Fluorescence Enhancement on Silver-Plated Plasma Micro-Nanostructured 3D Polymeric Microarray Substrates for Multiplex Mycotoxin Detection
Abstract
1. Introduction
2. Materials and Methods
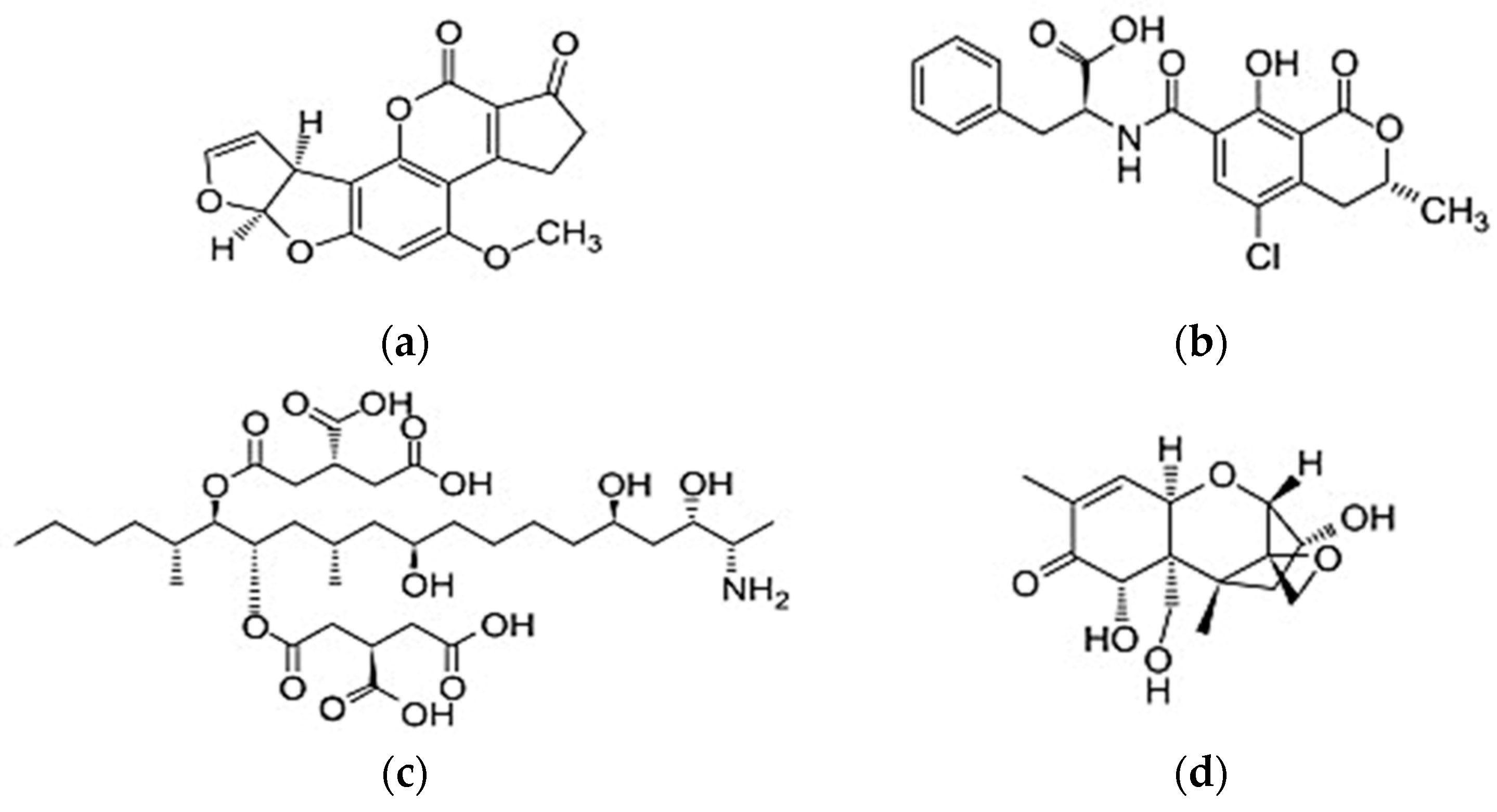
2.1. Reagents and Chemicals
2.2. Preparation of Aminosilane-Coated Glass Slides
2.3. Preparation of 3D Micro-Nanostructured Slides
2.4. Silver Film Deposition
2.5. Surface Characterization
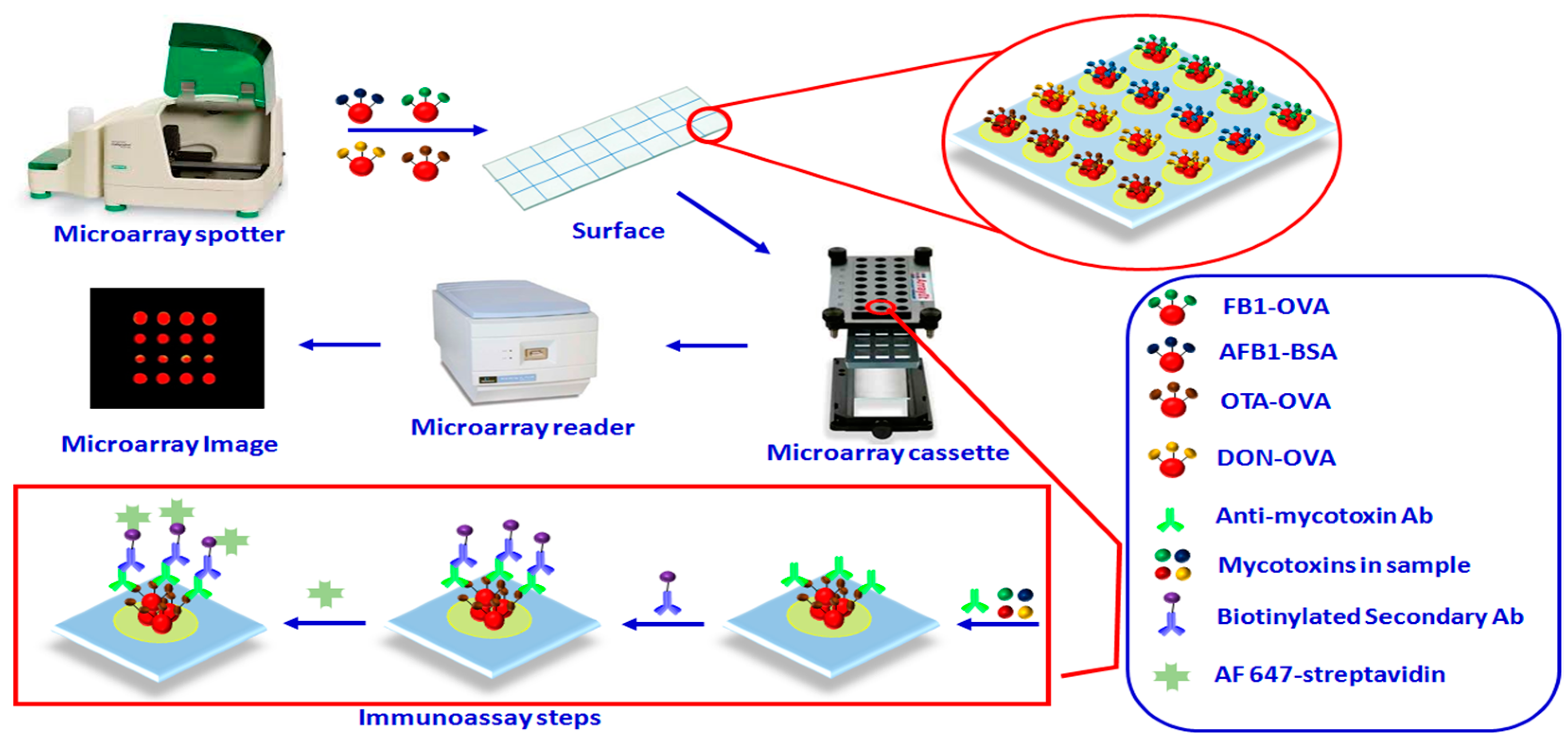
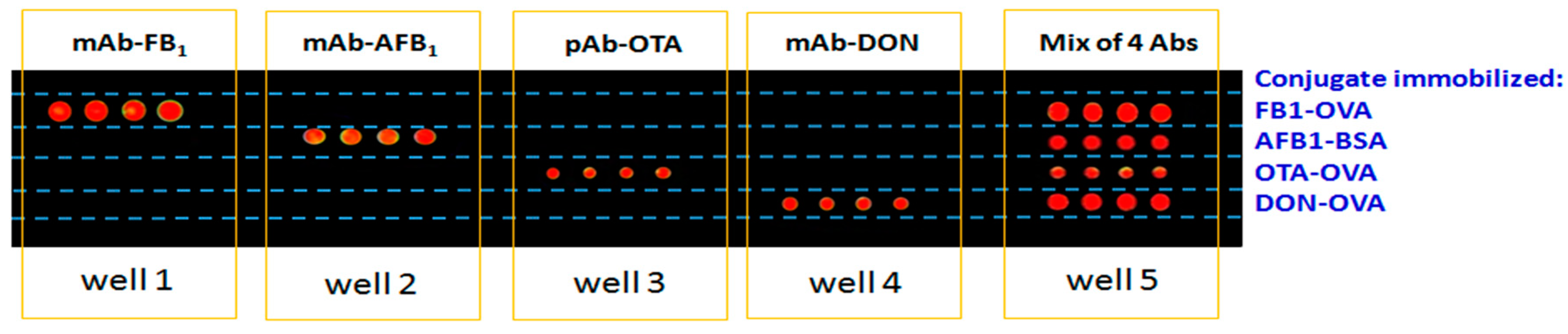
2.6. Microarray Fabrication and Detection
2.7. Analysis of Corn Samples
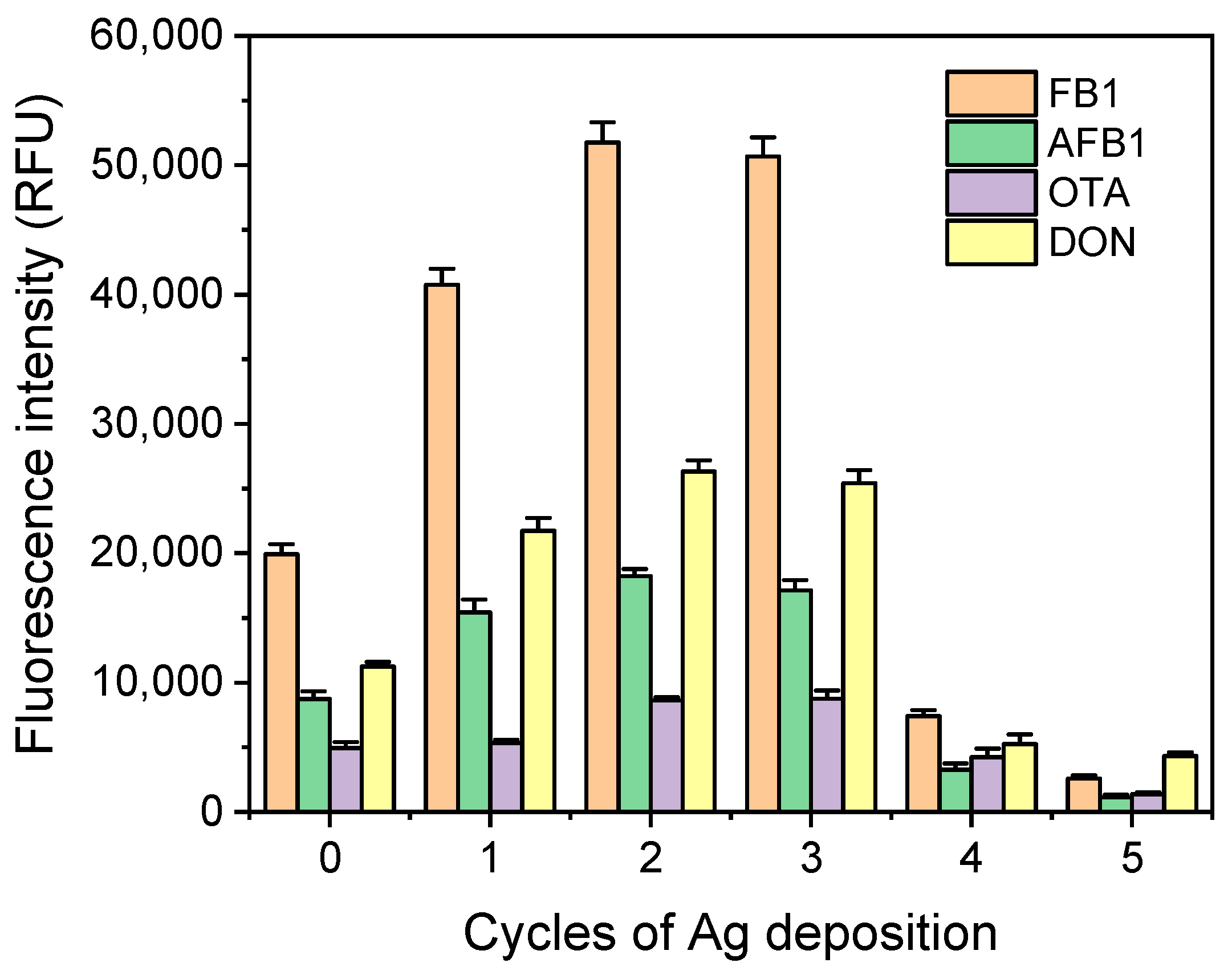
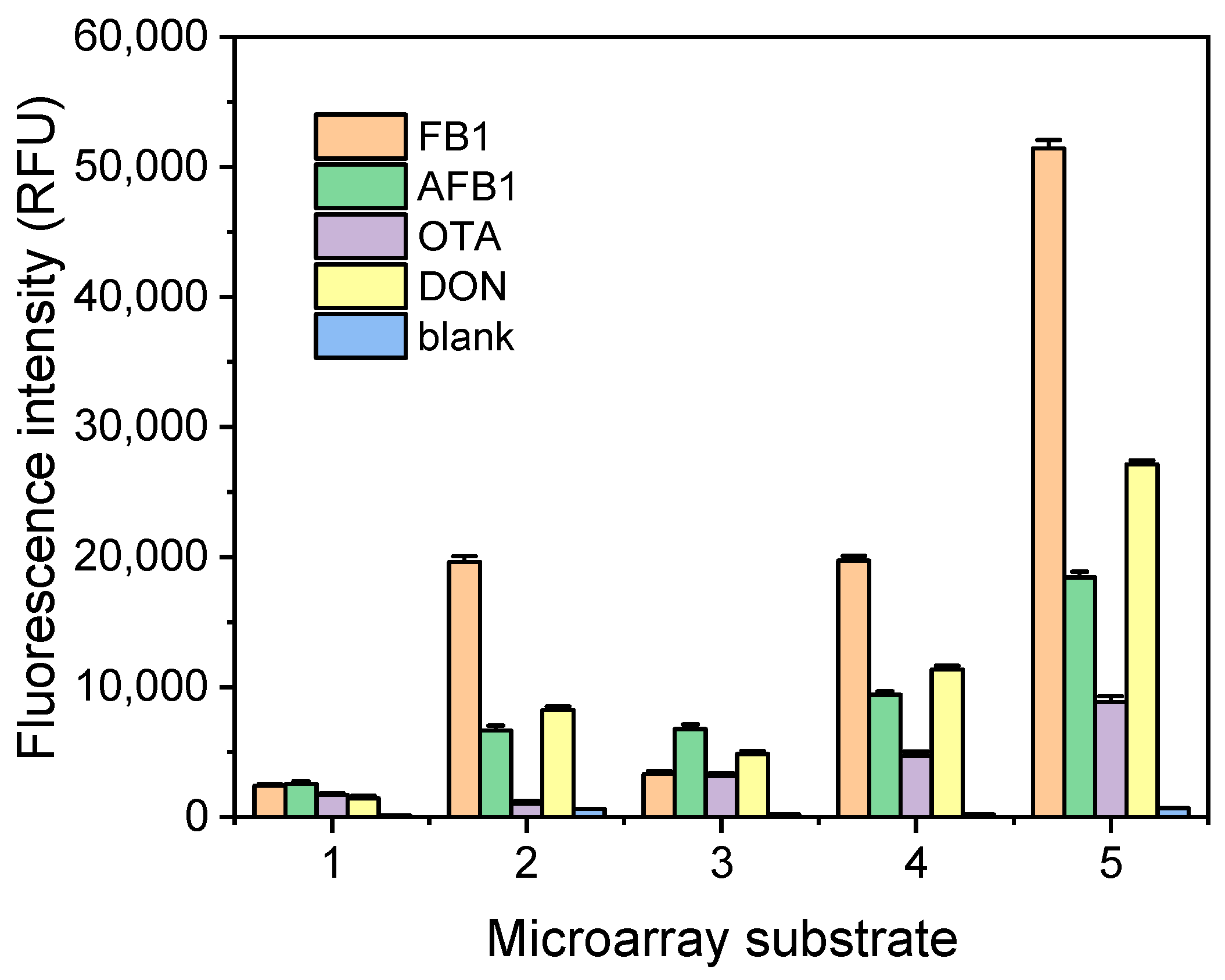
3. Results and Discussion
3.1. Plasmonic 3D Substrate Preparation
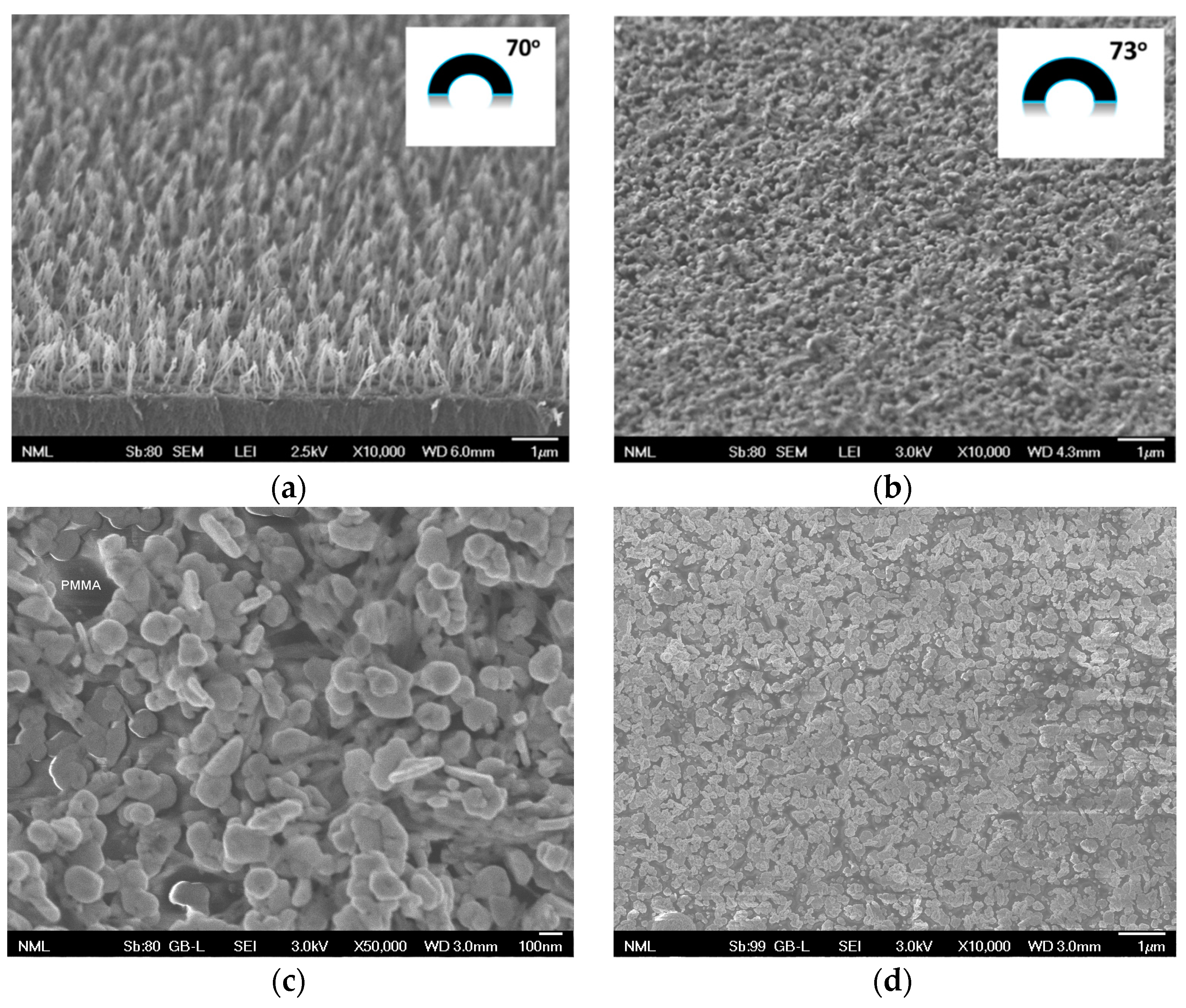
3.2. Plasmonic 3D Substrates Characterization
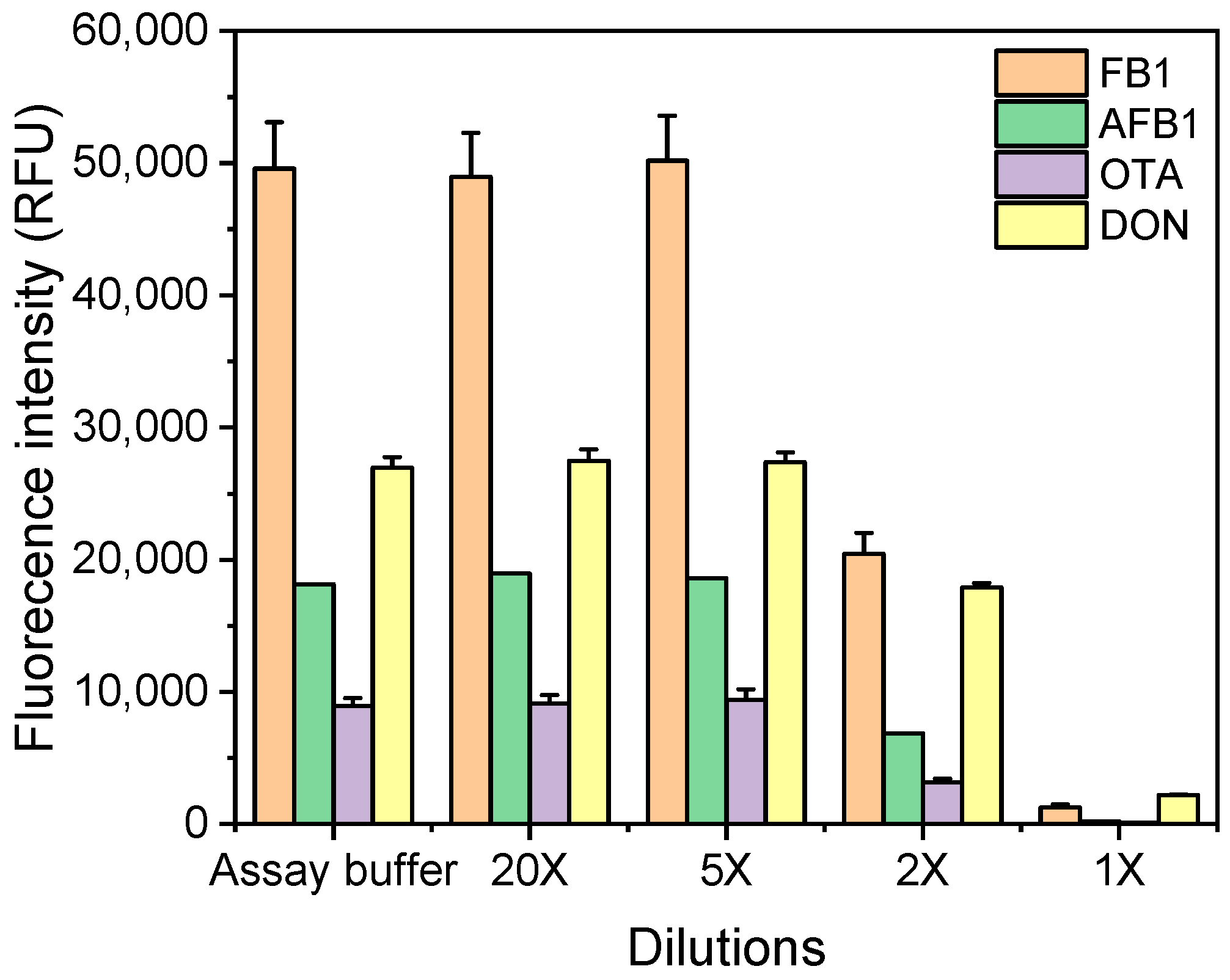
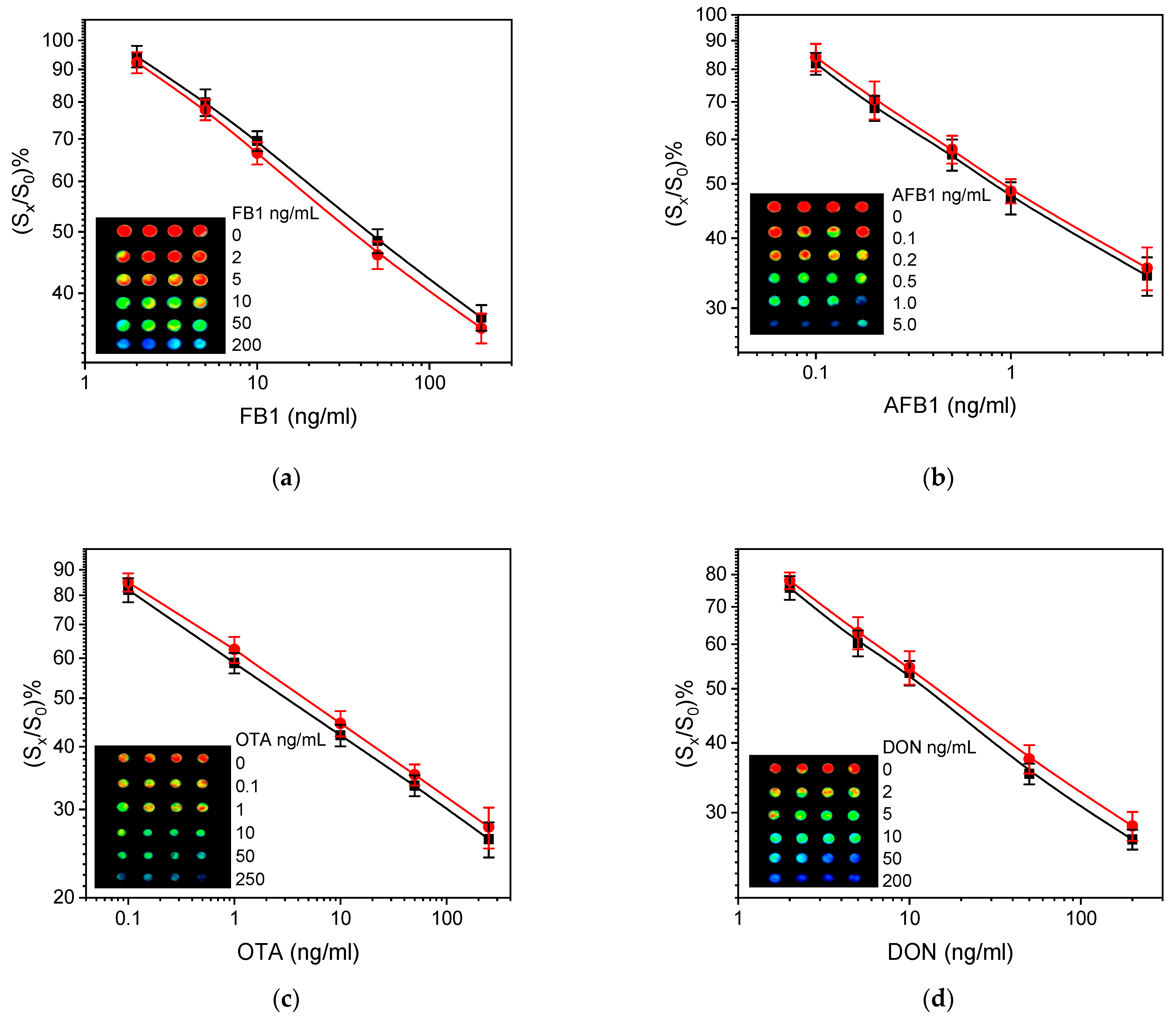
3.3. Analytical Performance of Multiplex Microarray
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, F.; Groopman, J.D.; Pestka, J.J. Public health impacts of foodborne mycotoxins. Annu. Rev. Food Sci. Technol. 2014, 5, 351–372. [Google Scholar] [CrossRef]
