Tetracycline Induces the Formation of Biofilm of Bacteria from Different Phases of Wastewater Treatment
Abstract
:1. Introduction
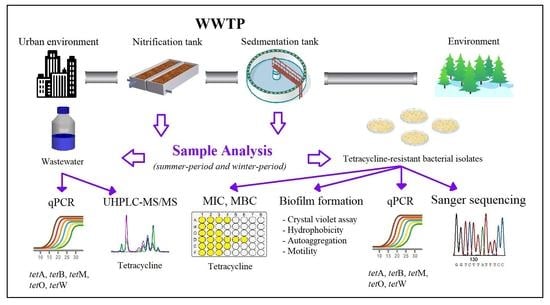
2. Materials and Methods
2.1. Sampling Sites
2.2. Determination of Tetracycline Content in Wastewater
2.3. DNA Extraction and Quantification of ARGs in Wastewater
2.4. Isolation of Bacterial Strains Resistant to Tetracycline
2.5. Characterization of Bacterial Strains
2.5.1. Growth Conditions and Standardized Inoculum Preparation
2.5.2. Enzymatic Activity
2.5.3. Crystal Violet Assay for Determination of Biofilm Formation in TSB
2.5.4. Crystal Violet Assay for Determination of Biofilm Formation in Wastewater
2.5.5. Autoaggregation Test
2.5.6. Hydrophobicity Test
2.5.7. Motility Test
2.6. Microdilution Method–Determination of Tetracycline MIC and MBC for Selected Isolates and the Influence of Sub-Inhibitory Tetracycline Concentrations on Growth and Biofilm Formation
2.7. Identification of Selected Isolates
2.8. Detection of ARGs of Selected Isolates
2.9. Statistical Analysis
3. Results and Discussion
3.1. Quantification of Tetracycline in Wastewater
3.2. Quantification of ARGs in Wastewater
3.3. Characterization of Bacterial Strains Isolated from Wastewater
3.3.1. Enzymatic Activity
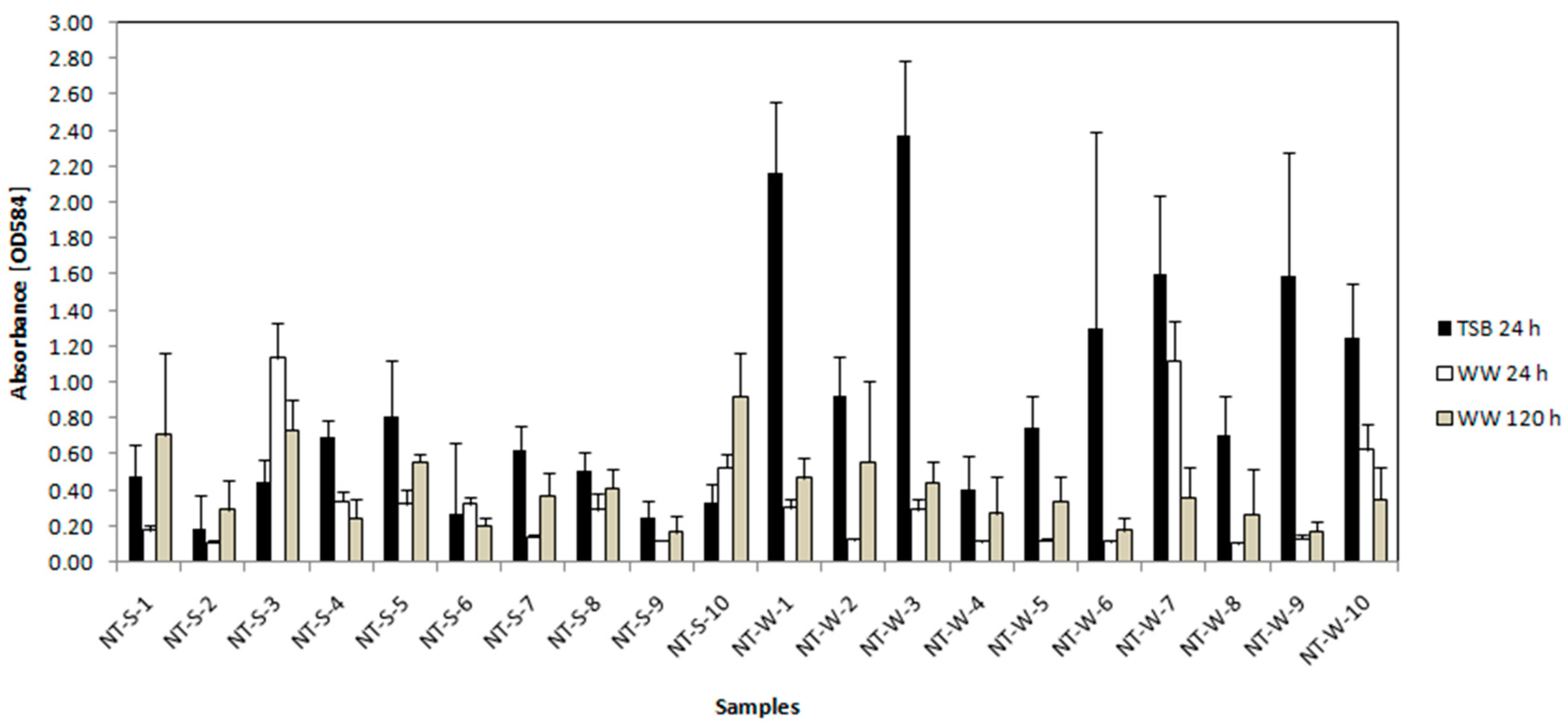
3.3.2. Crystal Violet Assay for Determination of Biofilm Formation
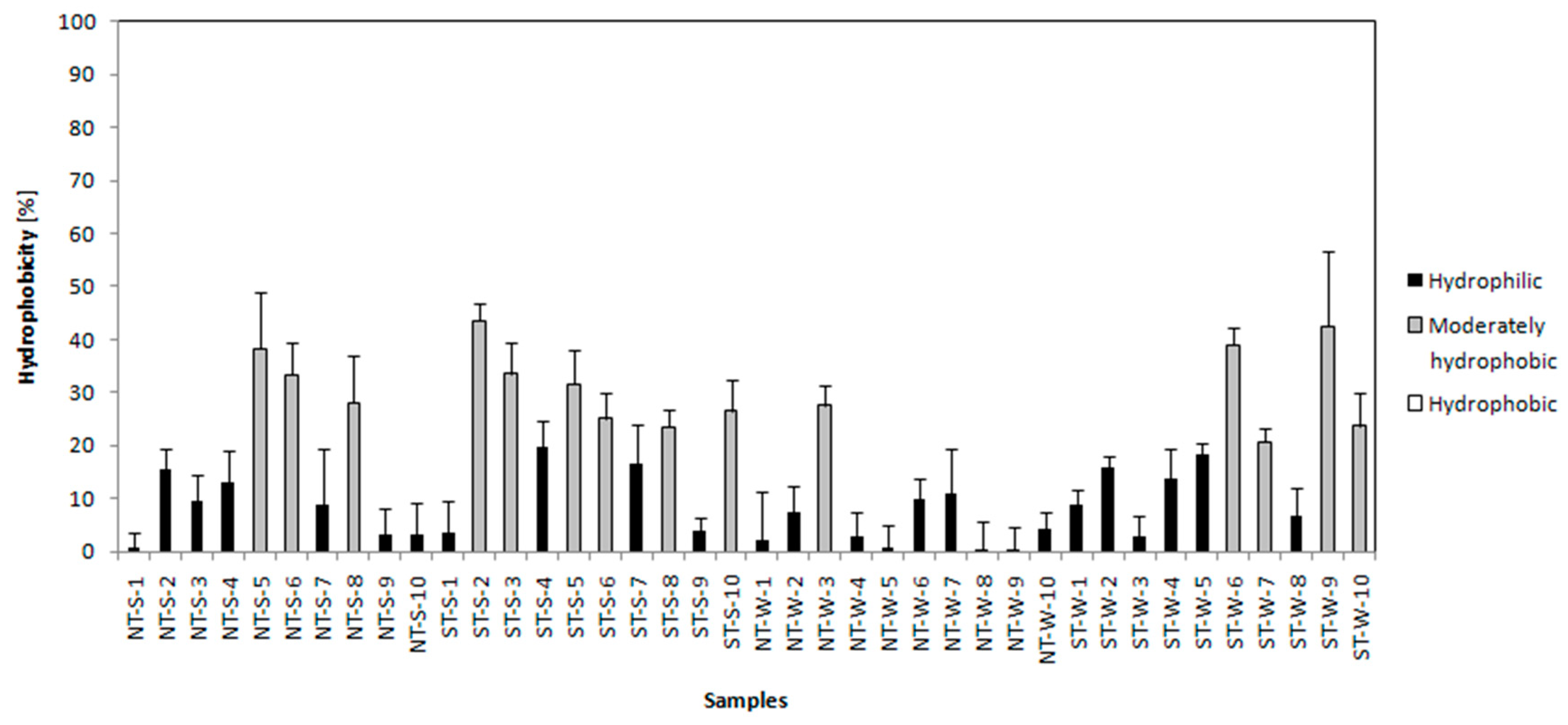
3.3.3. Autoaggregation Test, Hydrophobicity Test and Motility
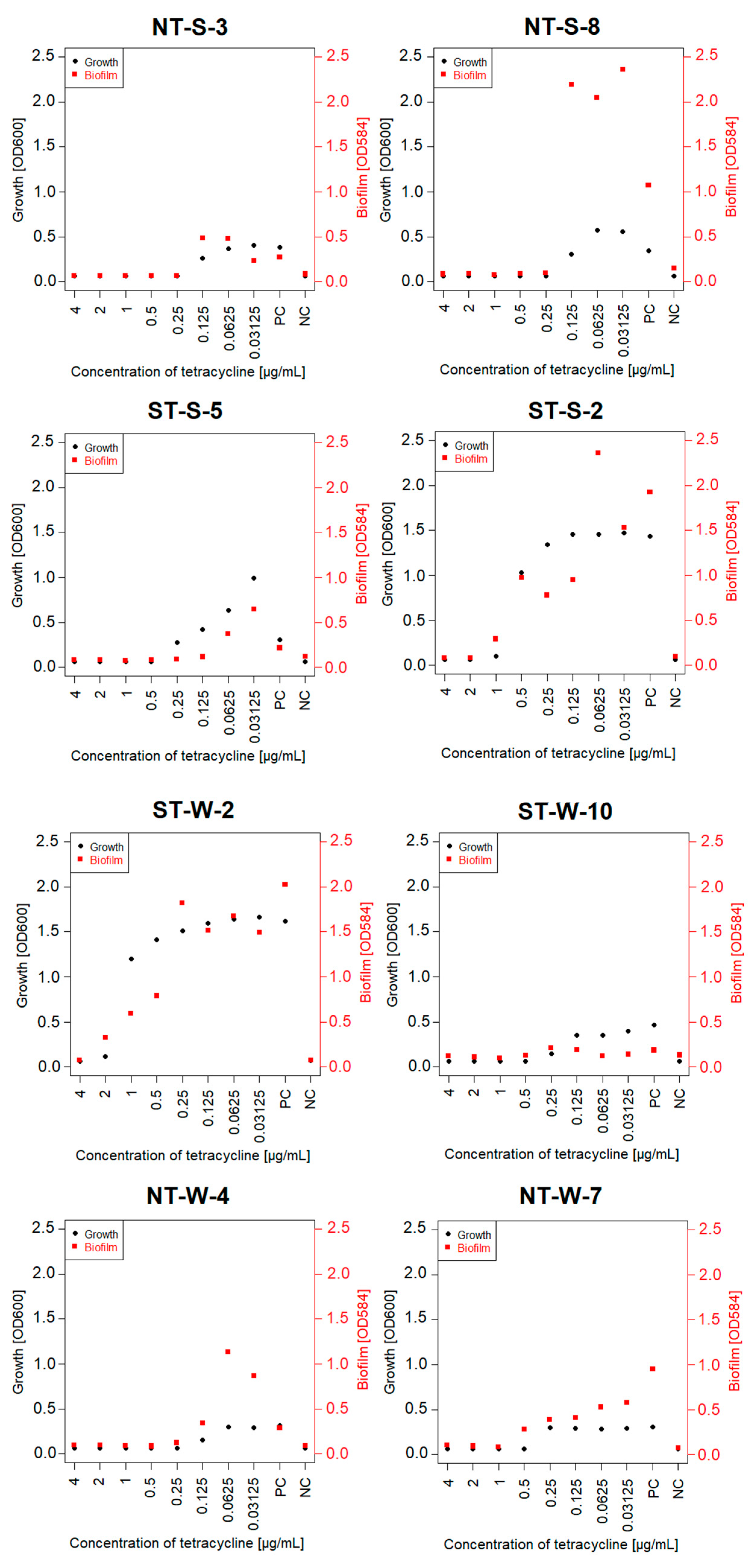
3.4. Tetracycline MIC and MBC for Selected Isolates and the Effect of Sub-Inhibitory Concentrations on Biofilm Formation
3.5. Identification of Selected Isolates and Detection of ARGs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Manaia, C.M.; Macedo, G.; Fatta-Kassinos, D.; Nunes, O.C. Antibiotic resistance in urban aquatic environments: Can it be controlled? Appl. Microbiol. Biotechnol. 2016, 100, 1543–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlin, C.; Bonot, S.; Courtois, S.; Block, J.-C. Persistence and dissemination of the multiple-antibiotic-resistance plasmid pB10 in the microbial communities of wastewater sludge microcosms. Water Res. 2011, 45, 2897–2905. [Google Scholar] [PubMed]
- Cyprowski, M.; Stobnicka-Kupiec, A.; Ławniczek-Wałczyk, A.; Bakal-Kijek, A.; Gołofit-Szymczak, M.; Górny, R.L. Anaerobic bacteria in wastewater treatment plant. Int. Arch. Occup. Environ. Health 2018, 91, 571–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonomo, R.A.; Szabo, D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006, 43, S49–S56. [Google Scholar] [CrossRef] [Green Version]
- Kaskhedikar, M.; Chhabra, D. Multiple drug resistance in Aeromonas hydrophila isolates of fish. Food Microbiol. 2010, 28, 157–168. [Google Scholar]
- Silge, A.; Schumacher, W.; Rösch, P.; Da Costa Filho, P.A.; Gérard, C.; Popp, J. Identification of water-conditioned Pseudomonas aeruginosa by Raman microspectroscopy on a single cell level. Syst. Appl. Microbiol. 2014, 37, 360–367. [Google Scholar] [CrossRef]
- Zhang, Y.; Marrs, C.F.; Simon, C.; Xi, C. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total Environ. 2009, 407, 3702–3706. [Google Scholar] [CrossRef]
