Breast Cancer Tumor Classification Using a Bag of Deep Multi-Resolution Convolutional Features
Abstract
1. Introduction
2. Background
2.1. BreakHis Breast Cancer Histopathological Image Dataset
2.2. Convolutional Neural Networks (CNNs)
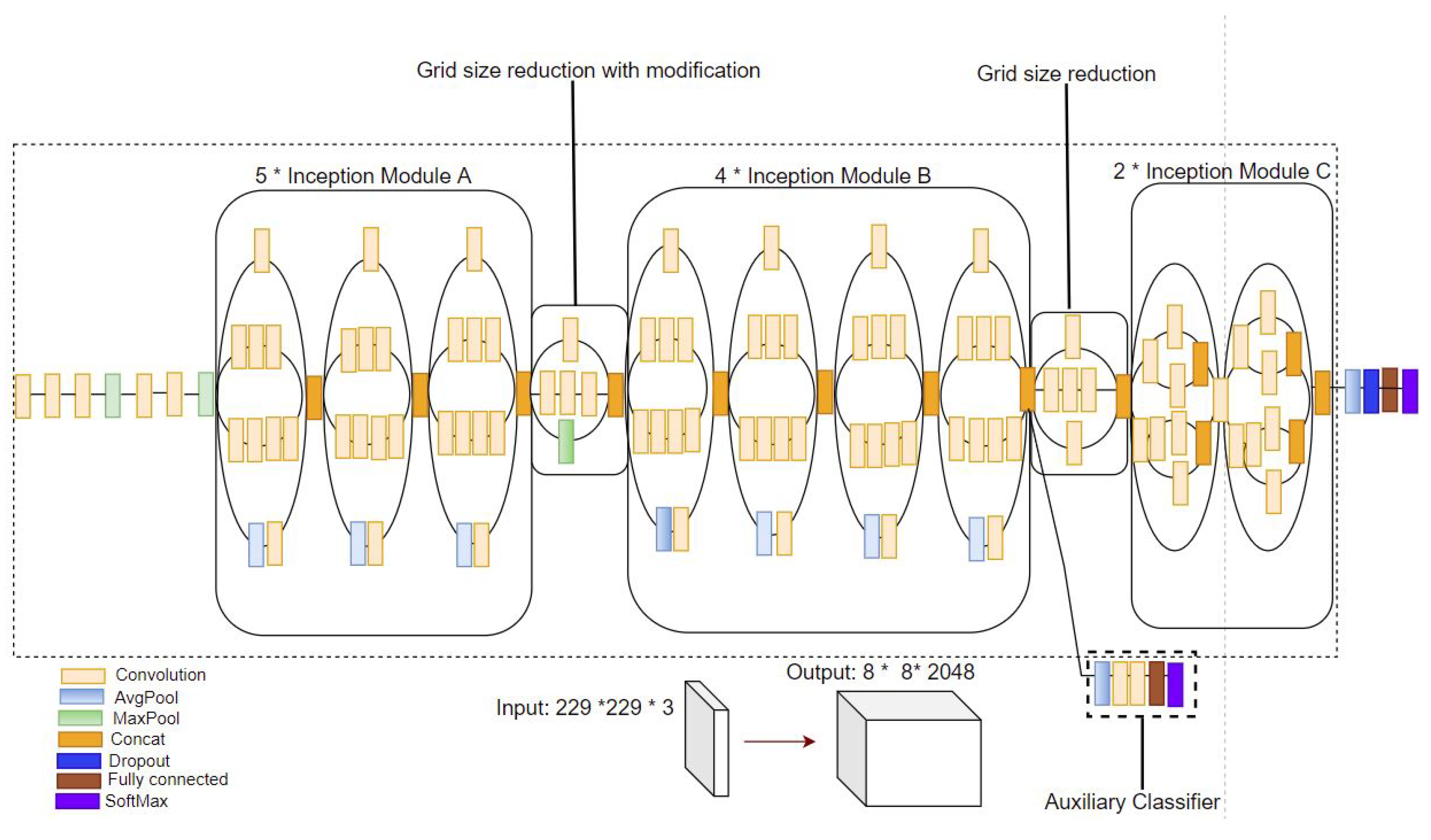
2.3. Pre-Trained CNNs for Deep Image Feature Extraction
3. Materials and Methods
3.1. Step 1: Histopathological Image Pre-Processing
3.2. Step 2: Deep Multi-Resolution Feature Extraction Using Pre-Trained CNNs
3.3. Step 3: Global Pooling of Features to Create BoDMCF
3.4. Step 4: BoDMCF Classification Using SVM
4. Evaluation and Results
4.1. Evaluation Metrics
4.2. Baseline State-of-the-Art CNN Image Classification Architecture
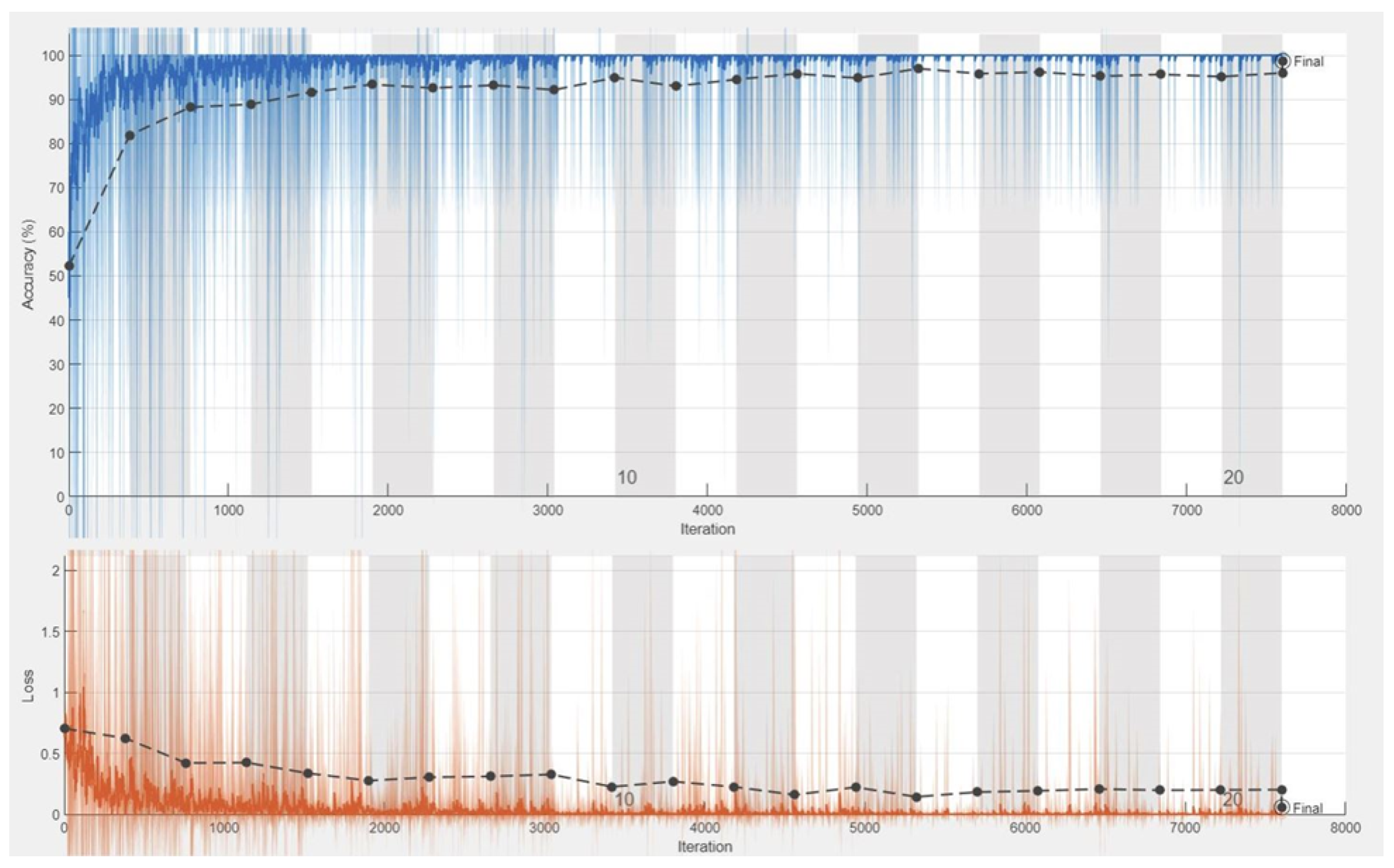
4.3. Experiments
| F. Extractors | Size | Bytes | Acc. | Sens. | Spec. | AUC | F1-Sc. | TPR | FPR | MCC | Kappa | Prec. