Abstract
Chinese hamster ovary (CHO) cells are the most commonly used cell lines in biopharmaceutical manufacturing. Genome-scale metabolic models have become a valuable tool to study cellular metabolism. Despite the presence of reference global genome-scale CHO model, context-specific metabolic models may still be required for specific cell lines (for example, CHO-K1, CHO-S, and CHO-DG44), and for specific process conditions. Many integration algorithms have been available to reconstruct specific genome-scale models. These methods are mainly based on integrating omics data (i.e., transcriptomics, proteomics, and metabolomics) into reference genome-scale models. In the present study, we aimed to investigate the impact of time points of transcriptomics integration on the genome-scale CHO model by assessing the prediction of growth rates with each reconstructed model. We also evaluated the feasibility of applying extracted models to different cell lines (generated from the same parental cell line). Our findings illustrate that gene expression at various stages of culture slightly impacts the reconstructed models. However, the prediction capability is robust enough on cell growth prediction not only across different growth phases but also in expansion to other cell lines.
1. Introduction
Genome-scale metabolic models are valuable tools to describe cellular metabolism that link metabolic genes, proteins, reactions, and metabolites [1]. One widely used computational approach for genome-scale models is flux balance analysis (FBA) that estimates an optimal intracellular metabolic flux distribution, using an assumption of pseudo-steady state, the stoichiometry of metabolic network, and a defined biological objective function [2,3,4,5]. In the mammalian cell culture process, genome-scale metabolic models offer a mechanistic link between genotype and metabolic phenotype [4,6,7,8], for example, in the investigation of the effects of gene expression change on metabolic pathways and the corresponding cell growth, protein biosynthesis, or byproduct secretion [9]. With many other examples, it has shown that the systems biology models can help obtain novel insights and guide further experiments [6,10,11].
Several studies have indicated that not all genes or enzymes are active in a given cell line or culture condition [6,12]. In this regard, it is required to create context-specific (i.e., cell line-specific or condition-specific) models. Cell line- or culture condition-based omics data are integrated into existing reference models to generate a subset of the genome-scale network that represents the metabolism of specific cell lines or environmental conditions [13,14]. Several recent reviews and studies have comprehensively compared and evaluated computational approaches that are available for omics data integration to reconstruct models [9,15,16]. Among those, GIMME (Gene Inactivity Moderated by Metabolism and Expression) is a widely used tool that can perform both model reconstruction and flux prediction based on gene expression data [17].
Chinese hamster ovary (CHO) cells are the predominant mammalian cell type used for the production of monoclonal antibody (mAb) [18,19,20]. A consensus genome-scale model, iCHO1766, was reconstructed for CHO cells in 2016 [7]. This model contains 6663 metabolic reactions, 4456 metabolites, and 1766 metabolic genes. Cell line-specific genome-scale metabolic models (e.g., CHO-K1, CHO-S, and CHO-DG44) were subsequently extracted from iCHO1766 [6,7]. In the GIMME approach, the active metabolic genes or associated reactions are identified based on their absolute expression value and a user-defined threshold. However, only the gene expression level at a single time point (under steady-state) is considered for integration. This way has ignored the fact of dynamic systems in cell culture processes. In batch or fed-batch cultures, CHO cells are undergoing culture condition changes, including environmental variation (e.g., pH) and altered substrate availability [21]. To adapt to a changing environment, it is expected that cells can undergo gene expression changes [22]. Therefore, it raises the question of whether the varied gene expression in the cell culture will influence the context-specific metabolic model reconstruction, which in turn will affect the model application in simulating cell physiology and identifying engineering targets to enhance production. Another concern is the feasibility of applying reconstructed context-specific models to different cell lines (generated from the same parental cell line), which may be of great interest to industrial scientists who work on the cell line development.
There has been little attention given to the above questions. For this purpose, we performed a series of systematic assessment in this study using three mAb-producing cell lines to understand the effect of time-series transcriptomic data on the robustness of genome-scale models. First, we analyzed the overall change in the dynamic cell culture transcriptome. Genome-scale models were then extracted by integrating transcriptomic data from each culture stage into a reference model. Next, we compared the extracted models between different growth phases for each cell line, in terms of predictive accuracy and network content. Finally, we investigated the feasibility of models derived from one cell line data for the prediction in other cell lines.
2. Materials and Methods
2.1. Experimental Data
Three glutamine synthetase (GS) CHO cell lines producing different mAbs were used in this study and respectively termed CHO-A (to produce the mAbs of Adalimumab), CHO-B (Bevacizumab), and CHO-R (Rituximab). These three cell lines were derived from the same parental cell line. The detailed information on cell culture experiments can be found in the previous publication [23]. Cells were cultured in shake flasks for seven days with an inoculation density of 2 × 105 cells/mL and a working volume of 50 mL. A commercial media CD FortiCHO (ThermoFisher Scientific, Waltham, MA, USA) was used for these three cell lines. For each cell line, triplicate flasks were conducted in batch mode. Time-series gene expression profiles were determined daily using RNA-sequencing (RNA-Seq) from day 3 to day 6 [23]. A total of four data points of gene expression were collected. Metabolic genes in the CHO model were selected from BiGG database [24] or published genome-scale CHO model [7]. More detailed information about the processing of RNA-Seq can be found in a previously published manuscript [23].
Amino acid concentrations were measured using a HPLC system (Shimadzu, Kyoto, Japan) connected with a triple quadrupole mass spectrometer (Shimadzu, Kyoto, Japan) operated in positive electrospray ionization mode. A column Scherzo SM-C18 (3 µm, 100 × 3 mm, IMTAKT) was used for the component separation. Amino acid standard mixtures were separated using an Intrada Amino Acid column (100 × 3 mm, 3 µm, IMTAKT, Japan). Mobile phase A (aqueous) consisted of 0.3% v/v formic acid in acetonitrile; mobile phase B (organic) consisted of 20% v/v acetonitrile in 100 mM ammonium format. The chromatographic separation was accomplished by different gradients of mobile A and mobile B phase as follows: the mobile phase was started at 20% B for 4 min, increased from 20% B to 100% B with a linear gradient for 10 min, then maintained at 100% B for 2 min. The mobile phase was returned to 20% B and continued for 2 min. LC conditions included column temperature at 37 °C, injection volume of 10 μL, and mobile phase flow rate of 0.6 mL/min. More detailed information about the quantification of amino acids by LC-MS/MS can be found in reference [25].
2.2. Time-Series Transcriptomics and Extracellular Metabolomics Data Analysis
The time-series gene expression was analyzed via ANOVA (analysis of variance) in MATLAB (R2016a, MathWorks, Natick, MA, USA) to determine whether the gene expression during cell culture had significant change among different time points. A p-value < 0.001 was considered to be statistically significant in this work.
The measured time-series extracellular metabolomics was used to calculate specific uptake or secretion rates by using the following equations (Equations (1)–(3)).
where , , , are the viable cell density (VCD), product (e.g., lactate, NH4, mAb or other by-products in the supernatant), nutrient concentration (e.g., glucose, glutamine, glutamate, and other amino acids) and time, respectively. The , , represent the specific cell growth rate, secretion, and uptake rate of the extracellular metabolites, respectively. The specific mAb productivity of phase I in CHO-A was set as 100% to normalize productivity within this study.
2.3. Stoichiometric Models and Flux Balance Analysis
The genome-scale metabolic model can be represented by a stoichiometric matrix () of size (where and mean the number of metabolites and reactions, respectively) and a vector of reaction fluxes () [26,27]. Under the assumption of a quasi-steady state, the mass balance equations are given by [3]. Flux balance analysis was used to estimate the intracellular metabolic fluxes by the optimization of a defined objective function using linear programming. During the exponential growth phase, the optimal flux distribution was obtained using the objective function of maximizing biomass [7]. The calculated uptake and secretion rates of metabolites (as described in Section 2.2) were served as experimental metabolic constraints. The Constraint-Based Reconstruction and Analysis (COBRA) toolbox was used for FBA analysis in this study [28,29]. Gurobi solver was selected to solve linear programming optimization problems [30].
2.4. Model Extraction
We used the GIMME method [17] to reconstruct cell line- or condition-specific models in this work. This method uses linear programming and metabolic objective functions (e.g., cell growth) to create context-specific reconstruction while minimizing the inconsistency between fluxes and gene expression data [17,31]. First, a user-defined threshold of gene expression (e.g., a single threshold value of 1) was utilized to determine which gene is considered active or not. In this study, the threshold was set to 1, which means that genes with a normalized TPM (transcripts per million) value greater than 1 were considered as active. Then, the non-active reactions were removed from the reference model and the new context-specific model was created while optimizing the objective function. The GIMME algorithm was complemented in COBRA in MATLAB (Matlab 2016a; Mathworks, Natick, MA, USA). The constraints applied to the models and the RNA-Seq data used in this study can be available by contacting authors.