- Faleye, A.C.; Adegoke, A.A.; Ramluckan, K.; Bux, F.; Stenström, T.A. Antibiotic Residue in the Aquatic Environment: Status in Africa. Open Chem. 2018, 16, 890–903. [Google Scholar] [CrossRef]
- Sanganyado, E.; Gwenzi, W. Antibiotic resistance in drinking water systems: Occurrence, removal, and human health risks. Sci. Total Environ. 2019, 669, 785–797. [Google Scholar] [CrossRef]
- Jałowiecki, L.; Żur, J.; Chojniak, J.; Ejhed, H.; Płaza, G. Properties of Antibiotic-Resistant Bacteria Isolated from Onsite Wastewater Treatment Plant in Relation to Biofilm Formation. Curr. Microbiol. 2018, 75, 639–649. [Google Scholar] [CrossRef] [Green Version]
- Smole Možina, S.; Klančnik, A.; Raspor, P. Mechanisms of microbial resistance in biofilms. In Biofilms in Bioengineering (Microbiology Research Advances), 2nd ed.; Simões, M., Mergulhão, F., Eds.; Nova Science Publishers: New York, NY, USA, 2013; pp. 311–332. [Google Scholar]
- Salcedo, D.E.; Lee, J.H.; Ha, U.H.; Kim, S.P. The effects of antibiotics on the biofilm formation and antibiotic resistance gene transfer. Desalin. Water Treat. 2014, 54, 3582–3588. [Google Scholar] [CrossRef]
- Satoh, H.; Odagiri, M.; Ito, T.; Okabe, S. Microbial community structures and in situ sulphate-reducing and sulphur-oxidizing activities in biofilms developed on mortar specimens in a corroded sewer system. Water Res. 2009, 43, 4729–4739. [Google Scholar] [PubMed] [Green Version]
- Auguet, O.; Maite, P.; Guasch-Balcells, H.; Borrego, C.M.; Gutierrez, O. Implications of downstream nitrate dosage in anaerobic sewers to control sulfide and methane emissions. Water Res. 2015, 68, 522–532. [Google Scholar] [PubMed]
- Van, T.T.H.; Yidana, Z.; Smooker, P.M.; Coloe, P.J. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses. J. Glob. Antimicrob. Resist. 2020, 20, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Borghi, A.A.; Palma, M.S.A. Tetracycline: Production, waste treatment and environmental impact assessment. Braz. J. Pharm. Sci. 2014, 50, 25–40. [Google Scholar] [CrossRef]
- Javid, A.; Mesdaghinia, A.; Nasseri, S.; Mahvi, A.H.; Alimohammadi, M.; Gharibi, H. Assessment of tetracycline contamination in surface and groundwater resources proximal to animal farming houses in Tehran, Iran. J. Environ. Health Sci. Eng. 2016, 14, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Hubeny, J.; Buta, M.; Zieliński, W.; Harnisz, M.; Korzeniewska, E.; Nowrotek, M.; Płaza, G. The Prevalence of tet(A) and tet(M) Tetracycline Resistance Genes in Municipal Wastewater. J. Ecol. Eng. 2019, 20, 1–6. [Google Scholar] [CrossRef]
- Kang, Y.; Li, Q.; Yin, Z.; Shen, M.; Zhao, H.; Bai, Y.; Mei, L.; Hu, J. High diversity and abundance of cultivable tetracycline-resistant bacteria in soil following pig manure application. Sci. Rep. 2018, 8, 1489. [Google Scholar] [CrossRef] [Green Version]
- Granados-Chinchilla, F.; Rodríguez, C. Tetracyclines in Food and Feedingstuffs: From Regulation to Analytical Methods, Bacterial Resistance, and Environmental and Health Implications. J. Anal. Methods Chem. 2017, 2017, 1315497. [Google Scholar] [CrossRef]