- Streit, E.; Schwab, C.; Sulyok, M.; Naehrer, K.; Krska, R.; Schatzmayr, G. Multi mycotoxin screening reveals the occurrence of 139 different secondary metabolites in feed and feed ingredients. Toxins 2013, 5, 504–523. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, L.; Giovannoli, C.; Baggiani, C. Mycotoxin detection. Curr. Opin. Biotechnol. 2016, 37, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Klaric, M.S.; Rasic, D.; Peraica, M. Deleterious effects of mycotoxin combinations involving ochratoxin A. Toxins 2013, 5, 1965–1987. [Google Scholar] [CrossRef] [PubMed]
- Creppy, E.E.; Chiarappa, P.; Baudrimont, I.; Borracci, P.; Moukha, S.; Carratu, M.R. Synergistic effects of fumonisinB1 and ochratoxin A: Are in vitro cytotoxicity data predictive of in vivo acute toxicity? Toxicology 2004, 201, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, R.; Guy, P.A. The pivotal role of mass spectrometry in determining the presence of chemical contaminants in food raw materials. Mass Spectrom. Rev. 2011, 30, 1073–1095. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hu, W.; Zhang, Q.; Li, P.; Li, C. Competitive immunoassays using antigen microarrays. Methods Mol. Biol. 2016, 1368, 237–247. [Google Scholar]
