Design and Development of an Intelligent Chlorophyll Content Detection System for Cotton Leaves
Abstract
1. Introduction
2. Overall System Design
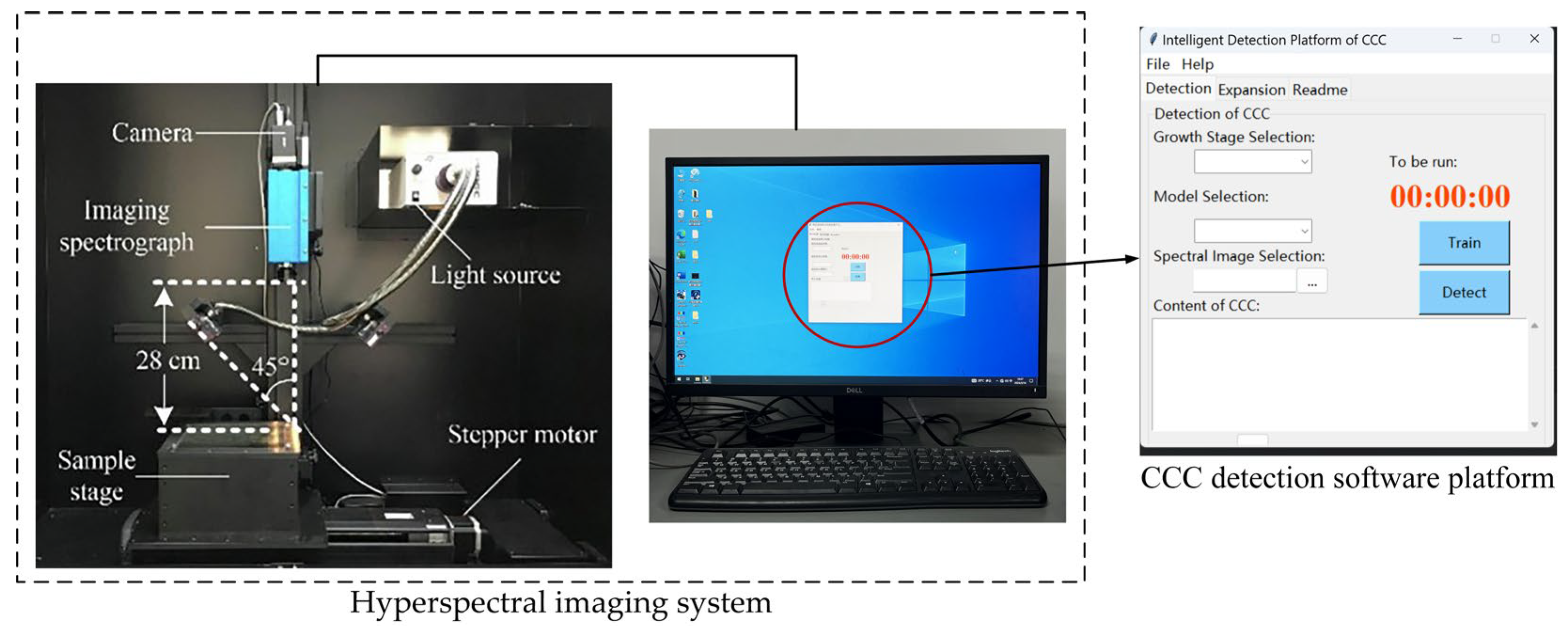
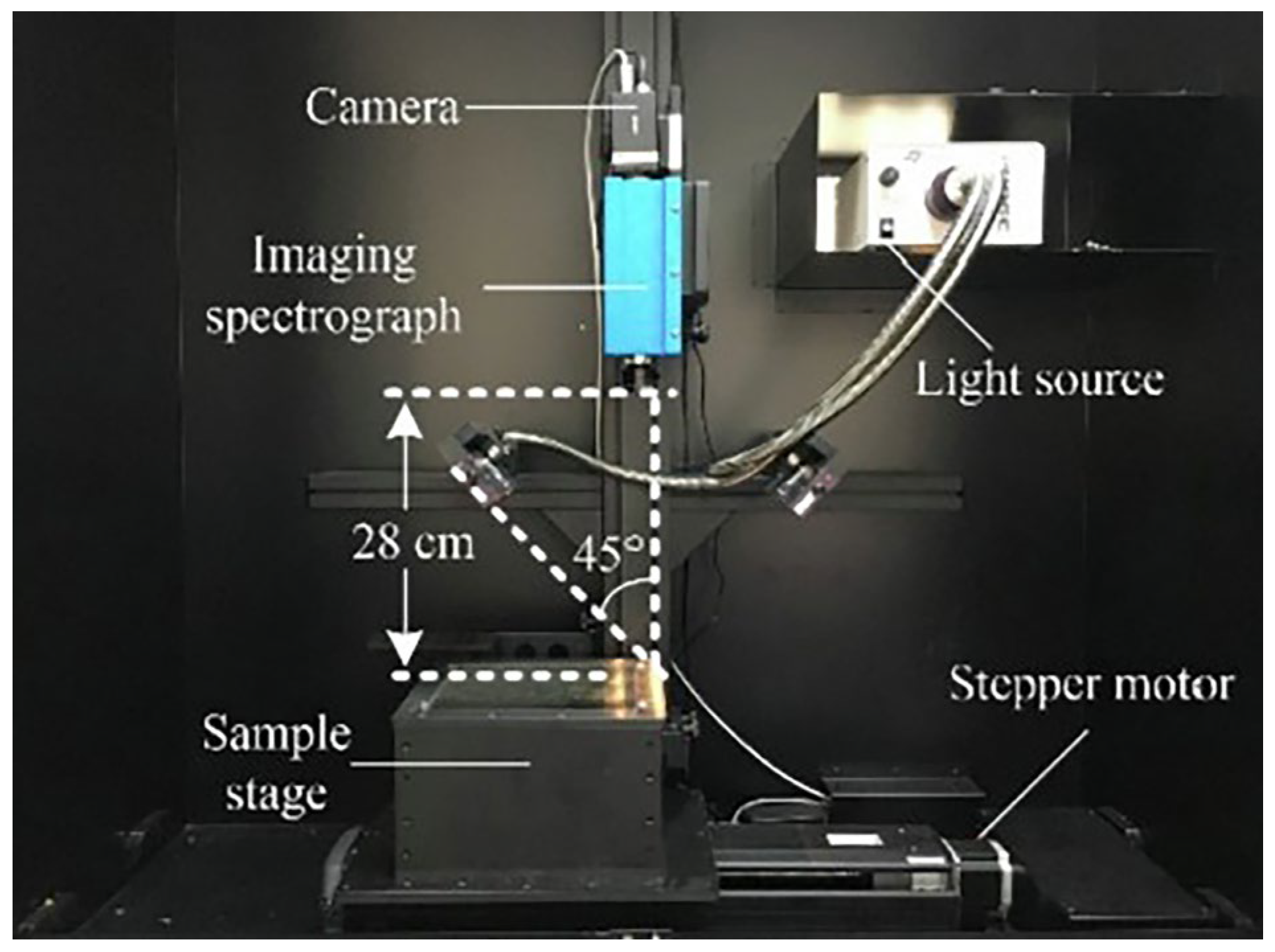
2.1. Hardware System Composition and Working Principle
2.1.1. Hyperspectral Image Acquisition Module
2.1.2. Correction of Hyperspectral Images
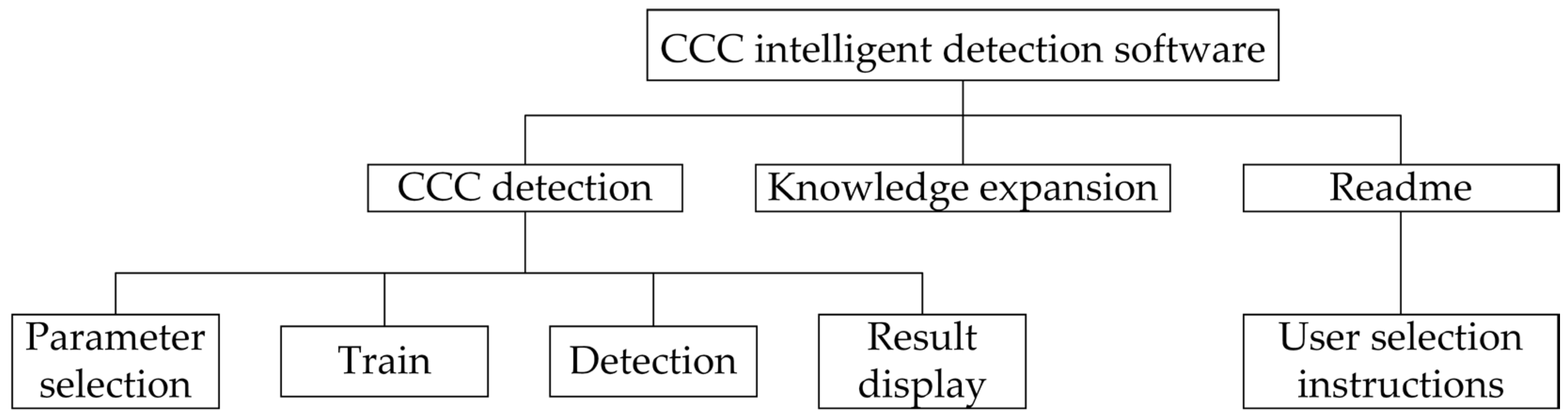
2.2. Software Design
2.2.1. Software Functional Architecture