- Daghrir, R.; Drogui, P. Tetracycline antibiotics in the environment: A review. Environ. Chem. Lett. 2013, 11, 209–227. [Google Scholar] [CrossRef]
- Deblonde, T.; Cossu-Leguille, C.; Hartemann, P. Emerging pollutants in wastewater: A review of the literature. Int. J. Hyg. Environ. Health 2011, 214, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.; Olson, N.D.; Raspanti, G.A.; Rosenberg Goldstein, R.E.; Gibbs, S.G.; Sapkota, A.; Sapkota, A.R. Antibiotic Concentrations Decrease during Wastewater Treatment but Persist at Low Levels in Reclaimed Water. Int. J. Environ. Res. Public Health 2017, 14, 668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasparrini, A.J.; Markley, J.L.; Kumar, H.; Wang, B.; Fang, L.; Irum, S.; Symister, C.T.; Wallace, M.; Burnham, C.-A.D.; Andleeb, S.; et al. Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun. Biol. 2020, 3, 241. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Nomura, N.; Suzuki, S. Biofilms: Hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 2020, 96, fiaa031. [Google Scholar] [CrossRef]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef] [Green Version]
- Svobodová, K.; Semerád, J.; Petráčková, D.; Novotný, Č. Antibiotic Resistance in Czech Urban Wastewater Treatment Plants: Microbial and Molecular Genetic Characterization. Microb. Drug Resist. 2018, 24, 830–838. [Google Scholar] [CrossRef]
- Stach, J.E.M.; Bathe, S.; Clapp, J.P.; Burns, R.G. PCR-SSCP comparison of 16SrDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol. Ecol. 2001, 36, 139–151. [Google Scholar] [CrossRef]
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of Antibiotic Resistance Genes and Bacterial Community Composition in a River Influenced by a Wastewater Treatment Plant. PLoS ONE 2013, 8, E78906. [Google Scholar] [CrossRef]
- Boerjesson, S.; Mattsson, A.; Lindgren, P.E. Genes encoding tetracycline resistance in a full-scale municipal wastewater treatment plant investigated during one year. J. Water Health 2010, 8, 247–256. [Google Scholar] [CrossRef]
- Rathnayake, I.U.; Hargreaves, M.; Huygens, F. Antibiotic resistance and virulence traits in clinical and environmental Enterococcus faecalis and Enterococcus faecium isolates. System. Appl. Microbiol. 2012, 35, 326–333. [Google Scholar] [CrossRef]
- Nord, C.-E.; Sjöberg, L.; Wadström, T.; Wretlind, B. Characterization of three Aeromonas and nine Pseudomonas species by extracellular enzymes and haemolysins. Med. Microbiol. Immunol. 1975, 161, 79–87. [Google Scholar] [CrossRef]
- Rollof, J.; Hedstrom, S.A.; Nilsson-Ehle, P. Lipolytic aktivity of Staphylococcus aureus strains from disseminated and localized infections. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. 1987, 95, 109–113. [Google Scholar]
- Igarashi, T.; Aritake, S.; Yasumoto, T. Mechanisms underlying the hemolytic and ichthyotoxic activities of maitotoxin. Nat. Toxins 1999, 7, 71–79. [Google Scholar] [CrossRef]