- Liu, Y.; Li, C.M. Advanced immobilization and amplification for high performance protein chips. Anal. Lett. 2012, 45, 130–155. [Google Scholar] [CrossRef]
- Krizkova, S.; Heger, Z.; Zalewska, M.; Moulick, A.; Adam, V.; Kizek, R. Nanotechnologies in protein microarrays. Nanomedicine 2015, 10, 2743–2755. [Google Scholar] [CrossRef]
- Sauer, U. Analytical protein microarrays: Advancements towards clinical applications. Sensors 2017, 17, 256. [Google Scholar] [CrossRef] [PubMed]
- Tsougeni, K.; Tserepi, A.; Boulousis, G.; Constantoudis, V.; Gogolides, E. Tunable poly(dimethylsiloxane) topography in O2 or Ar plasmas for controlling surface wetting properties and their ageing. Jpn. J. Appl. Phys. 2007, 46, 744. [Google Scholar] [CrossRef]
- Vourdas, N.; Tserepi, A.; Gogolides, E. Nanotextured super-hydrophobic transparent poly(methyl methacrylate) surfaces using high-density plasma processing. Nanotechnology 2007, 18, 125304. [Google Scholar] [CrossRef]
- Tsougeni, K.; Petrou, P.S.; Tserepi, A.; Kakabakos, S.E.; Gogolides, E. Nano-texturing of poly(methyl methacrylate) polymer using plasma processes and applications in wetting control and protein adsorption. Microelectron. Eng. 2009, 86, 1424–1427. [Google Scholar] [CrossRef]