2.2.2. Development Environment for CCC Intelligent Detection Software
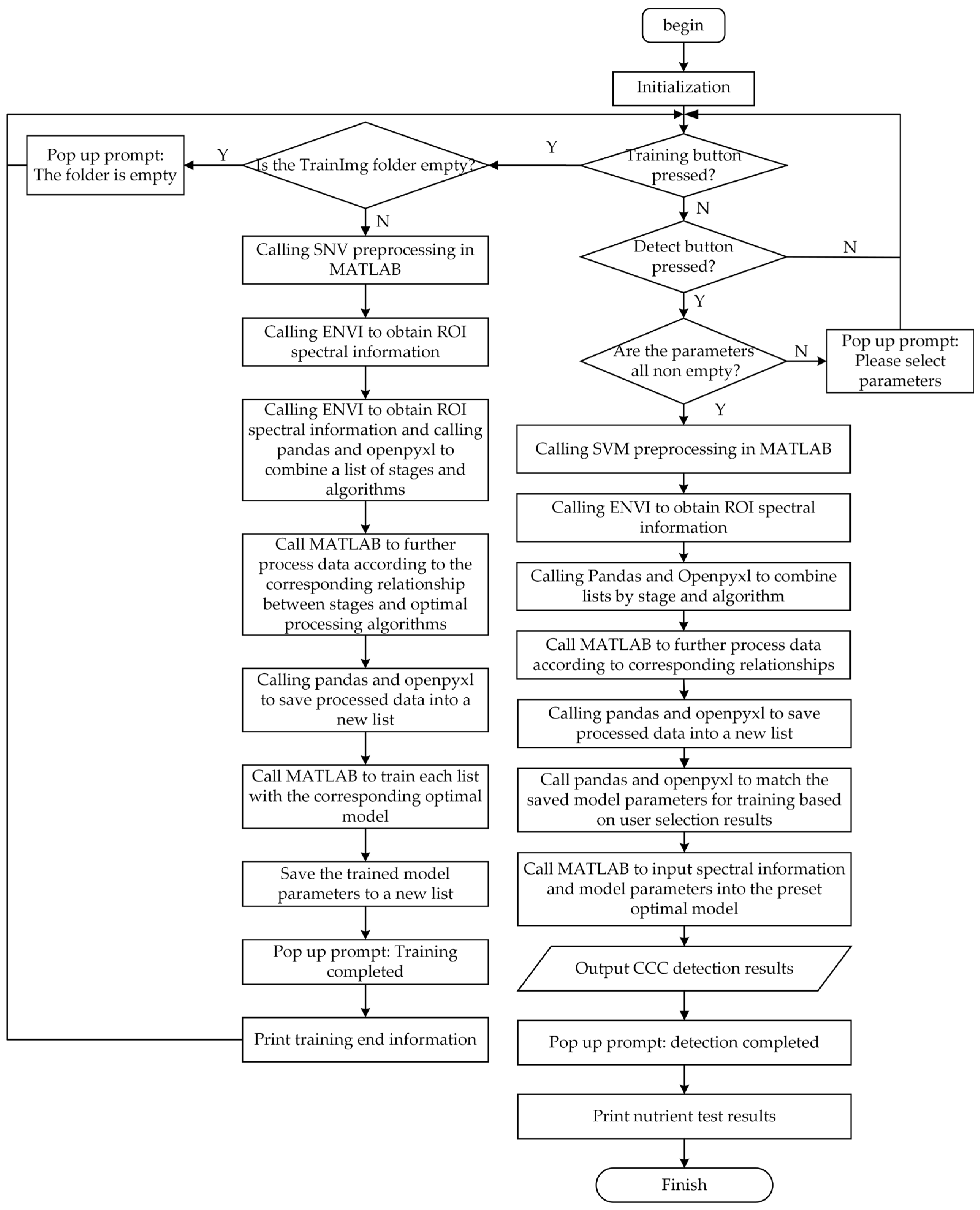
2.2.3. Workflow Design of Detection Software
3. Staged Detection Method for Chlorophyll in Cotton Leaves
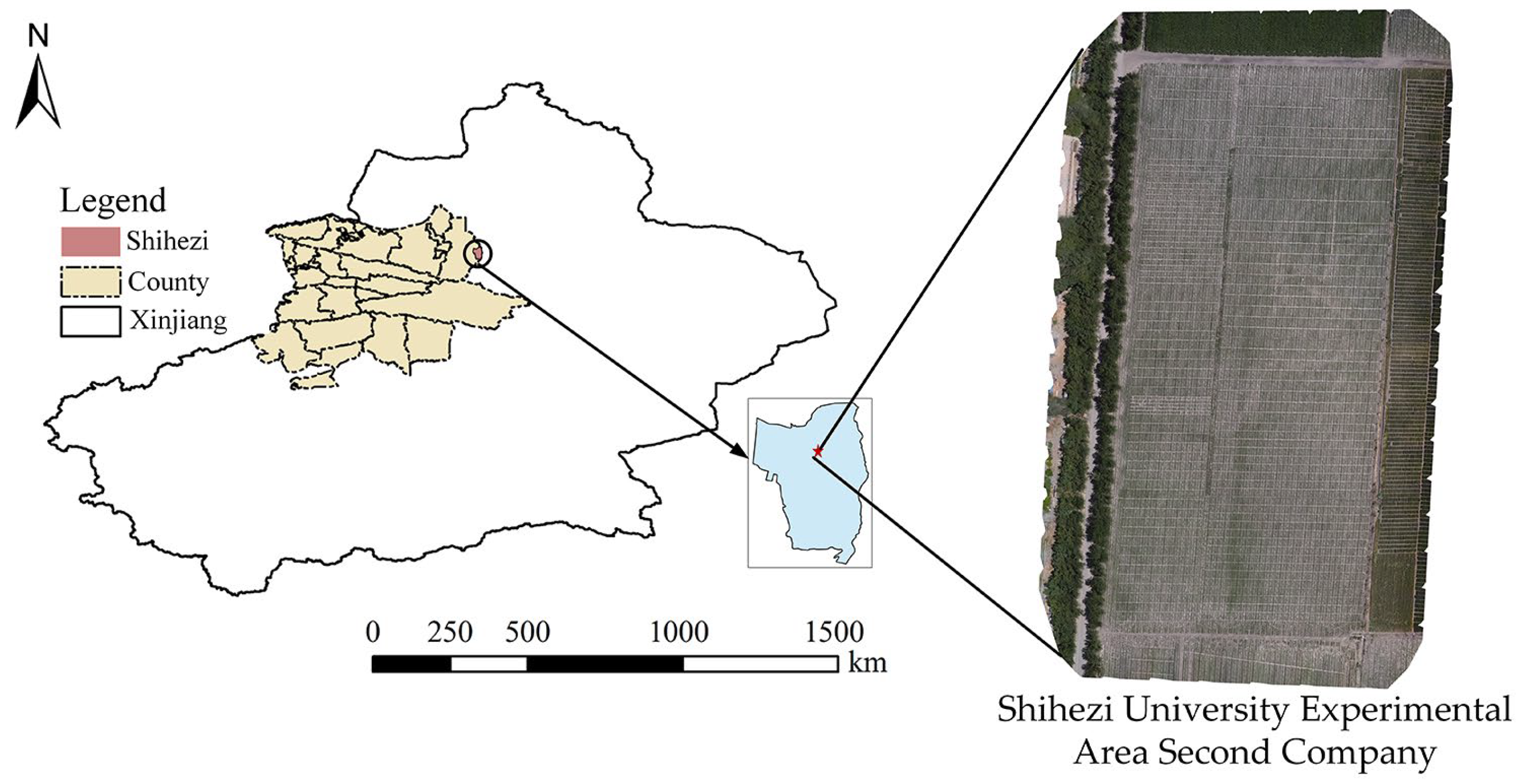
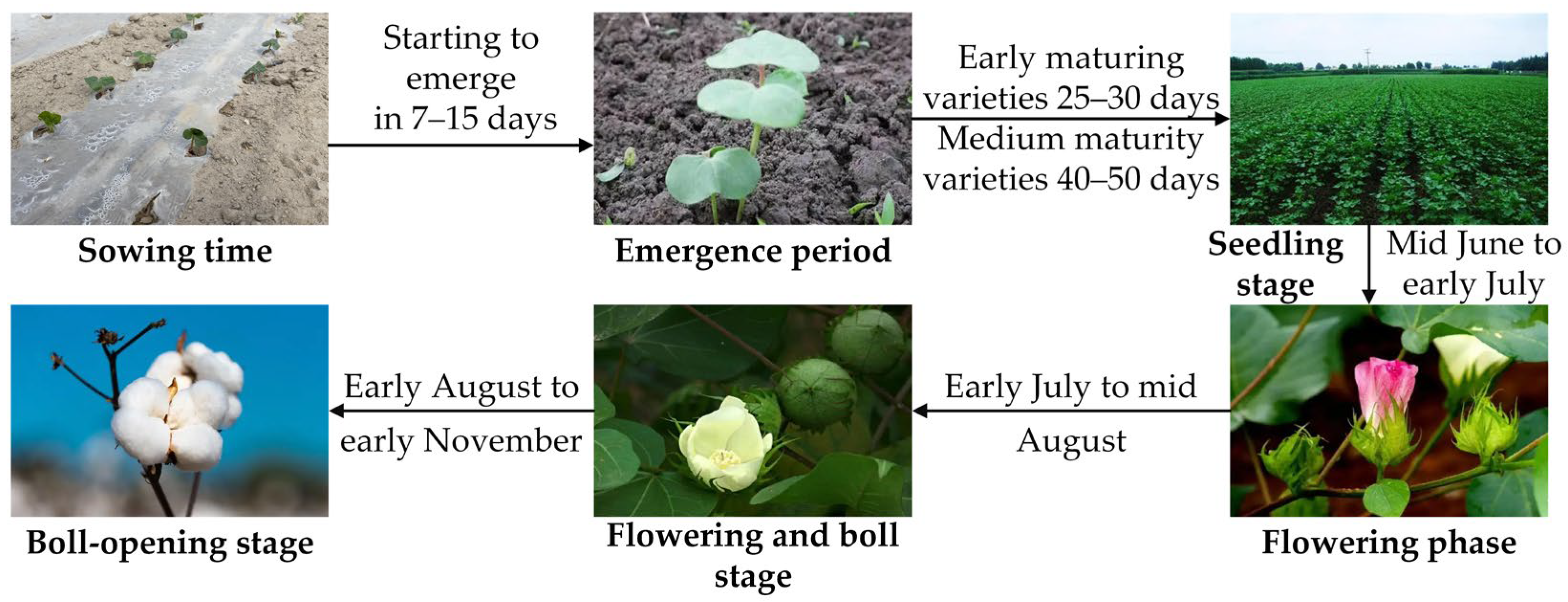
3.1. Sampling Location
3.2. Processing of Hyperspectral Images
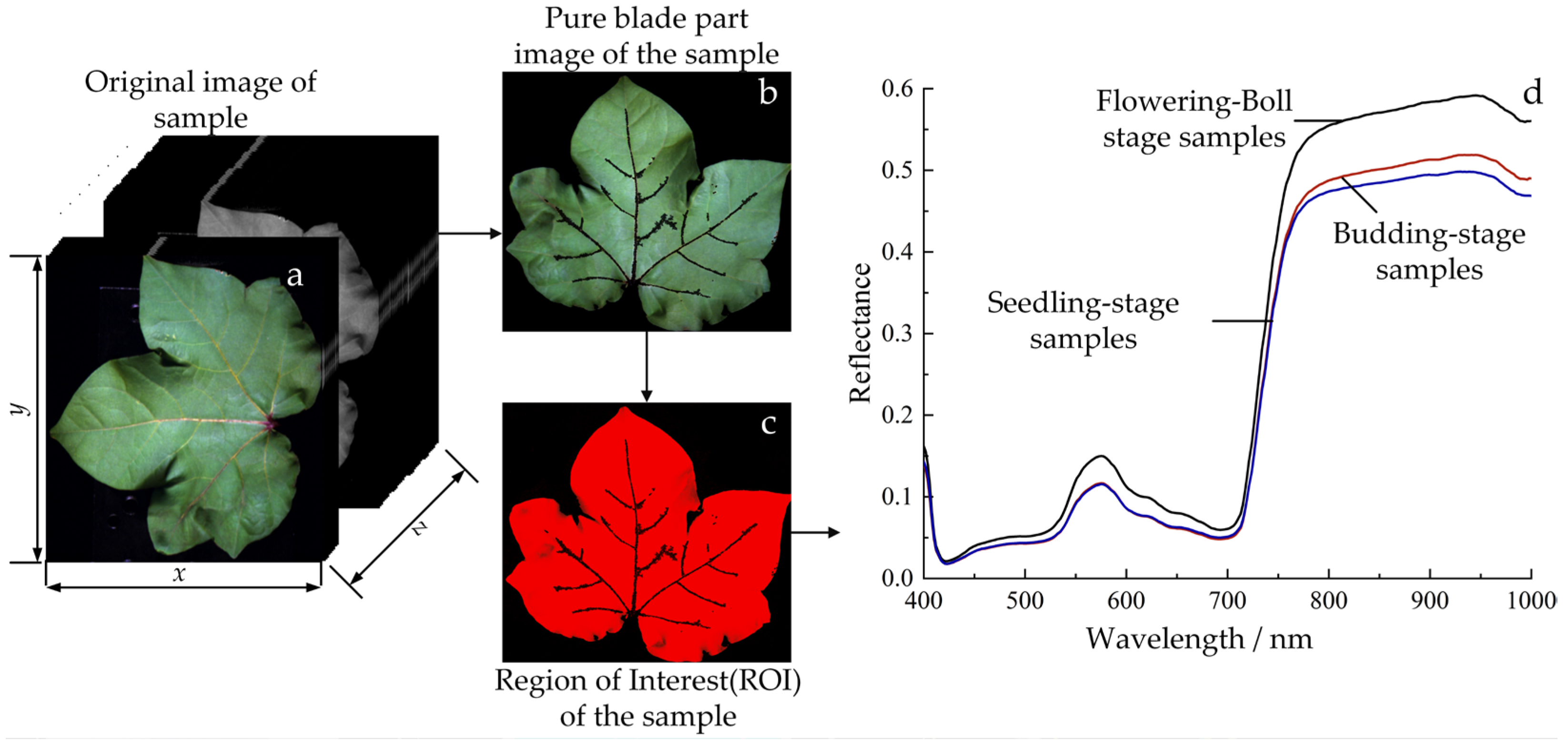
3.2.1. Hyperspectral Image Segmentation and the Extraction of Region of Interest Information
3.2.2. Preprocessing of Hyperspectral Information
3.3. Establishment and Evaluation of Chlorophyll Content Detection Model for Cotton Leaves
3.3.1. Model Accuracy Evaluation Criteria
3.3.2. Screening of Spectral Preprocessing Methods
3.3.3. Quantitative Prediction Model for CCC Based on SSA-BPNN and 1DCNN
4. Application Testing of Intelligent CCC Testing System
4.1. Accuracy Test for System Detection
4.2. Test of System Detection Speed
5. Conclusions
- (1)
- Based on hyperspectral imaging technology, the spectral information of cotton leaf samples at the seedling stage, budding stage, and flowering-boll stage was obtained. The performance of the partial least squares regression model established by the original spectrum and the spectra pretreated by different methods was compared, and SG was determined as the optimal spectral pretreatment method.