3. Results
3.1. CHO Cell Cultures and Dynamics
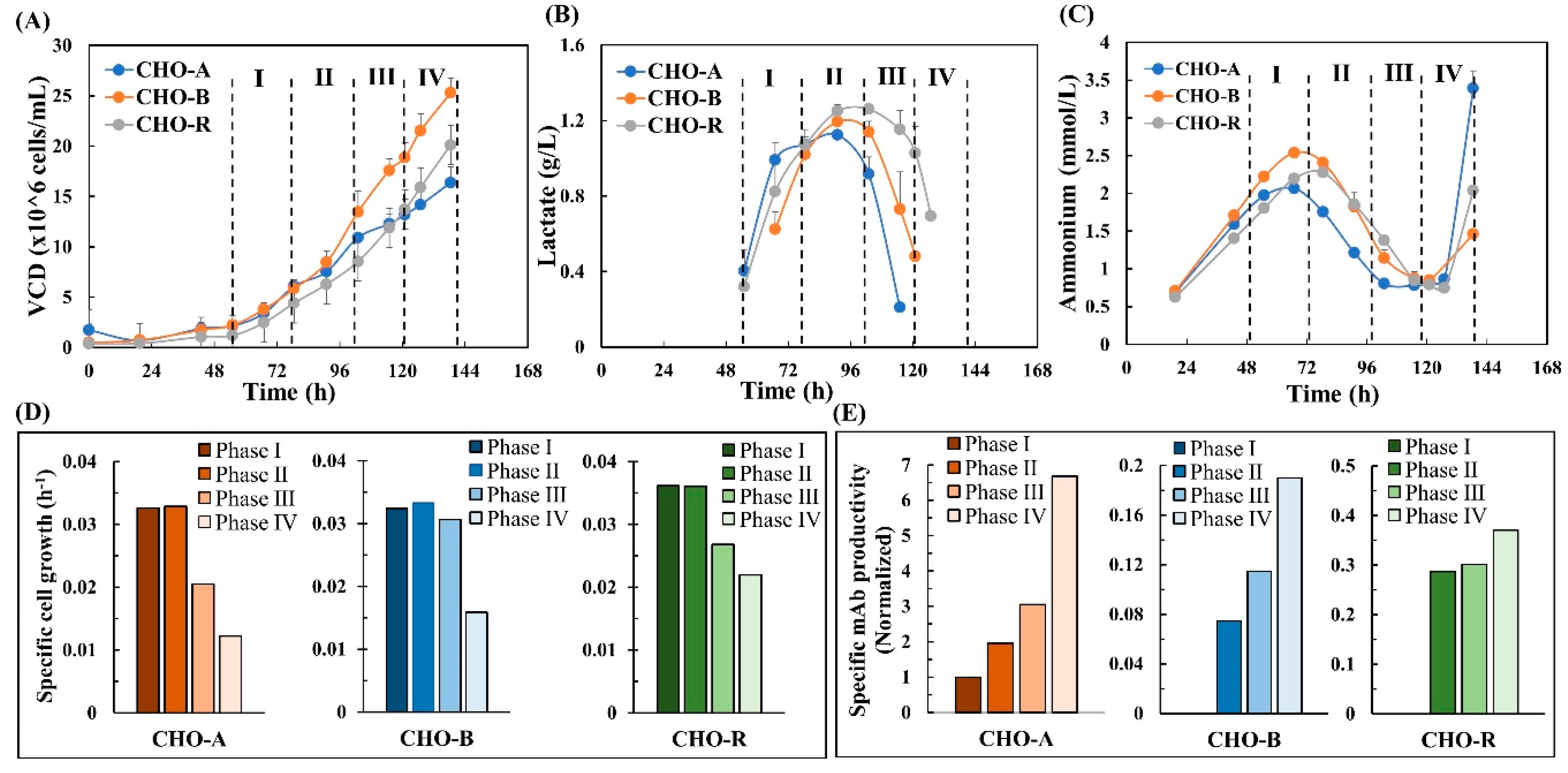
The daily measured viable cell density, lactate, and ammonium concentrations are shown in Figure 1. Other metabolite data are not shown but their specific rates were calculated and shown in Section 3.3. In this study, the genome-scale modeling focuses on the exponential growth phase. According to the cell growth and metabolic profiles (Figure 1A–C), the exponential phase can be divided into four sub-phases, including phase I (day 3), phase II (day 4), phase III (day 5) and phase IV (day 6). The specific rates of cell growth and mAb production are shown in Figure 1D,E. Clearly, variation was seen in these activities across the four phases.
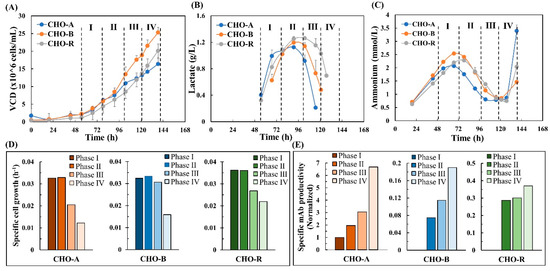
Figure 1.
Cell culture data. (A) Time-series viable cell density. (B) Time-series lactate concentration. (C) Time-series ammonium concentration. (D) Dynamic specific cell growth. (E) Dynamic specific mAb productivity. The specific mAb productivity of phase I in CHO-A was set as 100% to normalize productivity within this study.
3.2. Time-Series Transcriptomics
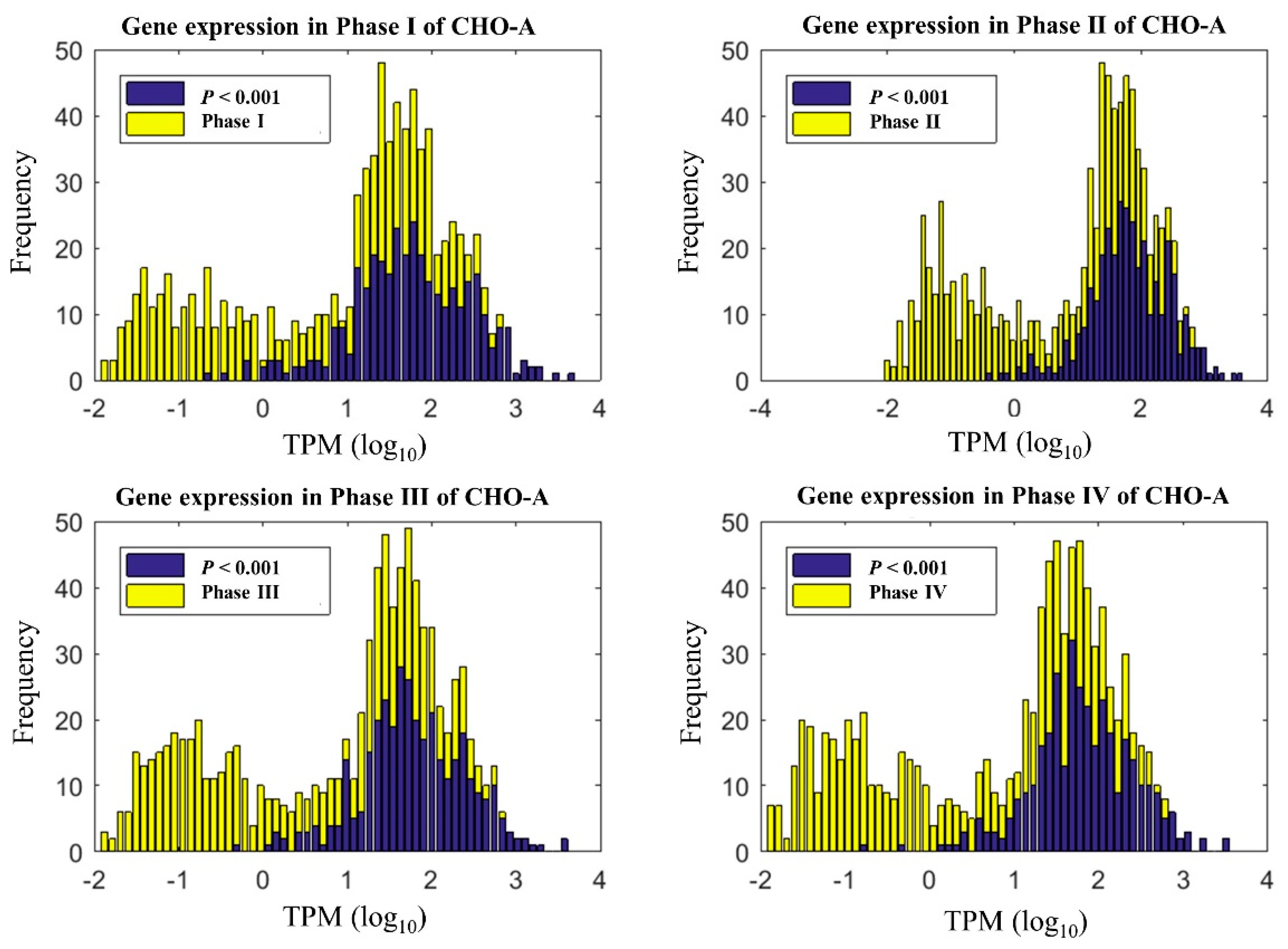
Here, we first analyzed the time-series transcriptomics and determined whether a significant change has occurred among different time points. In this experiment, gene expression was analyzed for cells by extracting total RNA and measuring transcript reads using RNA-Seq. Cell samples were collected at 67 h (day 3), 91 h (day 4), 115 h (day 5), and 127 h (day 6), respectively. Out of 26520 genes in global RNA-Seq data, we focused on the metabolic genes associated with the global CHO genome-scale model. As a result, 1728 genes in our datasets can be mapped to the global CHO genome-scale model [7]. With ANOVA analysis, CHO-A, CHO-B, and CHO-R cultures respectively had 352, 214, and 244 genes that were significantly changed among the four culture phases (p-value < 0.001). This represents a portion of genes that are 20.4% (352/1728 genes) in CHO-A, 12.4% (214/1728 genes) in CHO-B, and 14.1% (244/1728 genes) in CHO-R, respectively. As an example, the distribution of all metabolic genes in CHO-A over their expression level is shown (in yellow) in Figure 2. The dynamically varying genes are shown in dark blue, and it was seen that a significant portion of these genes were highly expressed. Given the dynamic variation seen at transcriptome during the exponential growth phase, it is necessary for us to investigate the effect of time-series transcriptomics on the context-specific genome-scale model reconstruction.
