Tammela, P.; Ilas, J.; Kikelj, D. New N-phenyl-4,5-dibromopyrrolamides and N-Phenylindolamides as ATPase inhibitors of DNA gyrase. Eur. J. Med. Chem. 2016, 117, 197–211. [Google Scholar] [CrossRef]
- Kaul, M.; Mark, L.; Zhang, Y.; Parhi, A.K.; Lyu, Y.L.; Pawlak, J.; Saravolatz, S.; Saravolatz, L.D.; Weinstein, M.P.; LaVoie, E.J.; et al. TXA709, an FtsZ-Targeting Benzamide Prodrug with Improved Pharmacokinetics and Enhanced In Vivo Efficacy against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2015, 59, 4845–4855. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, Z.; Li, X.; Song, Y.; Kang, J.; Yin, D.; Gao, Y.; Shi, N.; Duan, J. Mutations in gyrB play an important role in ciprofloxacin-resistant Pseudomonas aeruginosa. Infect. Drug Resist. 2019, 12, 261–272. [Google Scholar] [CrossRef]
- Cabrera, R.; Fernandez-Barat, L.; Vazquez, N.; Alcaraz-Serrano, V.; Bueno-Freire, L.; Amaro, R.; Lopez-Aladid, R.; Oscanoa, P.; Munoz, L.; Vila, J.; et al. Resistance mechanisms and molecular epidemiology of Pseudomonas aeruginosa strains from patients with bronchiectasis. J. Antimicrob. Chemother. 2022, 77, 1600–1610. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Goli, H.R.; Nahaei, M.R.; Rezaee, M.A.; Hasani, A.; Samadi Kafil, H.; Aghazadeh, M.; Sheikhalizadeh, V. Contribution of mexAB-oprM and mexXY (-oprA) efflux operons in antibiotic resistance of clinical Pseudomonas aeruginosa isolates in Tabriz, Iran. Infect. Genet. Evol. 2016, 45, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Devkota, M.D.; Pokhrel, B.M.; Banjara, M.R. Detection of bla(NDM-1,)mcr-1 and MexB in multidrug resistant Pseudomonas aeruginosa isolated from clinical specimens in a tertiary care hospital of Nepal. BMC Microbiol. 2023, 23, 153. [Google Scholar] [CrossRef] [PubMed]
- Abed, W.H.; Kareem, S.M. Molecular detection of gyrA and mexA genes in Pseudomonas aeruginosa. Mol. Biol. Rep. 2021, 48, 7907–7912. [Google Scholar] [CrossRef] [PubMed]
- Glavier, M.; Puvanendran, D.; Salvador, D.; Decossas, M.; Phan, G.; Garnier, C.; Frezza, E.; Cece, Q.; Schoehn, G.; Picard, M.; et al. Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex. Nat. Commun. 2020, 11, 4948. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).