- (2)
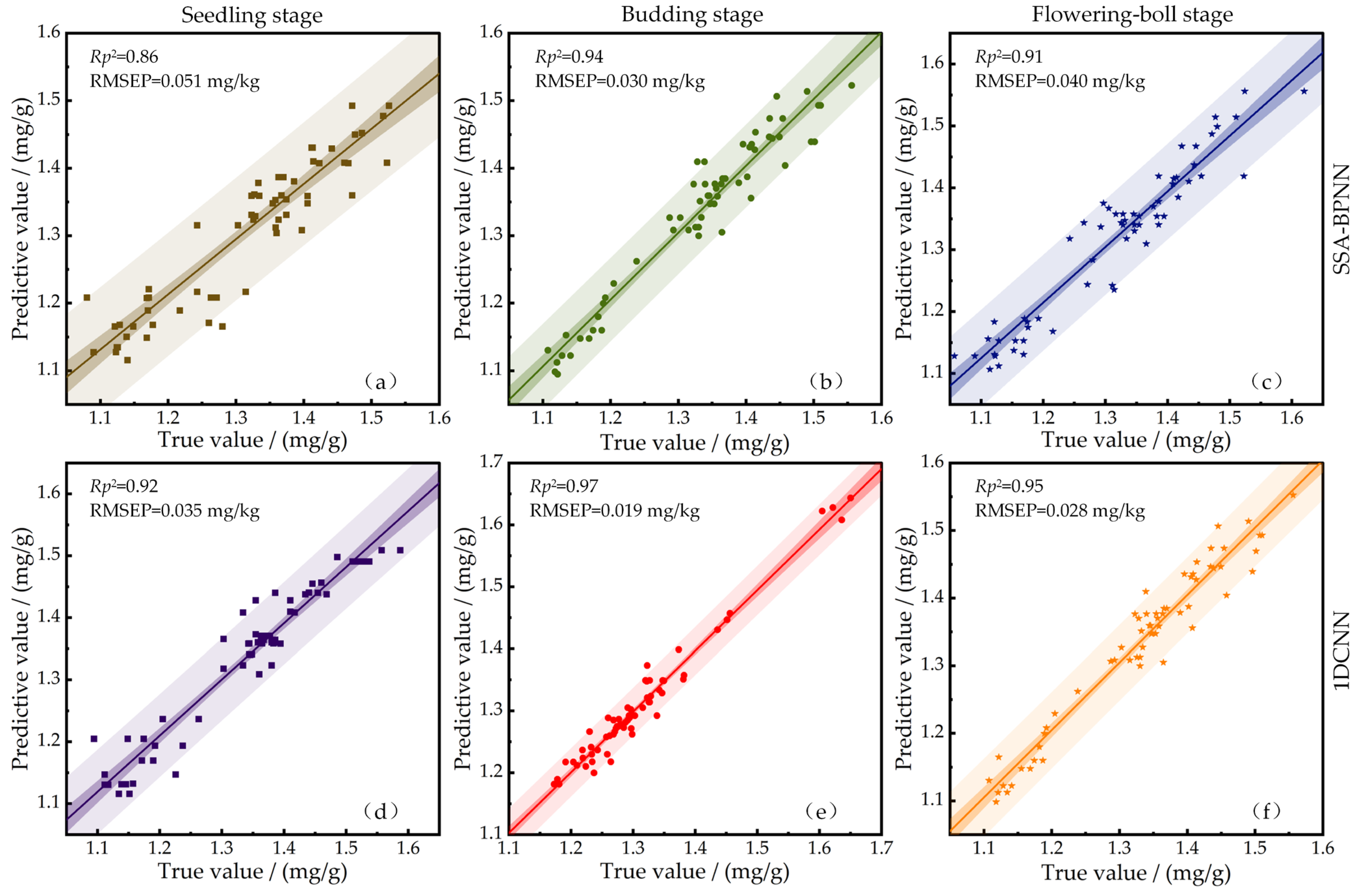
- Based on the spectral information processed by SG, the chlorophyll detection models of cotton leaves at different growth stages (seedling stage, budding stage, flowering-boll stage) using SSA-BPNN and 1DCNN were established. The results showed that the of the CCC detection model established by the 1DCNN algorithm at the seedling stage, germination stage, and flowering stage were 0.92, 0.97, and 0.95; RMSEP were 0.035 mg/g, 0.019 mg/g, and 0.028 mg/g; and RPD were 3.56, 5.54, and 4.58, respectively. In the early stage, the team mixed the spectral data of the leaves of cotton at three stages. After preprocessing and band screening, they established the CCC prediction models based on SSA-BPNN, GA-BPNN, and PSO-BPNN, respectively [41]. The performance of these models was significantly inferior to that of the SSA-BPNN established based on the full band of the three stages and also had a considerable gap compared with 1DCNN. It indicates that staged detection is more accurate, and this study can become the preferred solution for the embedded model of the intelligent detection system.
- (3)
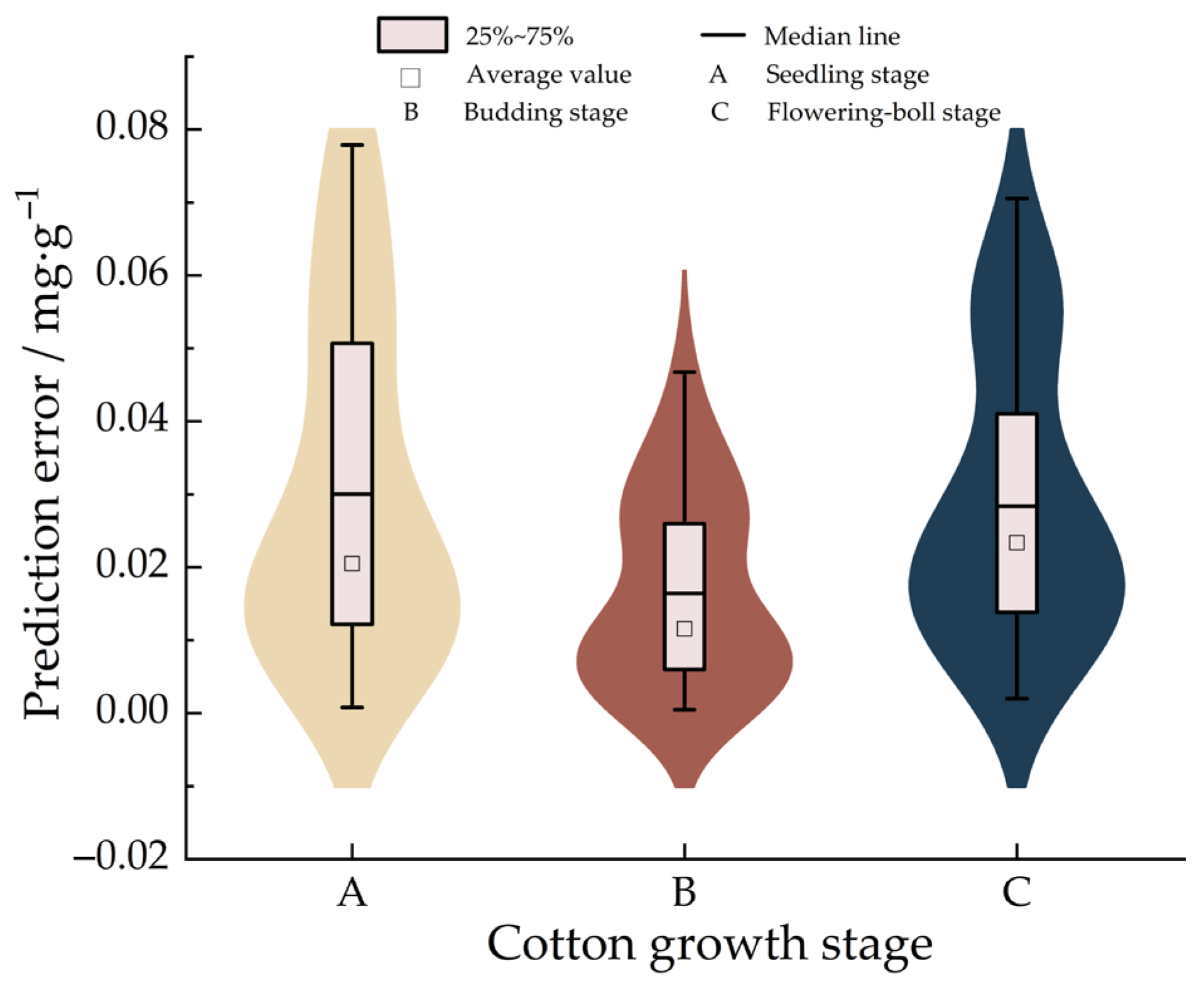
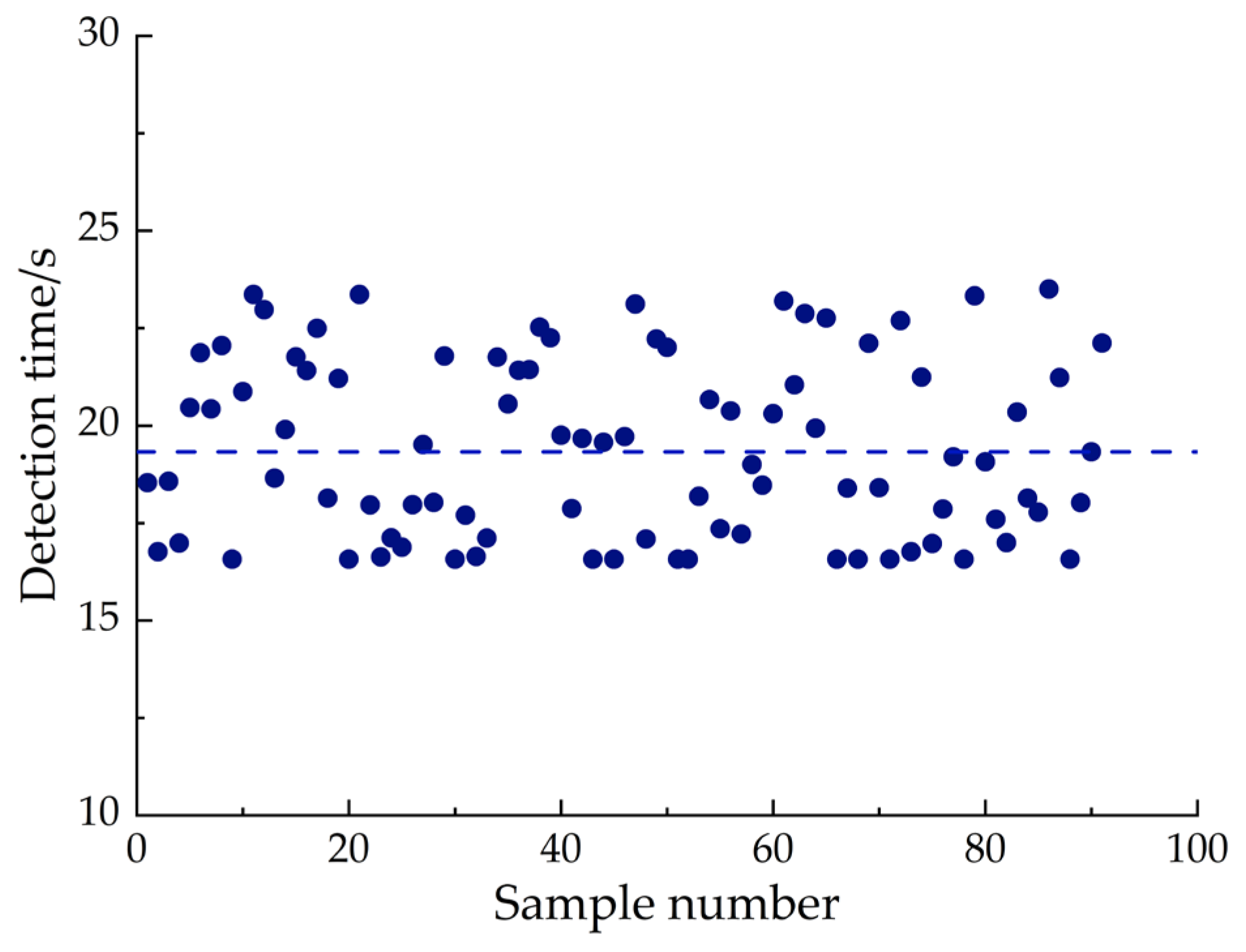
- In this study, the detection accuracy and detection time of the system are used as performance evaluation indexes to test the application of the CCC intelligent detection system. The results show that the average detection accuracy of CCC by using the intelligent detection system is more than 95%, and the detection accuracy rate of chlorophyll in cotton leaves is superior to that of the chlorophyll detection systems for wheat [42] and corn [43,44] plants developed by relevant scholars. Among them, the detection accuracy rate is the highest in the germination stage, reaching 99.133%, which has a relatively high detection accuracy rate. The coefficient of variation of the detection error is 1.606% to 2.778%, and the detection performance of the system is stable. At the same time, the average detection time of each sample detected by the system is only 19.33 s, and the detection efficiency is high. Based on the above evaluation, the CCC intelligent detection system can meet the requirements of CCC detection.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nijs, I.; Behaeghe, T.; Impens, I. Leaf nitrogen content as a predictor of photosynthetic capacity in ambient and global change conditions. J. Biogeogr. 1995, 22, 177–183. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Walthall, C.L.; Kim, M.S.; Colstoun, E.; Iii, M.M. Estimating corn leaf chlorophyll concentration from leaf and canopy reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Li, S.T.; Liu, X.Y.; He, P. Analyses on nutrient requirements in current agriculture production in China. J. Plant Nutr. Fertil. 2017, 23, 1416–1432. [Google Scholar]
- Wei, Q.; Zhang, B.Z.; Wei, Z. Estimation of canopy chlorophyll contention winter wheat by UAV multispectral remote sensing. J. Triticeae Crop. 2020, 40, 8. [Google Scholar]
- Shah, S.H.; Angel, Y.; Houborg, R.; Ali, S.; Mccabe, M.F. A random forest machine learning approach for the retrieval of leaf chlorophyll content in wheat. Remote Sens. 2019, 11, 920. [Google Scholar] [CrossRef]
- Deng, S.Q.; Zhao, Y.; Bai, X.Y.; Li, X.; Sun, Z.D.; Liang, J.; Cheng, S. Inversion of chlorophyll and leaf area index for winter wheat based on UAV image segmentation. Trans. Chin. Soc. Agric. Eng. 2022, 38, 136–145. [Google Scholar]
- Xiao, Q.L.; Tang, W.T.; Zhang, C.; Zhou, L.; Feng, L.; Shen, J.X.; Yan, T.Y.; Gao, P.G.; He, Y.; Wu, N. Spectral preprocessing combined with deep transfer learning to evaluate chlorophyll content in cotton leaves. Plant Phenomics 2022, 2022, 9813841. [Google Scholar] [CrossRef]