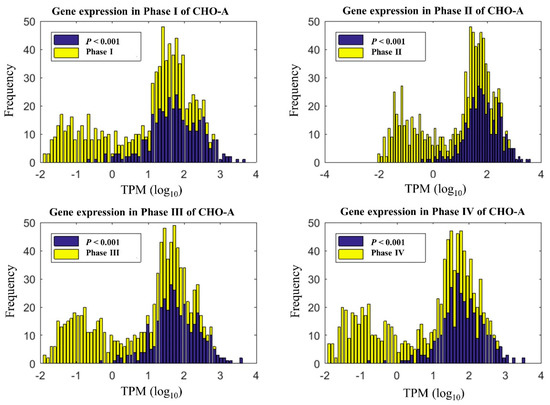
Figure 2.
Time-series gene expression under different phases in the CHO-A cell line. The genes of a p-value < 0.001 were considered to be statistically significant.
3.3. Extracellular Metabolomics and Metabolite Uptake and Secretion Rates
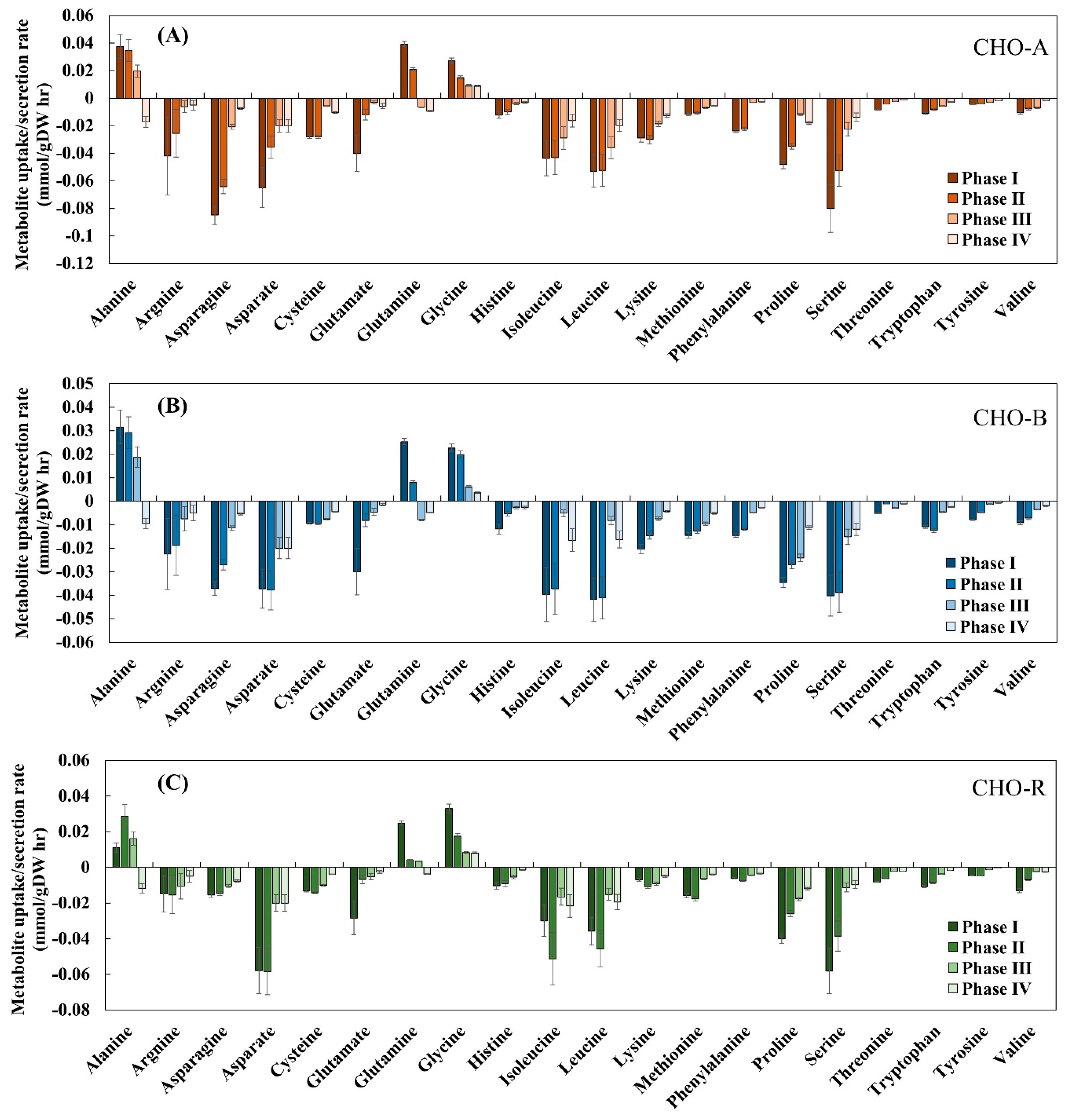
The specific rates of metabolite uptake and secretion were calculated using daily measured extracellular metabolomics. The calculated rates at each growth phase for all the three cell lines are shown in Figure 3. As seen in each cell line, the specific rates of all amino acids tended to decline from phase I (day 3) to phase IV (day 6), a phenomenon concurrent with the reduced cell growth over time. Alanine and glutamine were exceptional. Specifically, alanine was first produced before phase III and then reduced. This trend could be due to the production of alanine from pyruvate before phase III and later in phase IV alanine was converted back to pyruvate to maintain the flow to the TCA cycle [32]. It has been known that glutamine is important to generate TCA cycle intermediates such as α-ketoglutarate [33]. In the culture for GS-CHO cells, glutamine was not supplemented and was only intracellularly synthesized by GS activity. In this work, glutamine was shifted from production to consumption, indicating that at the early phase of cell culture, glutamate along with ammonium was utilized to produce glutamine. Taken together, these calculated metabolite uptakes and secretion rates revealed metabolic differences among the four culture phases, in line with the time-series transcriptomic difference seen in Section 3.2. All these calculated specific rates at each phase along with transcriptome were then used with the GIMME method to extract context-specific models from the reference CHO model.
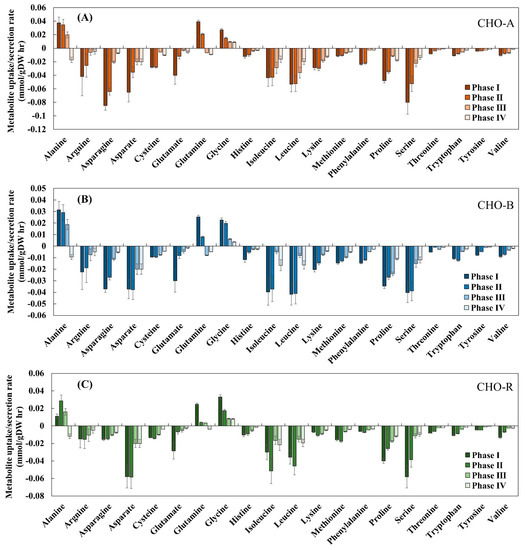
Figure 3.
Experimentally measured specific uptake and secretion rates among different culture phases in CHO-A (A), CHO-B (B), and CHO-R (C).