- Kurinčič, M.; Jeršek, B.; Klančnik, A.; Smole Možina, S.; Fink, R.; Dražić, R.; Raspor, P.; Bohinc, K. Effects of natural antimicrobials on bacterial cell hydrophobicity, adhesion, and zeta potential. Arh. Hig. Rada Toksikol. 2016, 67, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sterniša, M.; Klančnik, A.; Smole Možina, S. Spoilage Pseudomonas biofilm with Escherichia coli protection in fish meat at 5 °C. J. Sci. Food Agric. 2019, 99, 4635–4641. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.; Chaves-López, C.; Serio, A.; Goffredo, E.; Goga, B.T.C.; Paparella, A. Influence of Incubation Conditions on Biofilm Formation by Pseudomonas Fluorescens Isolated from Dairy Products and Dairy Manufacturing Plants. Ital. J. Food Saf. 2016, 5, 5793. [Google Scholar] [CrossRef]
- Kadam, T.A.; Rupa, L.; Balhal, D.K.; Totewad, N.D.; Gyananath, G. Determination of the degree of hydrophobicity—A technique to assess bacterial colonization on leaf surface and root region of lotus plant. Asian J. Exp. Sci. 2009, 23, 135–139. [Google Scholar]
- O’May, C.; Tufenkji, N. The Swarming Motility of Pseudomonas aeruginosa Is Blocked by Cranberry Proanthocyanidins and Other Tannin-Containing Materials. Appl. Environ. Microbiol. 2011, 77, 3061–3067. [Google Scholar] [CrossRef] [Green Version]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Hong, Y.; Park, J.; Sharma, V.K.; Cho, S. Sulfonamides and tetracyclines in livestock wastewater. Chemosphere 2013, 91, 888–894. [Google Scholar] [CrossRef]
- Jia, A.; Xiao, Y.; Hu, J.; Asami, M.; Kunikane, S. Simultaneous determination of tetracyclines and their degradation products in environmental waters by liquid chromatography–electrospray tandem mass spectrometry. J. Chromatogr. A 2009, 1216, 4655–4662. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Xia, Y.; Liu, L.; Li, A.-D.; Yang, Y.; Zhang, T. Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing. Microbiome 2019, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Flaherty, E.; Borrego, C.M.; Balcázar, J.L.; Cummins, E. Human exposure assessment to antibiotic-resistant Escherichia coli through drinking water. Sci. Total Environ. 2018, 616–617, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Schwarz, S. Tetracycline and Chloramphenicol Resistance Mechanisms. In Antimicrobial Drug Resistance: Mechanisms of Drug Resistance, 2nd ed.; Mayers, D.G., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing AG: Cham, Switzerland, 2009; Volume 1, pp. 183–193. [Google Scholar]
- Aminov, R.I.; Garrigues-Jeanjean, N.; Mackie, R.I. Molecular Ecology of Tetracycline Resistance: Development and Validation of Primers for Detection of Tetracycline Resistance Genes Encoding Ribosomal Protection Proteins. Appl. Environ. Microbiol. 2001, 67, 22–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connell, S.R.; Tracz, D.M.; Nierhaus, K.H.; Taylor, D.E. Ribosomal Protection Proteins and Their Mechanism of Tetracycline Resistance. Antimicrob. Agents Chemother. 2003, 47, 3675–3681. [Google Scholar] [CrossRef] [Green Version]
- Czekalski, N.; Gascón Díez, E.; Burgmann, H. Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J. 2014, 8, 1381–1390. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, X.; Huang, H.; Zhang, J. Prevalence of Antibiotic Resistance Genes and Their Association with Antibiotics in a Wastewater Treatment Plant: Process Distribution and Analysis. Water 2019, 11, 2495. [Google Scholar] [CrossRef] [Green Version]