- Yang, P.Q.; van der Tol, C.; Campbell, P.K.E.; Middleton, E.M. Fluorescence correction vegetation index (FCVI): A physically based reflectance index to separate physiological and non-physiological information in far-red sun-induced chlorophyll fluorescence-Science Direct. Remote Sens. Environ. 2020, 240, 111676. [Google Scholar] [CrossRef]
- Li, Y.J.; Ma, B.X.; Li, C.; Yu, G.W. Accurate prediction of soluble solid content in dried Hami jujube using SWIR hyperspectral imaging with comparative analysis of models. Comput. Electron. Agric. 2022, 193, 106655. [Google Scholar] [CrossRef]
- Katja, B.; Jochem, V.; Féret, J.B.; Tobias, H.; Matthias, W.; Wolfram, M.; Gustau, C.V. Retrieval of aboveground crop nitrogen content with a hybrid machine learning method. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102174. [Google Scholar]
- Zhou, Y.J.; Zheng, J. Inversion Model Design of Chlorophyll a Based on BP Neural Network and Remote Sensing Image. Innov. Comput. 2020, 675, 531–538. [Google Scholar]
- An, T.; Yu, S.Y.; Huang, W.Q.; Li, G.L.; Tian, X.; Fan, S.X.; Dong, C.W.; Zhao, C.J. Robustness and accuracy evaluation of moisture prediction model for black tea withering process using hyperspectral imaging. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 269, 120791. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.S. Bifurcation curves of positive steady-state solutions for a reaction–diffusion problem of lake eutrophication. J. Math. Anal. Appl. 2016, 449, 1708–1724. [Google Scholar] [CrossRef]
- Zhang, Q.; Smith, D.W.; Baxter, C.W. Artificial neural networks: A tool with significant potential in environmental engineering and science-introduction. J. Environ. Eng. Sci. 2004, 3, III–IV. [Google Scholar]
- Zhang, Z.H. A gentle introduction to artificial neural networks. Ann. Transl. Med. 2016, 4, 370. [Google Scholar] [CrossRef]
- Wei, X.G.; Wu, L.L.; Ge, D.; Yao, M.Z.; Bai, Y.K. Prediction of the maturity of greenhouse grapes based on imaging technology. Plant Phenomics 2022, 2022, 9753427. [Google Scholar] [CrossRef]
- Sun, B.Y.; Chang, Q.R.; Liu, M.Y. Inversion chlorophyll mass fraction in winter wheat canopy by hyperspectral reflectance. Acta Agric. Boreali Occident. Sin. 2017, 26, 552–559. [Google Scholar]
- Li, Y.Y.; Chang, Q.R.; Liu, X.Y. Estimation of maize leaf SPAD value based on hyperspectrum and BP neural network. Trans. Chin. Soc. Agric. Eng. 2016, 32, 135–142. [Google Scholar]
- Zhou, Y.; Lu, A.J.; Liu, X. Prediction of chlorophyll a content in water body based on BP neural network with improved Genetic Algorithm. Electron. Test 2022, 37–42. [Google Scholar] [CrossRef]
- Yang, T.; Lu, J.; Liao, F.; Qi, H.; Tian, Y. Retrieving potassium levels in wheat blades using normalised spectra. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102412. [Google Scholar] [CrossRef]
- Wang, G.L.; Wang, W.J.; Cheng, K.; Liu, X.; Zhao, J.G.; Li, H.; Guo, E.H.; Li, Z.W. Hyperspectral imaging combined with back propagation neural network optimized by sparrow search algorithm for predicting gelatinization properties of millet flour. Food Sci. 2022, 43, 65–70. [Google Scholar]
- Bauriegel, E.; Giebel, A.; Geyer, M.; Schmidt, U.; Herppich, W.B. Early detection of fusarium infection in wheat using Hyper-Spectral imaging. Comput. Electron. Agric. 2011, 75, 304–312. [Google Scholar] [CrossRef]
- Yang, L.; Wu, Y.Q.; Wang, J.L. Research on recurrent neural network. J. Comput. Appl. 2018, 38, 1–6+26. [Google Scholar]