3.4. Model Extraction for Each Cell Line
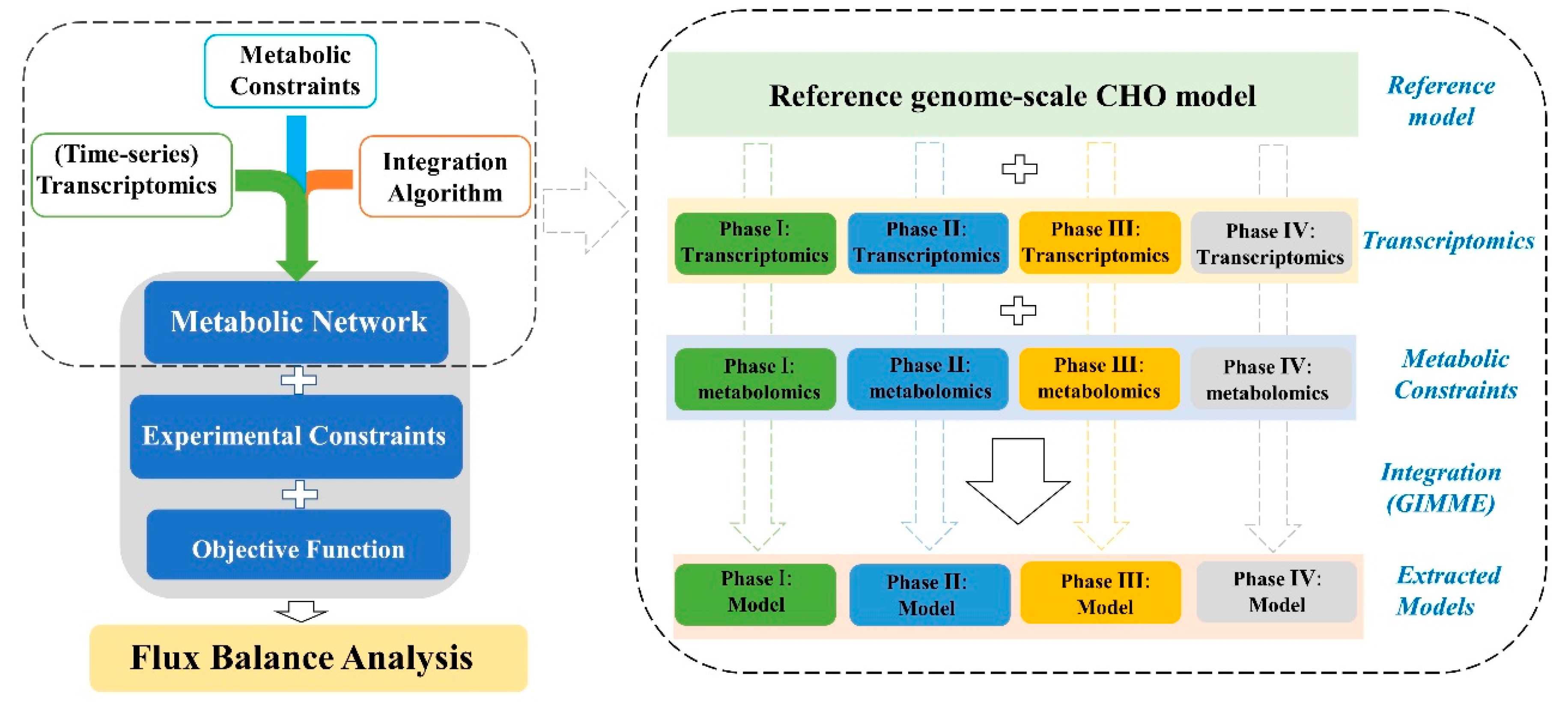
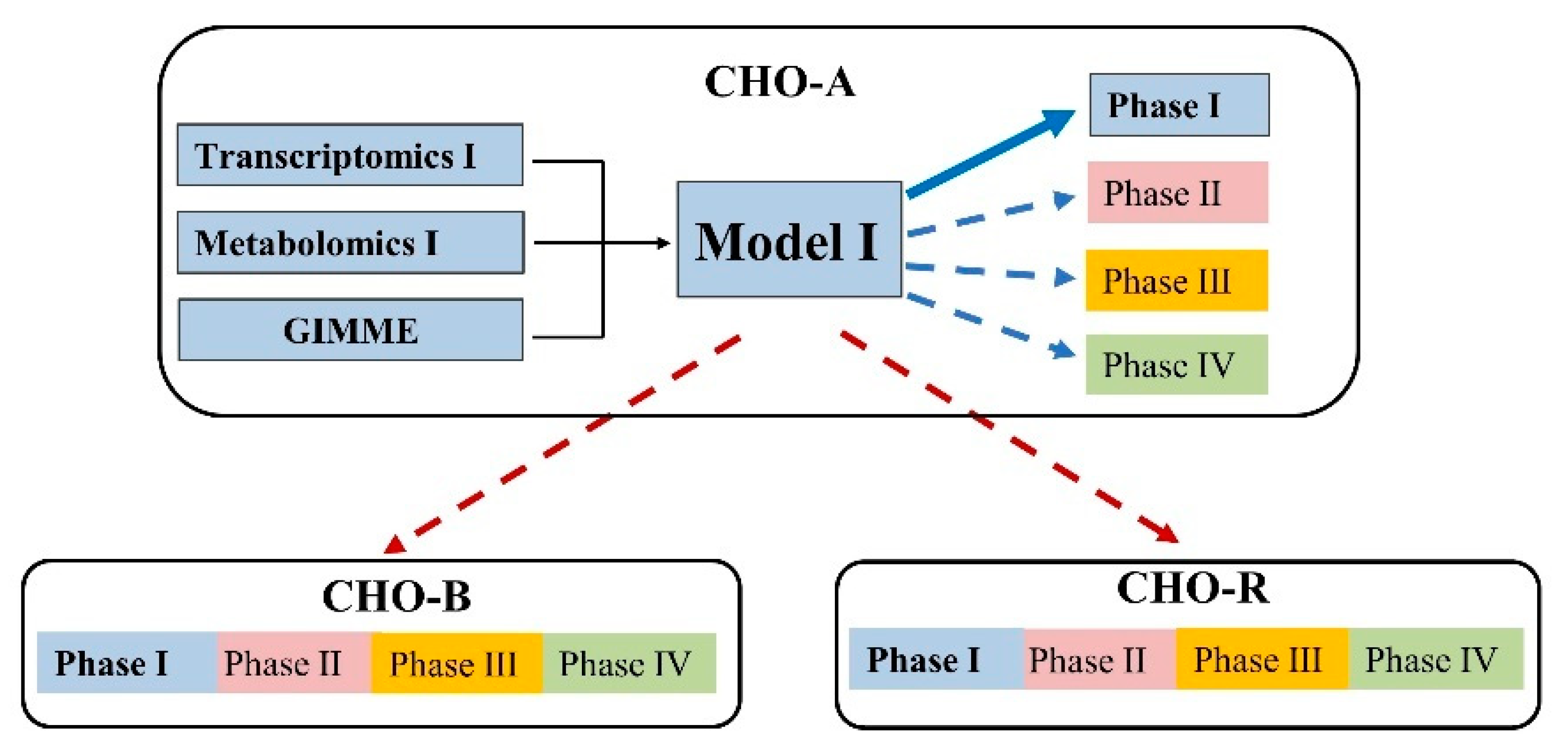
The reconstruction of context-specific models for each cell line followed the steps as described in Figure 4. The published global CHO genome-scale model was served as the reference model for all the reconstructions [7]. Each new model was subtracted from the reference model by the integration algorithm GIMME and based on the specific-stage omics data. The experimentally measured specific metabolic rates (as shown in Figure 3) were used as model constraints while maximizing cell growth was defined as the objective function. For all the reconstructed models, the functions of biomass and mAb biosynthesis will be maintained by force all the time.
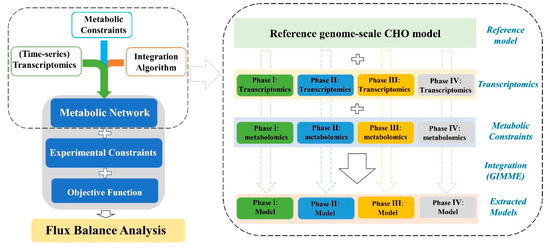
Figure 4.
The framework to formulate different genome-scale metabolic models by the integration of available omics data and using computational algorithm GIMME.