- Miettinen, I.T.; Vartiainen, T.; Martikainen, P.J. Bacterial enzyme activities in ground water during bank infiltration of lake water. Water Res. 1996, 30, 2495–2501. [Google Scholar] [CrossRef]
- Carpa, R. Enzymological research on soils from different environments. Ann. Romanian Soc. Cell Biol. 2011, XVI, 44–48. [Google Scholar]
- Ramnath, L.; Sithole, B.; Govinden, R. Classification of lipolytic enzymes and their biotechnological applications in the pulping industry. Can. J. Microbiol. 2017, 63, 179–192. [Google Scholar] [CrossRef]
- McDaniel, C.S.; Wales, M.E.; Carvajal, J.C. Enzymatic Antimicrobial and Antifouling Coating and Polymeric Materials. U.S. Patent 0238811 A1, 24 September 2009. [Google Scholar]
- Yigit, N.; Aktas, E.; Dagistan, S.; Ayyildiz, A. Investigating Biofilm Production, Coagulase and Hemolytic Activity in Candida Species Isolated from Denture Stomatitis Patients. Eurasian J. Med. 2011, 43, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Coelho, S.M.O.; Reinoso, E.; Pereira, I.A.; Soares, L.C.; Demo, M.; Bogni, C.; Souza, M.M.S. Virulence factors and antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis in Rio de Janeiro. Pes. Vet. Bras. 2009, 29, 369–374. [Google Scholar] [CrossRef]
- Tielen, P.; Kuhn, H.; Rosenau, F.; Jaeger, K.-E.; Flemming, H.-C.; Wingender, J. Interaction between extracellular lipase LipA and the polysaccharide alginate of Pseudomonas aeruginosa. BMC Microbiol. 2013, 13, 159. [Google Scholar] [CrossRef] [Green Version]
- Abdulina, D.R.; Kurmakova, I.N.; Bondar, E.S.; Demchenko, N.R.; Bondarenko, A.I. Seasonal Dynamics of Bacteria in Corrosive Biofilms Formed on the Surface of Wastewater Treatment Plants. J. Water Chem. Technol. 2019, 41, 44–51. [Google Scholar] [CrossRef]
- Li, D.; Qi, R.; Yang, M.; Zhang, Y.; Yu, T. Bacterial community characteristics under longterm antibiotic selection pressures. Water Res. 2011, 45, 6063–6073. [Google Scholar] [PubMed]
- Solano, C.; Echeverz, M.; Lasa, I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 2014, 18, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277. [Google Scholar] [CrossRef]
- Krasowska, A.; Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell Infect. Microbiol. 2014, 4, 112. [Google Scholar] [CrossRef] [Green Version]
- Simões, L.C.; Simões, M.; Vieira, M.J. Biofilms interactions between distinct bacterial genera isolated from drinking water. Appl. Environ. Microbiol. 2007, 73, 6192–6200. [Google Scholar] [CrossRef] [Green Version]
- Penesyan, A.; Nagy, S.S.; Kjelleberg, S.; Gillings, M.R.; Paulsen, I.T. Rapid microevolution of biofilm cells in response to antibiotics. NPJ Biofilms Microbiomes 2019, 5, 34. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.-J.; Hu, H.-Y.; Lu, S.-Q.; Li, Y.; Tang, F.; Lu, Y.; Wei, B. Monitoring and Evaluation of Antibiotic-Resistant Bacteria at a Municipal Wastewater Treatment Plant in China. Environ. Int. 2012, 42, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Obayiuwana, A.; Ogunjobi, A.; Yang, M.; Ibekwe, M. Characterization of Bacterial Communities and Their Antibiotic Resistance Profiles in Wastewaters Obtained from Pharmaceutical Facilities in Lagos and Ogun States, Nigeria. Int. J. Environ. Res. Public Health 2018, 15, 1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehm, A.; Steiner, S.; Zaehringer, F.; Casanova, A.; Hamburger, F.; Ritz, D.; Keck, W.; Ackermann, M.; Shirmer, T.; Jenal, U. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 2009, 72, 1500–1516. [Google Scholar] [CrossRef] [PubMed]