- Tsougeni, K.; Vourdas, N.; Tserepi, A.; Gogolides, E.; Cardinaud, C. Mechanisms of oxygen plasma nanotexturing of organic polymer surfaces: From stable super hydrophilic to super hydrophobic surfaces. Langmuir 2009, 25, 11748–11759. [Google Scholar] [CrossRef] [PubMed]
- Vourdas, N.E.; Vlachopoulou, M.-E.; Tserepi, A.; Gogolides, E. Nano-textured polymer surfaces with controlled wetting and optical properties using plasma processing. Int. J. Nanotechnol. 2009, 6, 196–207. [Google Scholar] [CrossRef]
- Tsougeni, K.; Tserepi, A.; Constantoudis, V.; Gogolides, E.; Petrou, P.S.; Kakabakos, S.E. Plasma nanotextured PMMA surfaces for protein arrays: Increased protein binding and enhanced detection sensitivity. Langmuir 2010, 26, 13883–13891. [Google Scholar] [CrossRef] [PubMed]
- Tsougeni, K.; Ellinas, K.; Koukouvinos, G.; Petrou, P.S.; Tserepi, A.; Kakabakos, S.E.; Gogolides, E. Three-dimensional (3D) plasma micro-nanotextured slides for high performance biomolecule microarrays: Comparison with epoxy-silane coated glass slides. Colloid Surf. B Biointerfaces 2018, 165, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Nagl, S.; Schaeferling, M.; Wolgbeis, O.S. Fluorescence analysis in microarray technology. Microchim. Acta 2005, 151, 1–21. [Google Scholar] [CrossRef]