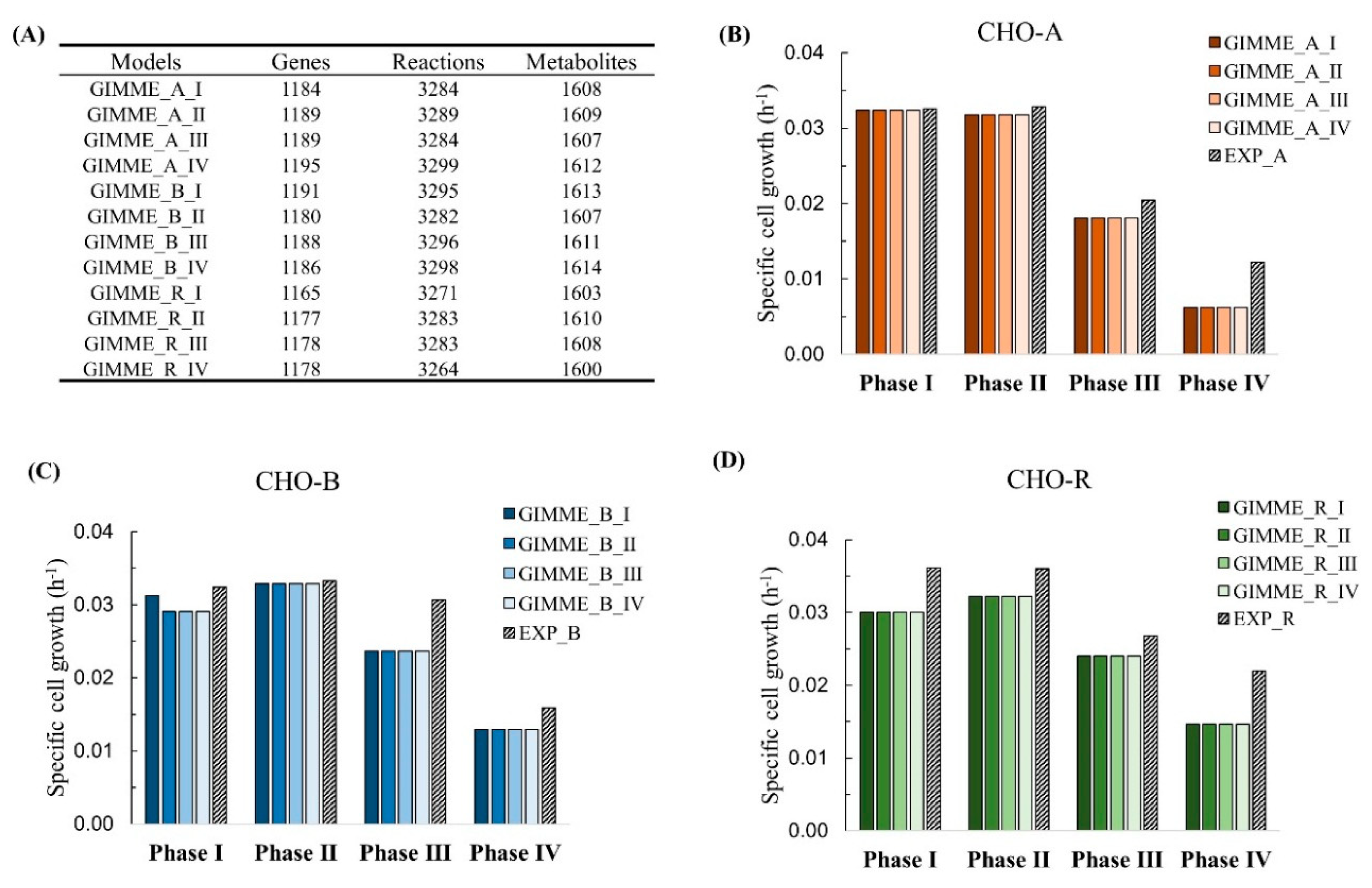
As a result, the context-specific model for each growth phase was extracted from the reference model. The ideal metabolic model should be robust and able to predict cell growth for all culture phases. It also means that a model should not be too sensitive to experimental constraints (derived from extracellular metabolomics) under different conditions; otherwise, the resulting models are likely to be overfitting to the provided experimental constraints. To demonstrate the robustness of the model extracted by the integration of transcriptomics at each phase, we tested each single-phase model to predict cell growth for the whole culture not exclusively to its own phase. The strategy of analysis is presented at the top part in Figure 5. For example, the extracted model with gene expression and experimental constraints from phase I in CHO-A was applied to predict the cell growth from phase I to phase IV in CHO-A (as described in Figure 5). The tests were done with each of the CHO-A, CHO-B, and CHO-R. The results of model predictions are shown in Figure 6. It turned out that, although the number of metabolic genes, metabolites, and reactions was varied by integrating the gene expression at different phases (Figure 6A), the model prediction results were similar and comparable with experimental values using any model (Figure 6B–D). Thus, the prediction performance of cell growth was not significantly perturbed by the gene dynamics. It indicates that the extracted model built from any growth phase could provide an equal opportunity to interpret the phenotype within the same cell line.
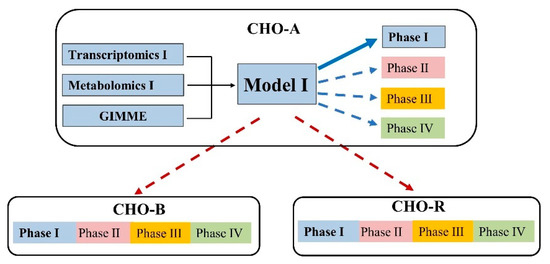
Figure 5.
Evaluation of the applicability of the extracted model under multiple conditions (with the same cell line) and multiple cell lines. Here CHO-A models were used as an example. The same evaluation process was repeated in CHO-B and CHO-R.
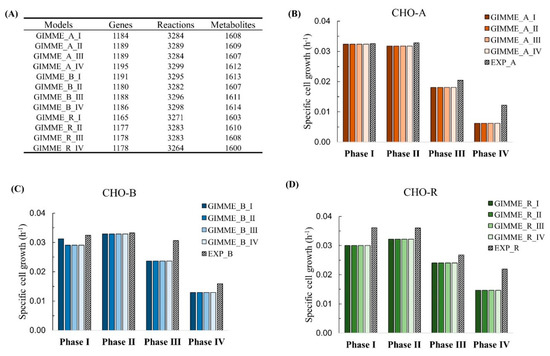
Figure 6.
Extracted genome-scale models for each phase by the integration of time-series gene expression using GIMME. (A) Network content for all the extracted GIMME models. Cell growth prediction in CHO-A (B), CHO-B (C), and CHO-R (D).
3.5. Comparison of the Network Content Among Extracted Models
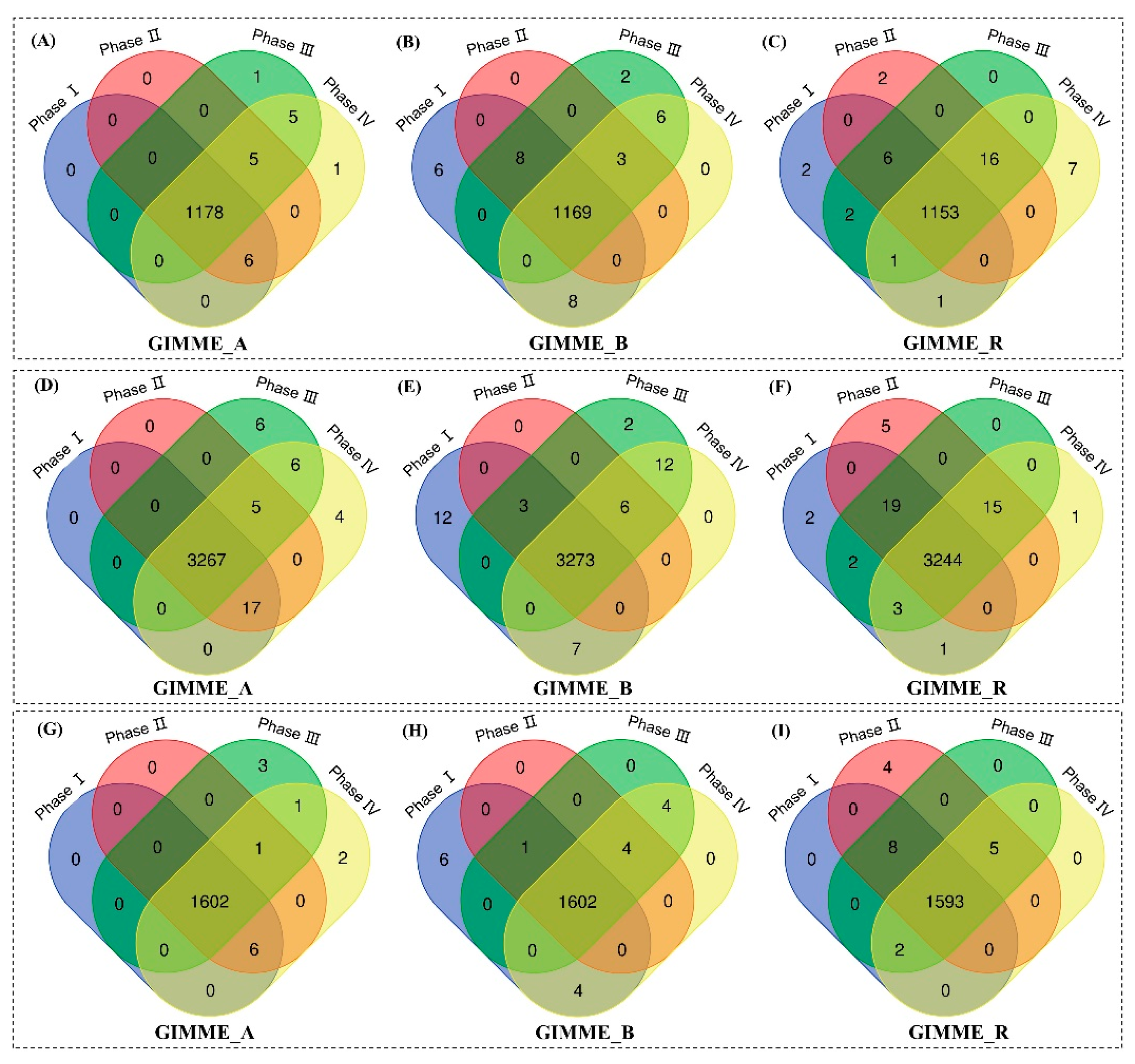
To further illustrate the network content, we applied the Venn diagram analysis to compare the models extracted at different growth phases. Figure 7 reveals the intersection of genes, reactions, and metabolites from each extracted model. In CHO-A, the intersection includes 1178 genes (99% of total genes in all four CHO-A models), 3267 reactions (98.8% of total reactions in all CHO-A models), and 1602 metabolites (99.7% of total metabolites in all CHO-A models). In CHO-B, this includes 1169 genes, 3273 reactions, 1602 metabolites; and in CHO-R, this includes 1153 genes, 3244 reactions, 1593 metabolites. It was thus shown that the network content of extracted models has high similarity. Here, we call this shared metabolic content information to be the core network for each cell line.