- Gao, P. Study on Real-Time Diagnosis of Verticillium Wilt of Cotton Based on Image Recognition; Shihezi University: Shihezi, China, 2019. [Google Scholar]
- De Cesaro Júnior, T.; Rieder, R. Automatic identification of insects from digital images: A Survey. Comput. Electron. Agric. 2020, 178, 105784. [Google Scholar] [CrossRef]
- Acquarelli, J.; Van, L.T.; Gerretzen, J. Convolutional neural networks for vibrational spectroscopic data analysis. Anal. Chim. Acta 2017, 954, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.H.; Xu, H.L.; Huang, G.Q.; Wang, B. Spinach freshness detection based on hyperspectral image and deep learning method. Trans. Chin. Soc. Agric. Eng. 2019, 35, 277–284. [Google Scholar]
- Wu, N.; Zhang, C.; Bai, X.; Du, X.; He, Y. Discrimination of chrysanthemum varieties using hyperspectral imaging combined with a deep convolutional neural network. Molecules 2018, 23, 2831. [Google Scholar] [CrossRef]
- Conrad, A.O.; Li, W.; Lee, D.Y.; Wang, G.L.; Bonello, P. Machine learning-based presymptomatic detection of rice sheath blight using spectral profiles. Plant Phenomics 2020, 2020, 8954085. [Google Scholar] [CrossRef]
- Hctor, P.C.; Karina, J.V.; Mario, G.; Omar, F.U.; Beln, A.L. Determination of moisture in rice grains based on visible spectrum analysis. Agronomy 2022, 12, 3021. [Google Scholar] [CrossRef]
- Colovic, M.; Yu, K.; Todorovic, M.; Cantore, V.; Hamze, M.; Albrizio, R.; Anna, M.S. Hyperspectral vegetation indices to assess water and nitrogen status of sweet maize crop. Agronomy 2022, 12, 2181. [Google Scholar] [CrossRef]
- Milas, A.S.; Romanko, M.; Reil, P.; Abeysinghe, T.; Marambe, A. The importance of leaf area index in mapping chlorophyll content of corn under different agricultural treatments using uav images. Int. J. Remote Sens. 2018, 39, 5415–5431. [Google Scholar] [CrossRef]
- Chen, B.Q.; Zhang, Y.; Wang, K. A dynamic prediction method for cotton growth and yield in Xinjiang based on APSIM. J. Agric. Mach. 2025, 56, 82–90. [Google Scholar]
- Bai, E.L.; Zhang, Z.Q.; Guo, Z.C. Cotton color detection method based on machine vision. J. Text. 2024, 45, 36–43. [Google Scholar]
- Da Rosa, C.N.; Pereira Filho, W.; Bremer, U.F.; Putzke, J.; De Andrade, A.M.; Kramer, G.; Hillebrand, F.L.; De Jesus, J.B. Spectral behavior of vegetation in Harmony Point, Nelson Island, Antarctica. Biodivers. Conserv. 2022, 31, 1867. [Google Scholar] [PubMed]
- Geelen, B.; Blanch, C.; Gonzalez, P.; Tack, N.; Lambrechts, A. A tiny, vis-nir snapshot multispectral camera. In Proceedings of the Advanced Fabrication Technologies for Micro/Nano Optics & Photonics VIII, San Francisco, CA, USA, 7–12 February 2015; Volume 9374. [Google Scholar]
- Ali, A.M.; Thind, H.S.; Sharma, S.; Varinderpal-Singh. Prediction of dry direct-seeded rice yields using chlorophyll meter, leaf color chart and greenseeker optical sensor in northwestern India. Field Crop. Res. 2014, 161, 11–15. [Google Scholar] [CrossRef]
- Cao, Q.; Miao, Y.X.; Li, F.; Gao, X.W.; Liu, B.; Lu, D.J. Developing a new crop circle active canopy sensor-based precision nitrogen management strategy for winter wheat in north China plain. Precis. Agric. 2017, 18, 2–18. [Google Scholar] [CrossRef]
- Siedliska, A.; Baranowski, P.; Zubik, M.; Mazurek, W.; Sosnowska, B. Detection of fungal infections in strawberry fruit by vnir/swir hyperspectral imaging. Postharvest Biol. Technol. 2018, 139, 115–126. [Google Scholar] [CrossRef]
- Zahra, J.; Amirhossein, A.; Philippe, C.; Michael, B.C. Monitoring the coefficient of variation: A literature review. Comput. Ind. Eng. 2021, 162, 107600. [Google Scholar]