- May, T.; Ito, A.; Okabe, S. Induction of Multidrug Resistance Mechanism in Escherichia coli Biofilms by Interplay between Tetracycline and Ampicillin Resistance Genes. Antimicrob. Agents Chemother. 2009, 53, 4628–4639. [Google Scholar] [CrossRef] [Green Version]
- Talagrand-Reboul, T.; Jumas-Bilak, E.; Lamy, B. The Social Life of Aeromonas through Biofilm and Quorum Sensing Systems. Front. Microbiol. 2017, 8, 37. [Google Scholar] [CrossRef]
- Barati, A.; Ghaderpour, A.; Chew, L.L.; Bong, C.W.; Thong, K.L.; Chong, V.C.; Chai, L.C. Isolation and Characterization of Aquatic-Borne Klebsiella pneumoniae from Tropical Estuaries in Malaysia. Int. J. Environ. Res. Public Health 2016, 13, 426. [Google Scholar] [CrossRef] [Green Version]
- Mahapatra, A.; Padhi, N.; Mahapatra, D.; Bhatt, M.; Sahoo, D.; Jena, S.; Dash, D.; Chayani, N. Study of Biofilm in Bacteria from Water Pipelines. J. Clin. Diagn. Res. 2015, 9, DC09–DC11. [Google Scholar] [CrossRef]
- Emami, S.; Nikokar, I.; Ghasemi, Y.; Ebrahimpour, M.; Ebrahim-Saraie, H.S.; Araghian, A.; Faezi, S.; Farahbakhsh, M.; Rajabi, A. Antibiotic Resistance Pattern and Distribution of pslA Gene Among Biofilm Producing Pseudomonas aeruginosa Isolated From Waste Water of a Burn Center. Jundishapur J. Microbiol. 2015, 8, e23669. [Google Scholar] [CrossRef] [Green Version]
- Brink, A.J.; Coetzee, J.; Clay, C.G.; Sithole, S.; Richards, G.A.; Poirel, L.; Nordmann, P. Emergence of New Delhi metallo-beta-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa. J. Clin. Microbiol. 2012, 50, 525–527. [Google Scholar] [CrossRef] [Green Version]
- Luczkiewicz, A.; Kotlarska, E.; Artichowicz, W.; Tarasewicz, K.; Fudala-Ksiaze, S. Antimicrobial resistance of Pseudomonas spp. isolated from wastewater and wastewater-impacted marine coastal zone. Environ. Sci. Pollut. Res. 2015, 22, 19823–19834. [Google Scholar] [CrossRef] [Green Version]
- Stoesser, N.; Mathers, A.J.; Moore, C.E.; Day, N.P.; Crook, D.W. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect. Dis. 2016, 16, 285–286. [Google Scholar] [CrossRef] [Green Version]
- King, T.L.B.; Schmidt, S.; Essack, S.Y. Antibiotic resistant Klebsiella spp. from hospital, hospital effluents and wastewater treatment plants in the uMgungundlovu District, KwaZulu-Natal, South Africa. Sci. Total Environ. 2020, 712, 135550. [Google Scholar] [CrossRef] [PubMed]



| Sample | Lipase Activity | Lecithinase Activity | Hemolytic Activity | Motility (Swimming) |
|---|---|---|---|---|
| NT-S-1 | + | + | α | + |
| NT-S-2 | + | + | γ | − |
| NT-S-3 | + | + | β | + |
| NT-S-4 | + | + | γ | − |
| NT-S-5 | + | + | β | + |
| NT-S-6 | + | + | γ | + |
| NT-S-7 | + | − | γ | − |
| NT-S-8 | + | − | γ | + |