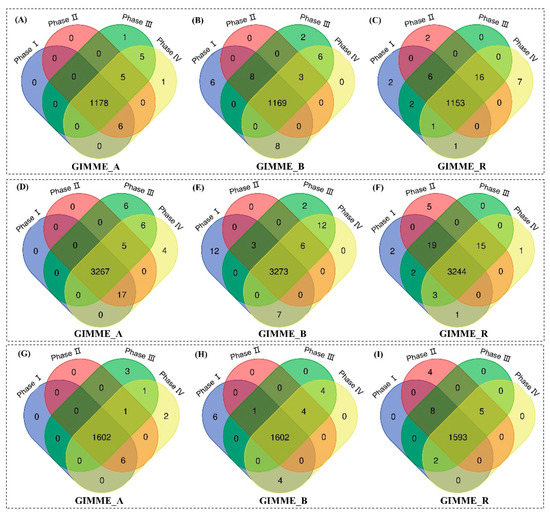
Figure 7.
Comparison of model contents between different growth phases for CHO-A, CHO-B, and CHO-R cell lines in terms of genes (A–C), reactions (D–F) and metabolites (G–I).
3.6. Evaluate the Feasibility of Applying Extracted Models to Different Cell Lines
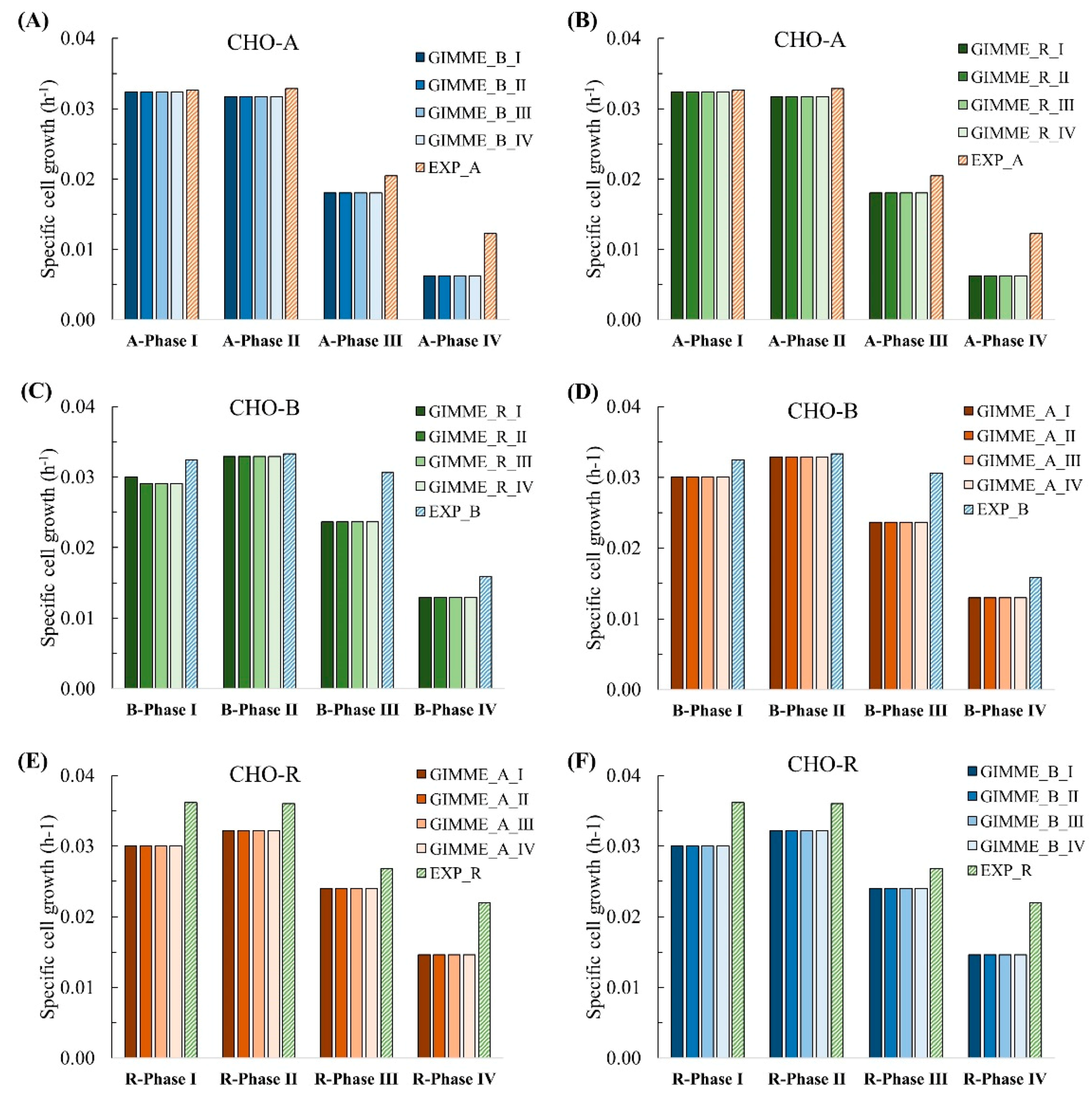
In the end, we evaluated the feasibility of extracted models for predicting cell growth for other cell lines, as shown in the bottom part of Figure 5. As a result, the cell growth for CHO-A was predictable using the extracted CHO-B (Figure 8A) or CHO-R models (Figure 8B). These predicted cell growth results were all close to the experimental results. Similar results were also observed vice versa to the other two cell lines, shown in Figure 8C,D for CHO-B and Figure 8D–F for CHO-R, respectively. The results suggested that each extracted model had captured the core reactions linked to biomass and antibody production and is robust to experimental constraints from multiple conditions in different cell lines.
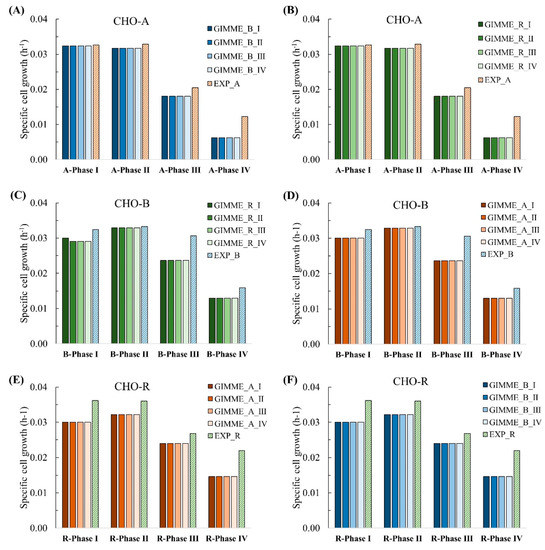
Figure 8.
Expand the application of extracted models built from time-series gene expression and GIMME method to multiple cell lines. Apply the CHO-B models (A) and CHO-R models (B) on CHO-A data; apply the CHO-R models (C) and CHO-A models (D) on CHO-B data; apply the CHO-A models (E) and CHO-B models (F) on CHO-R data.
We also analyzed the core genes, metabolites, and reactions commonly accumulated among different cell lines (CHO-A, CHO-B, and CHO-R) (data not shown). It was found that the CHO-A, CHO-B, and CHO-R GIMME models have a commonality of 1152 genes, 3242 reactions, and 1592 metabolites, as shared by all the 12 models of 4 phases in 3 cell lines (as seen in Figure 6A). It was thus revealed that all the extracted models integrated with time-series transcriptomics are closely related. It is reasonable because these 3 cell lines are generated from the same parental cell line.
4. Discussion
In this study, we analyzed the dynamics of the time-series transcriptomic data in mAb-producing cell cultures and built context-specific models of three different CHO cell lines by integrating omics data at specific growth phases. Our findings illustrate that gene expression at various stages of culture slightly impacts reconstructed models in terms of the number of metabolic genes, metabolites, and reactions. However, the prediction capability of models extracted from any dynamic period of culture is robust enough to predict the whole cell culture, not only across different growth phases but also in expansion to other cell lines. Since the experimental data of three cell lines used in this study have similar metabolic behavior and cell growth profile, it will be worthwhile to evaluate the extracted models that developed for cell lines with different metabolic behaviors in the future.