- Yu, S.Y.; Bu, H.R.; Hu, X.; Dong, W.C.; Zhang, L.X. Establishment and Accuracy Evaluation of Cotton Leaf Chlorophyll Content Prediction Model Combined with Hyperspectral Image and Feature Variable Selection. Agronomy 2023, 13, 2120. [Google Scholar] [CrossRef]
- Yang, Z.Q.; Li, G.Q.; Zhang, Y.M.; Su, W.B.; Wu, J.Z.; Duan, T.C.; Qi, L. Plant Chlorophyll a and Chlorophyll b Nondestructive Testing System: Research and Development. Chin. Agric. Sci. Bull. 2021, 37, 147–152. [Google Scholar]
- Zhang, C.Q.; Liu, M.S.; Chao, J.Y.; Tang, B.; Li, M.Z.; Sun, H. Contact-based Crop Chlorophyll Detection System Based on Feature Wavelengths. Trans. Chin. Soc. Agric. Mach. 2024, 55, 255–262. [Google Scholar]
- Wang, N.; Li, Z.; Li, J.M.; Zhang, Y.; Sun, H.; Li, M.Z. Design and Development of Intelligent Plant Chlorophyll Index Detection System. Trans. Chin. Soc. Agric. Mach. 2025. Available online: http://kns.cnki.net/kcms/detail/11.1964.S.20230911.2037.048.html (accessed on 21 June 2025).

| Functional Module | Configuration Information | |
|---|---|---|
| Hardware platform | Operating system | Windows 11 23H2 (64 bit) |
| CPU | AMD Ryzen 5 5600X 3.70 GHz (Advanced Micro Devices (AMD), Santa Clara, MO, USA) | |
| Memory | 32.0 GB (DDR4 3600 MHz) | |
| Graphics card | GeForce RTX 3060Ti 8GD6X (Corporation, Santa Clara, MO, USA) | |
| Central control platform | Programming language | Python 3.11.5 |
| Standard library | Tkinter, matlab.engine, pyenvi, pandas, openpyxl | |
| Spectral information extraction | External software | ENVI 5.3 |
| Model training detection | External software | MATLAB 2018b |
| Model Name | Growth Stage | RE (%) | RMSEP (mg/g) | RPD | |
|---|---|---|---|---|---|
| Seedling stage | RAW | 1.71 | 0.80 | 0.039 | 2.37 |
| SG | 1.61 | 0.84 | 0.035 | 2.43 | |
| 1D | 1.64 | 0.80 | 0.036 | 2.39 | |
| Budding stage | RAW | 1.66 | 0.85 | 0.034 | 2.46 |
| SG | 1.25 | 0.89 | 0.029 | 2.79 | |
| 1D | 1.32 | 0.88 | 0.030 | 2.72 | |
| Flowering-boll stage | RAW | 1.81 | 0.82 | 0.043 | 2.36 |
| SG | 1.35 | 0.87 | 0.032 | 2.62 | |
| 1D | 1.43 | 0.85 | 0.032 | 2.56 |
| Model Name | Growth Stage | RE (%) | RMSEP (mg/g) | RPD | |
|---|---|---|---|---|---|
| Seedling stage | SSA-BPNN | 1.26 | 0.86 | 0.051 | 2.62 |
| 1DCNN | 1.16 | 0.92 | 0.035 | 3.56 | |
| Budding stage | SSA-BPNN | 1.04 | 0.94 | 0.030 | 4.16 |
| 1DCNN | 0.89 | 0.97 | 0.019 | 5.54 | |
| Flowering-boll stage | SSA-BPNN | 1.20 | 0.91 | 0.040 | 3.23 |
| 1DCNN | 0.96 | 0.95 | 0.028 | 4.58 |
| Growth Stage | True Values (mg/g) | Predicted Values (mg/g) | Standard Deviation (mg/g) | Prediction Accuracy (%) | Coefficient of Variation (%) |
|---|---|---|---|---|---|
| Seedling stage | 1.354 | 1.355 | 0.038 | 96.998 | 2.778 |
| Budding stage | 1.280 | 1.275 | 0.020 | 98.357 | 1.606 |
| Flowering-boll stage | 1.352 | 1.355 | 0.034 | 97.164 | 2.506 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, W.; Zhang, L.; Hu, X.; Yu, S. Design and Development of an Intelligent Chlorophyll Content Detection System for Cotton Leaves. Processes 2025, 13, 2329. https://doi.org/10.3390/pr13082329
Wei W, Zhang L, Hu X, Yu S. Design and Development of an Intelligent Chlorophyll Content Detection System for Cotton Leaves. Processes. 2025; 13(8):2329. https://doi.org/10.3390/pr13082329
Chicago/Turabian StyleWei, Wu, Lixin Zhang, Xue Hu, and Siyao Yu. 2025. "Design and Development of an Intelligent Chlorophyll Content Detection System for Cotton Leaves" Processes 13, no. 8: 2329. https://doi.org/10.3390/pr13082329
APA StyleWei, W., Zhang, L., Hu, X., & Yu, S. (2025). Design and Development of an Intelligent Chlorophyll Content Detection System for Cotton Leaves. Processes, 13(8), 2329. https://doi.org/10.3390/pr13082329