| NT-S-9 | + | + | γ | + |
| NT-S-10 | + | + | β | + |
| ST-S-1 | − | − | γ | + |
| ST-S-2 | + | − | γ | + |
| ST-S-3 | + | + | β | ++ |
| ST-S-4 | − | − | γ | ++ |
| ST-S-5 | + | + | γ | + |
| ST-S-6 | + | − | γ | − |
| ST-S-7 | − | - | γ | − |
| ST-S-8 | + | + | γ | + |
| ST-S-9 | − | − | γ | − |
| ST-S-10 | + | − | γ | ++ |
| NT-W-1 | + | + | γ | ++++ |
| NT-W-2 | + | + | α | + |
| NT-W-3 | + | − | γ | + |
| NT-W-4 | + | + | γ | − |
| NT-W-5 | − | − | γ | − |
| NT-W-6 | − | − | γ | + |
| NT-W-7 | + | + | γ | + |
| NT-W-8 | − | − | γ | +++ |
| NT-W-9 | − | − | γ | − |
| NT-W-10 | + | + | β | ++ |
| ST-W-1 | + | − | γ | − |
| ST-W-2 | − | − | γ | + |
| ST-W-3 | + | − | γ | − |
| ST-W-4 | + | + | α | − |
| ST-W-5 | − | + | γ | ++ |
| ST-W-6 | − | − | γ | − |
| ST-W-7 | + | + | γ | ++ |
| ST-W-8 | + | + | γ | ++ |
| ST-W-9 | + | + | γ | + |
| ST-W-10 | + | + | α | + |
| Sample | Biofilm | Hydrophobicity d | Autoaggregation e | Motility f | |||
|---|---|---|---|---|---|---|---|
| TSB a | WW 24 h b | WW 120 h c | 5 h | 24 h | |||
| NT-S-1 | Weak | Weak | Weak | Hydrophilic | Moderate | Moderate | Positive c |
| NT-S-2 | Weak | Weak | Weak | Hydrophilic | Moderate | Moderate | Negative c |
| NT-S-3 | Weak | Moderate | Weak | Hydrophilic | Moderate | Moderate | Positive c |
| NT-S-4 | Weak | Weak | Weak | Hydrophilic | Weak | Moderate | Negative c |
| NT-S-5 | Weak | Weak | Weak | Moderately hydrophobic | Weak | Moderate | Positive c |
| NT-S-6 | Weak | Weak | Weak | Moderately hydrophobic | Weak | Moderate | Positive c |
| NT-S-7 | Weak | Weak | Weak | Hydrophilic | Moderate | Moderate | Negative c |
| NT-S-8 | Weak | Weak | Weak | Moderately hydrophobic | Moderate | Moderate | Positive c |
| NT-S-9 | Weak | Weak | Weak | Hydrophilic | Weak | Moderate | Positive c |
| NT-S-10 | Weak | Weak | Weak | Hydrophilic | Weak | Weak | Positive c |
| ST-S-1 | Weak | Weak a | Weak a | Hydrophilic b | Weak | Moderate | Positive |
| ST-S-2 | Weak | Weak a | Weak a | Moderately hydrophobic b | Weak | Moderate | Positive |
| ST-S-3 | Weak | Weak a | Weak a | Moderately hydrophobic b | Weak | Moderate | Moderately positive |
| ST-S-4 | Moderate | Weak a | Weak a | Hydrophilic b | Weak | Moderate | Moderately positive |
| ST-S-5 | NP | Weak a | NP a | Moderately hydrophobic b | Weak | Weak | Positive |
| ST-S-6 | Weak | Weak a | NP a | Moderately hydrophobic b | Moderate | Strong | Negative |
| ST-S-7 | Weak | Weak a | Weak a | Hydrophilic b | Weak | Moderate | Negative |
| ST-S-8 | NP | Weak a | NP a | Moderately hydrophobic b | Weak | Moderate | Positive |
| ST-S-9 | Weak | Weak a | Weak a | Hydrophilic b | Moderate | Moderate | Negative |
| ST-S-10 | Weak | Weak a | Weak a | Moderately hydrophobic b | Weak | Moderate | Moderately positive |
| NT-W-1 | Moderate | Weak | Weak | Hydrophilic e | Weak | Moderate | Strong positive |
| NT-W-2 | Weak | Weak | Weak | Hydrophilic e | Moderate | Moderate | Positive |
| NT-W-3 | Moderate | Weak | Weak | Moderately hydrophobic e | Moderate | Moderate | Positive |
| NT-W-4 | Weak | Weak | Weak | Hydrophilic e | Weak | Moderate | Negative |