It is important to note that flux balance analysis utilizes three elements during computation (Figure 4, left), including stoichiometry of metabolic network, model constraints (e.g., specific uptake or secretion rates), and optimization principle (i.e., objective function) [10,34]. One of the most basic model constraints imposed on genome-scale models is the nutrient availability and its uptake or secretion rate. Some studies have demonstrated that constraint selection could strongly influence the ability of growth rate prediction [35]. Therefore, accurate measurements of extracellular metabolomics are another critical requirement other than the reconstruction of the metabolic network, to predict accurate cell growth and the intracellular flux distributions. Given that the genome-scale models are inherently highly underdetermined (e.g., as seen in Figure 6A, the number of reactions is much larger than that of metabolites), the adequacy of metabolomic dataset constraining the derived models may be a concern for model performance. The study of model constraints is still an important subject, as there is no explicit understanding of how many constraints must be required for genome-scale models. Currently, specific uptake or secretion rates of amino acids, glucose, and glutamate (as introduced in Section 2.2) are the most common constraints, since the quantification of these metabolites is relatively easy to perform [6,7]. This study has thus applied these rates as the metabolic constraints under each phase. As seen in Figure 6, the results indicated that the divergent gene expression data only slightly impact the reconstructed models. However, theoretically more biological- and data-derived constraints could potentially contribute to more accurate model behaviors.
Other diverse constraints have also been applied to genome-scale models, beyond the metabolic constraints of determined uptake and production rates. For example, the elemental balance of carbon through intracellular reactions was introduced as additional constraints [36]. Sánchez et al. (2017) incorporated enzymatic constraints in a yeast genome-scale metabolic model to ensure each metabolic flux does not exceed its maximum capacity, significantly reducing flux variability of model simulations [37]. Additionally, several attempts of applying the fluxes obtained from 13C metabolic flux analysis to constrain the genome-scale model have been reported in other organisms [34].
To identify optimal solutions, FBA requires a biologically relevant objective function which represents the metabolic goal of the organism [38], such as maximizing cell growth rate and minimizing total flux. Although biomass production which describes the growth requirements of a cell is widely used in the flux balance analysis [39], several previous studies have demonstrated that flux distributions and predicted cell growth are sensitive to the changes in biomass composition [40,41]. It indicated that accurate representation of biomass is another key to improve the predictive capability of flux balance analysis. Zuñiga et al. (2018) took into account dynamic biomass compositions for various culture conditions and stages, by applying dynamic biomass objective functions in a genome-scale metabolic model of Chlorella vulgaris. The constraint model accurately predicted the growth rate under heterotrophic and photoautotrophic conditions [42]. Another study conducted by Sarkar et al. (2019) developed a diurnal flux balance model that enabled considerations of varying nature of metabolism and different biomass composition under diurnal variations in phototrophic organisms [43]. Lachance et al. (2019) recently developed a software package to define a biomass objective function from experimental data, for example, the multiple omic datasets. This data-driven approach can be used to generate condition- and cell line-specific biomass objective functions and thus improving the quality of new genome-scale models [44]. Schuetz et al. (2007) systematically evaluated various objective functions for predicting intracellular fluxes and highlighted that objective functions were likely experimental condition-dependent, and no single objective function can describe the flux states under all conditions [45]. Indeed, in this work, we observed that the model prediction in phase I and phase II is closer to experimental measurement, compared to phase III and phase IV (as seen in Figure 6 and Figure 8). This suggested that the single objective function of maximization cell growth might not precisely represent the cellular state in these two late growth phases. In this regard, defining objective functions is anticipated to be another critical topic to be studied in the field of FBA [38].
Transcriptomes are integrated for genome-scale model reconstruction by serving as a proxy for the activity levels of relevant enzymes [26]. However, it is well understood that transcript levels often represent the levels of their corresponding proteins poorly [46]. Therefore, it is expected that proteomic data integration can constrain genome-scale models more effectively. Several studies had integrated multi-omics into genome-scale models to constrain the solution space [7,26].
Additionally, other features beyond metabolism, e.g., secretory pathway capacity, can influence cell growth and recombinant protein productivity. In mammalian cells, more than 25% of synthesized proteins (including enzymes and antibodies) are exported through the secretory pathway [47]. Therefore, a detailed understanding of the secretory pathway is valuable to be taken into account of protein productivity. Recently, Gutierrez et al. (2020) integrated the core secretory pathways with the genome-scale metabolic model of CHO cells to capture the energetic and machinery demands of secreted proteins [47]. In the future, this new reconstructed model can be used to interrogate cellular activities more comprehensively and explore the critical features of productivity among different CHO cell lines.
In summary, we believe the study of three mAb-producing cell lines presented here can provide guidelines to other researchers on how to handle and incorporate transcriptomics data into genome-scale models, especially for cell line-specific model extraction. Additionally, the extracted models can be used to describe fluxes for physiological understanding, which in turn contribute to potential target identification for cell line engineering or clone selection. On the other hand, a well-developed model ultimately would be applied to predict the effect of perturbations of feeding strategies or media composition on cell phenotype and served as an efficient tool to guide experimental work in process development.
Author Contributions
Conceptualization, Z.H.; methodology, Z.H.; software, Z.H.; formal analysis, Z.H.; experiments, Z.H.; writing—original draft preparation, Z.H.; writing—review and editing, S.Y.; supervision, S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NSF/IUCRC AMBIC (1624718).
Acknowledgments
The authors gratefully thank Maurizio Cattaneo for the kind provision of cell-lines.
Conflicts of Interest
The authors declare no conflict of interest.
References
- O’Brien, E.J.; Monk, J.M.; Palsson, B.O. Using Genome-scale Models to Predict Biological Capabilities. Cell 2015, 161, 971–987. [Google Scholar] [CrossRef]
- Pandey, V.; Hadadi, N.; Hatzimanikatis, V. Enhanced flux prediction by integrating relative expression and relative metabolite abundance into thermodynamically consistent metabolic models. PLoS Comput. Biol. 2019, 15, e1007036. [Google Scholar] [CrossRef]
- Orth, J.D.; Thiele, I.; Palsson, B.O. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Calmels, C.; McCann, A.; Malphettes, L.; Andersen, M.R. Application of a curated genome-scale metabolic model of CHO DG44 to an industrial fed-batch process. Metab. Eng. 2019, 51, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Sou, S.N.; Sellick, C.; Lee, K.; Mason, A.; Kyriakopoulos, S.; Polizzi, K.M.; Kontoravdi, C. How does mild hypothermia affect monoclonal antibody glycosylation? Biotechnol. Bioeng. 2015, 112, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Yusufi, F.N.K.; Lakshmanan, M.; Ho, Y.S.; Loo, B.L.W.; Ariyaratne, P.; Yang, Y.; Ng, S.K.; Tan, T.R.M.; Yeo, H.C.; Lim, H.L.; et al. Mammalian Systems Biotechnology Reveals Global Cellular Adaptations in a Recombinant CHO Cell Line. Cell Syst. 2017, 4, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Hefzi, H.; Ang, K.S.; Hanscho, M.; Bordbar, A.; Ruckerbauer, D.; Lakshmanan, M.; Orellana, C.A.; Baycin-Hizal, D.; Huang, Y.; Ley, D.; et al. A Consensus Genome-scale Reconstruction of Chinese Hamster Ovary Cell Metabolism. Cell Syst. 2016, 3, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.; Herrgard, M.J.; Rocha, I. Stoichiometric Representation of Gene-Protein-Reaction Associations Leverages Constraint-Based Analysis from Reaction to Gene-Level Phenotype Prediction. PLoS Comput. Biol. 2016, 12, e1005140. [Google Scholar] [CrossRef]