- Tseng, M.L.; Huang, Y.W.; Hsiao, M.K.; Huang, H.W.; Chen, H.M.; Chen, Y.L.; Chu, C.H.; Chu, N.N.; He, Y.J.; Chang, C.M.; et al. Fast fabrication of an Ag nanostructure substrate using the femtosecond laser for broad-band and tunable plasmonic enhancement. ACS Nano 2012, 6, 5190–5197. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, J.; Lakowicz, J.R. Plasmon enhanced fluorescence from single fluorophores end-linked to gold nanorods. JACS 2010, 132, 5540–5541. [Google Scholar] [CrossRef]
- Larguinho, M.; Baptista, P.V. Gold and silver nanoparticles for clinical diagnostics—From genomics to proteomics. J. Proteom. 2012, 75, 2811–2823. [Google Scholar] [CrossRef]
- Gu, X.; Wu, Y.; Zhang, L.; Liu, Y.; Li, Y.; Yan, Y.; Wu, D. Hybrid magnetic nanoparticle/nanogold clusters and their distance-dependent metal-enhanced fluorescence effect via DNA hybridization. Nanoscale 2014, 6, 8681–8693. [Google Scholar] [CrossRef]
- Liu, C.; Meng, F.; Wang, B.; Zhang, L.; Cui, X. Plasmonicnanograting enhanced fluorescence for protein microarray analysis of carcinoembryonic antigen (CEA). Anal. Methods 2018, 10, 145–150. [Google Scholar] [CrossRef]