| NT-W-5 | Weak | Weak | Weak | Hydrophilic e | Weak | Moderate | Negative |
| NT-W-6 | Moderate | Weak | Weak | Hydrophilic e | Moderate | Moderate | Positive |
| NT-W-7 | Moderate | Moderate | Weak | Hydrophilic e | Weak | Moderate | Positive |
| NT-W-8 | Weak | Weak | Weak | Hydrophilic e | Moderate | Moderate | Medium positive |
| NT-W-9 | Moderate | Weak | Weak | Hydrophilic e | Weak | Moderate | Negative |
| NT-W-10 | Moderate | Weak | Weak | Hydrophilic e | Weak | Moderate | Moderately positive |
| ST-W-1 | Weak | Weak a | Weak a | Hydrophilic | Weak c | Moderate c | Negative |
| ST-W-2 | Weak | Weak a | Weak a | Hydrophilic | Weak c | Moderate c | Positive |
| ST-W-3 | Weak | Weak a | NP a | Hydrophilic | Weak c | Moderate c | Negative |
| ST-W-4 | Weak | NP a | Weak a | Hydrophilic | Weak c | Moderate c | Negative |
| ST-W-5 | Weak | Weak a | Weak a | Hydrophilic | Weak c | Moderate c | Moderately positive |
| ST-W-6 | Weak | Weak a | Weak a | Moderately hydrophobic | Weak c | Moderate c | Negative |
| ST-W-7 | Weak | Weak a | NP a | Moderately hydrophobic | Weak c | Moderate c | Moderately positive |
| ST-W-8 | Weak | Weak a | NP a | Hydrophilic | Weak c | Moderate c | Moderately positive |
| ST-W-9 | Weak | NP a | NP a | Moderately hydrophobic | Weak c | Moderate c | Positive |
| ST-W-10 | Weak | NP a | Weak a | Moderately hydrophobic | Weak c | Moderate c | Positive |
| Sample | MIC TET [µg/mL] | MBC TET [µg/mL] | Identification | tetA | tetB | tetM | tetO | tetW |
|---|---|---|---|---|---|---|---|---|
| NT-S-3 | 0.250 | 0.500 | Aeromonas salmonicida | – | + | – | – | + |
| NT-S-8 | 0.250 | 0.500 | Aeromonas salmonicida | – | – | – | – | + |
| ST-S-2 | 2.000 | 4.000 | Aeromonas salmonicida | – | – | – | – | + |
| ST-S-5 | 0.500 | 1.000 | Aeromonas sp. | – | – | – | – | + |
| NT-W-4 | 0.250 | 0.500 | Aeromonas sp. | – | – | – | – | + |
| NT-W-7 | 0.500 | 1.000 | Klebsiella pneumoniae | – | – | – | – | + |
| ST-W-2 | 4.000 | 8.000 | Pseudomonas sp. | – | – | – | – | + |
| ST-W-10 | 0.500 | 0.500 | Aeromonas sp. | – | – | – | – | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stachurová, T.; Malachová, K.; Semerád, J.; Sterniša, M.; Rybková, Z.; Smole Možina, S. Tetracycline Induces the Formation of Biofilm of Bacteria from Different Phases of Wastewater Treatment. Processes 2020, 8, 989. https://doi.org/10.3390/pr8080989
Stachurová T, Malachová K, Semerád J, Sterniša M, Rybková Z, Smole Možina S. Tetracycline Induces the Formation of Biofilm of Bacteria from Different Phases of Wastewater Treatment. Processes. 2020; 8(8):989. https://doi.org/10.3390/pr8080989
Chicago/Turabian StyleStachurová, Tereza, Kateřina Malachová, Jaroslav Semerád, Meta Sterniša, Zuzana Rybková, and Sonja Smole Možina. 2020. "Tetracycline Induces the Formation of Biofilm of Bacteria from Different Phases of Wastewater Treatment" Processes 8, no. 8: 989. https://doi.org/10.3390/pr8080989
APA StyleStachurová, T., Malachová, K., Semerád, J., Sterniša, M., Rybková, Z., & Smole Možina, S. (2020). Tetracycline Induces the Formation of Biofilm of Bacteria from Different Phases of Wastewater Treatment. Processes, 8(8), 989. https://doi.org/10.3390/pr8080989