- Lewis, N.E.; Nagarajan, H.; Palsson, B.O. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 2012, 10, 291–305. [Google Scholar] [CrossRef]
- Sha, S.; Huang, Z.; Wang, Z.; Yoon, S. Mechanistic modeling and applications for CHO cell culture development and production. Curr. Opin. Chem. Eng. 2018, 22, 54–61. [Google Scholar] [CrossRef]
- Chen, C.; Le, H.; Goudar, C.T. Integration of systems biology in cell line and process development for biopharmaceutical manufacturing. Biochemical Engineering Journal 2016, 107, 11–17. [Google Scholar] [CrossRef]
- Joshi, C.J.; Schinn, S.-M.; Richelle, A.; Shamie, I.; O’Rourke, E.J.; Lewis, N.E. StanDep: capturing transcriptomic variability improves context-specific metabolic models. bioRxiv 2019. [Google Scholar] [CrossRef]
- Richelle, A.; Chiang, A.W.T.; Kuo, C.C.; Lewis, N.E. Increasing consensus of context-specific metabolic models by integrating data-inferred cell functions. PLoS Comput. Biol. 2019, 15, e1006867. [Google Scholar] [CrossRef] [PubMed]
- Robaina Estevez, S.; Nikoloski, Z. Generalized framework for context-specific metabolic model extraction methods. Front. Plant. Sci 2014, 5, 491. [Google Scholar] [CrossRef] [PubMed]
- Hyduke, D.R.; Lewis, N.E.; Palsson, B.O. Analysis of omics data with genome-scale models of metabolism. Mol. Biosyst. 2013, 9, 167–174. [Google Scholar] [CrossRef]
- Machado, D.; Herrgard, M. Systematic evaluation of methods for integration of transcriptomic data into constraint-based models of metabolism. PLoS Comput. Biol. 2014, 10, e1003580. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.A.; Palsson, B.O. Context-Specific Metabolic Networks Are Consistent with Experiments. PLoS Comput. Biol. 2008, 4, e1000082. [Google Scholar] [CrossRef]
- Chen, C.; Wong, H.E.; Goudar, C.T. Upstream process intensification and continuous manufacturing. Curr. Opin. Chem. Eng. 2018, 22, 191–198. [Google Scholar] [CrossRef]
- Kotidis, P.; Jedrzejewski, P.; Sou, S.N.; Sellick, C.; Polizzi, K.; Del Val, I.J.; Kontoravdi, C. Model-based optimization of antibody galactosylation in CHO cell culture. Biotechnol. Bioeng. 2019, 116, 1612–1626. [Google Scholar] [CrossRef]
- Graham, R.J.; Bhatia, H.; Yoon, S. Consequences of trace metal variability and supplementation on Chinese hamster ovary (CHO) cell culture performance: A review of key mechanisms and considerations. Biotechnol. Bioeng. 2019, 116, 3446–3456. [Google Scholar] [CrossRef]
- Templeton, N.; Dean, J.; Reddy, P.; Young, J.D. Peak antibody production is associated with increased oxidative metabolism in an industrially relevant fed-batch CHO cell culture. Biotechnol. Bioeng. 2013, 110, 2013–2024. [Google Scholar] [CrossRef] [PubMed]
- Hernandez Bort, J.A.; Hackl, M.; Hoflmayer, H.; Jadhav, V.; Harreither, E.; Kumar, N.; Ernst, W.; Grillari, J.; Borth, N. Dynamic mRNA and miRNA profiling of CHO-K1 suspension cell cultures. Biotechnol. J. 2012, 7, 500–515. [Google Scholar] [CrossRef] [PubMed]
- Sha, S.; Bhatia, H.; Yoon, S. An RNA-seq based transcriptomic investigation into the productivity and growth variants with Chinese hamster ovary cells. J. Biotechnol. 2018, 271, 37–46. [Google Scholar] [CrossRef] [PubMed]
- BiGG Models. Available online: http://bigg.ucsd.edu/ (accessed on 10 March 2020).
- Park, S. A Metabolomic Approach to Understanding the Metabolic Link between Obesity and Diabetes. Mol. Cells 2015, 38, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Lee, D.Y.; Yoon, S. Quantitative intracellular flux modeling and applications in biotherapeutic development and production using CHO cell cultures. Biotechnol. Bioeng. 2017, 114, 2717–2728. [Google Scholar] [CrossRef]
- Quek, L.E.; Dietmair, S.; Kromer, J.O.; Nielsen, L.K. Metabolic flux analysis in mammalian cell culture. Metab. Eng. 2010, 12, 161–171. [Google Scholar] [CrossRef]
- Becker, S.A.; Feist, A.M.; Mo, M.L.; Hannum, G.; Palsson, B.O.; Herrgard, M.J. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat. Protoc. 2007, 2, 727–738. [Google Scholar] [CrossRef]
- Schellenberger, J.; Que, R.; Fleming, R.M.; Thiele, I.; Orth, J.D.; Feist, A.M.; Zielinski, D.C.; Bordbar, A.; Lewis, N.E.; Rahmanian, S.; et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 2011, 6, 1290–1307. [Google Scholar] [CrossRef]
- Gurobi Solver. Available online: https://www.gurobi.com/ (accessed on 10 March 2020).
- Cho, J.S.; Gu, C.; Han, T.H.; Ryu, J.Y.; Lee, S.Y. Reconstruction of context-specific genome-scale metabolic models using multiomics data to study metabolic rewiring. Curr. Opin. Syst. Biol. 2019, 15, 1–11. [Google Scholar] [CrossRef]
- Li, J.; Wong, C.L.; Vijayasankaran, N.; Hudson, T.; Amanullah, A. Feeding lactate for CHO cell culture processes: impact on culture metabolism and performance. Biotechnol. Bioeng. 2012, 109, 1173–1186. [Google Scholar] [CrossRef]
- Pereira, S.; Kildegaard, H.F.; Andersen, M.R. Impact of CHO Metabolism on Cell Growth and Protein Production: An Overview of Toxic and Inhibiting Metabolites and Nutrients. Biotechnol. J. 2018, 13, e1700499. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.G.; Kumar, V.S.; Weaver, D.; Ghosh, A.; Chubukov, V.; Mukhopadhyay, A.; Arkin, A.; Keasling, J.D. A Method to Constrain Genome-Scale Models with 13C Labeling Data. PLoS Comput. Biol. 2015, 11, e1004363. [Google Scholar] [CrossRef]
- Opdam, S.; Richelle, A.; Kellman, B.; Li, S.; Zielinski, D.C.; Lewis, N.E. A Systematic Evaluation of Methods for Tailoring Genome-Scale Metabolic Models. Cell Syst. 2017, 4, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Lularevic, M.; Racher, A.J.; Jaques, C.; Kiparissides, A. Improving the accuracy of flux balance analysis through the implementation of carbon availability constraints for intracellular reactions. Biotechnol. Bioeng. 2019, 116, 2339–2352. [Google Scholar] [CrossRef]
- Sanchez, B.J.; Zhang, C.; Nilsson, A.; Lahtvee, P.J.; Kerkhoven, E.J.; Nielsen, J. Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 2017, 13, 935. [Google Scholar] [CrossRef]
- Montezano, D.; Meek, L.; Gupta, R.; Bermudez, L.E.; Bermudez, J.C. Flux Balance Analysis with Objective Function Defined by Proteomics Data-Metabolism of Mycobacterium tuberculosis Exposed to Mefloquine. PLoS ONE 2015, 10, e0134014. [Google Scholar] [CrossRef]
- Feist, A.M.; Palsson, B.O. The biomass objective function. Curr. Opin. Microbiol. 2010, 13, 344–349. [Google Scholar] [CrossRef]
- Lakshmanan, M.; Long, S.; Ang, K.S.; Lewis, N.; Lee, D.-Y. On the impact of biomass composition in constraint-based flux analysis. bioRxiv 2019. [Google Scholar] [CrossRef]
- Dikicioglu, D.; Kirdar, B.; Oliver, S.G. Biomass composition: the “elephant in the room” of metabolic modelling. Metabolomics 2015, 11, 1690–1701. [Google Scholar] [CrossRef]
- Zuniga, C.; Levering, J.; Antoniewicz, M.R.; Guarnieri, M.T.; Betenbaugh, M.J.; Zengler, K. Predicting Dynamic Metabolic Demands in the Photosynthetic Eukaryote Chlorella vulgaris. Plant Physiol. 2018, 176, 450–462. [Google Scholar] [CrossRef]
- Sarkar, D.; Mueller, T.J.; Liu, D.; Pakrasi, H.B.; Maranas, C.D. A diurnal flux balance model of Synechocystis sp. PCC 6803 metabolism. PLoS Comput. Biol. 2019, 15, e1006692. [Google Scholar] [CrossRef] [PubMed]
- Lachance, J.C.; Lloyd, C.J.; Monk, J.M.; Yang, L.; Sastry, A.V.; Seif, Y.; Palsson, B.O.; Rodrigue, S.; Feist, A.M.; King, Z.A.; et al. BOFdat: Generating biomass objective functions for genome-scale metabolic models from experimental data. PLoS Comput. Biol. 2019, 15, e1006971. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, R.; Kuepfer, L.; Sauer, U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol. Syst. Biol. 2007, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Grosseholz, R.; Koh, C.C.; Veith, N.; Fiedler, T.; Strauss, M.; Olivier, B.; Collins, B.C.; Schubert, O.T.; Bergmann, F.; Kreikemeyer, B.; et al. Integrating highly quantitative proteomics and genome-scale metabolic modeling to study pH adaptation in the human pathogen Enterococcus faecalis. NPJ Syst. Biol. Appl. 2016, 2, 16017. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, J.M.; Feizi, A.; Li, S.; Kallehauge, T.B.; Hefzi, H.; Grav, L.M.; Ley, D.; Baycin Hizal, D.; Betenbaugh, M.J.; Voldborg, B.; et al. Genome-scale reconstructions of the mammalian secretory pathway predict metabolic costs and limitations of protein secretion. Nat. Commun. 2020, 11, 68. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).