- Li, H.; Wang, M.; Qiang, W.; Hu, H.; Li, W.; Xu, D. Metal-enhanced fluorescent detection for protein microarrays based on a silver plasmonic substrate. Analyst 2014, 139, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Todescato, F.; Antognoli, A.; Meneghello, A.; Cretaio, E.; Signorini, R.; Bozio, R. Sensitive detection of Ochratoxin A in food and drinks using metal-enhanced fluorescence. Biosens. Bioelectron. 2014, 57, 125–132. [Google Scholar] [CrossRef]
- Lee, H.J.; Ryu, D. Worldwide occurrence of mycotoxins in cereals and cereal-derived food products: Public health perspectives of their co-occurrence. J. Agric. Food Chem. 2017, 23, 7034–7051. [Google Scholar] [CrossRef] [PubMed]
- Papasarantos, I.; Klimentzou, P.; Koutrafouri, V.; Anagnostouli, M.; Zikos, C.; Paravatou-Petsotas, M.; Livaniou, E. Solid-phase synthesis of a biotin derivative and its application to the development of anti-biotin antibodies. Appl. Biochem. Biotechnol. 2010, 162, 221–232. [Google Scholar] [CrossRef]
- Perosa, F.; Carbone, R.; Ferrone, S.; Dammacco, F.F. Purification of human immunoglobulins by sequential precipitation with caprylic acid and ammonium sulphate. J. Immunol. Methods 1990, 128, 9–16. [Google Scholar] [CrossRef]
- Koukouvinos, G.; Petrou, P.S.; Misiakos, K.; Drygiannakis, D.; Raptis, I.; Goustouridis, D.; Kakabakos, S.E. A label-free flow-through immunosensor for determination of total- and free-PSA in human serum samples based on white-light reflectance spectroscopy. Sens. Actuator B 2015, 209, 1041–1048. [Google Scholar] [CrossRef]
| Mycotoxin | EU MRL in Corn (μg/kg) | Multiplex Microarray Linear Working Range (μg/kg) | Multiplex Microarray LoD (μg/kg) | Intra-Assay CVs | Inter-Assay CVs |
|---|---|---|---|---|---|
| FB1 | 500 | 10–400 | 4.0 | 3.6–5.6 | 5.1–8.9 |
| AFB1 | 2 | 0.5–20 | 0.2 | 2.9–6.1 | 4.6–8.5 |
| OTA | 3 | 0.5–400 | 0.2 | 3.7–7.2 | 5.5–9.1 |
| DON | 750 | 10–400 | 4.0 | 4.1–6.9 | 4.7–8.4 |
| Mycotoxin | Amount Added (ng/mL) | Amount Determined (ng/mL) | % Recovery |
|---|---|---|---|
| FB1 | 100 | 107 ± 6.0 | 107 ± 6.0 |
| 300 | 310 ± 16 | 103 ± 5.2 | |
| AFB1 | 0.45 | 0.42 ± 0.04 | 93.3 ± 9.5 |
| 10 | 11.1 ± 0.08 | 111 ± 7.2 | |
| OTA | 0.60 | 0.50 ± 0.04 | 83.3 ± 8.0 |
| 10 | 11.5 ± 0.38 | 115 ± 3.3 | |
| DON | 100 | 116 ± 5.4 | 116 ± 5.4 |
| 300 | 327 ± 22 | 109 ± 6.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koukouvinos, G.; Karachaliou, C.-E.; Kanioura, A.; Tsougeni, K.; Livaniou, E.; Kakabakos, S.E.; Petrou, P.S. Fluorescence Enhancement on Silver-Plated Plasma Micro-Nanostructured 3D Polymeric Microarray Substrates for Multiplex Mycotoxin Detection. Processes 2021, 9, 392. https://doi.org/10.3390/pr9020392
Koukouvinos G, Karachaliou C-E, Kanioura A, Tsougeni K, Livaniou E, Kakabakos SE, Petrou PS. Fluorescence Enhancement on Silver-Plated Plasma Micro-Nanostructured 3D Polymeric Microarray Substrates for Multiplex Mycotoxin Detection. Processes. 2021; 9(2):392. https://doi.org/10.3390/pr9020392
Chicago/Turabian StyleKoukouvinos, Georgios, Chrysoula-Evangelia Karachaliou, Anastasia Kanioura, Katerina Tsougeni, Evangelia Livaniou, Sotirios Elias Kakabakos, and Panagiota Sotirios Petrou. 2021. "Fluorescence Enhancement on Silver-Plated Plasma Micro-Nanostructured 3D Polymeric Microarray Substrates for Multiplex Mycotoxin Detection" Processes 9, no. 2: 392. https://doi.org/10.3390/pr9020392
APA StyleKoukouvinos, G., Karachaliou, C.-E., Kanioura, A., Tsougeni, K., Livaniou, E., Kakabakos, S. E., & Petrou, P. S. (2021). Fluorescence Enhancement on Silver-Plated Plasma Micro-Nanostructured 3D Polymeric Microarray Substrates for Multiplex Mycotoxin Detection. Processes, 9(2), 392. https://doi.org/10.3390/pr9020392