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Efficientnetb0 $ Resnet50 | Train_features 5536 × 3328 Test_features 2373 × 3328 | Train_features 73,695,232 Test_features 31,589,376 | 0.9966 | 0.9987 | 0.9662 | 0.9972 | 0.9946 | 0.9987 | 0.0043 | 0.9922 | 0.8378 | 0.9907 |
| Efficientnetb0 $ Inception-V3 | Train_features 5536 × 2816 Test_features 2373 × 2816 | Train_features 62,357,504 Test_features 26,729,472 | 0.9962 | 0.9933 | 0.9699 | 0.9352 | 0.9939 | 0.9933 | 0.0025 | 0.9912 | 0.9352 | 0.9946 |
| Resnet50 $ Inception-V3 | Train_features 5536 × 3584 Test_features 2373 × 3584 | Train_features 79,364,096 Test_features 34,019,328 | 0.9979 | 0.9960 | 0.9705 | 0.9974 | 0.9966 | 0.9960 | 0.0012 | 0.9951 | 0.8375 | 0.9973 |
| BoDMCF | Train_features 5536 × 4864 Test_features 2373 × 4864 | Train_features 107,708,416 Test_features 46,169,088 | 0.9992 | 0.9987 | 0.9797 | 0.9990 | 0.9987 | 0.9987 | 0.0006 | 0.9980 | 0.8368 | 0.9987 |
| Authors | Models | Accuracy |
|---|---|---|
| M. Amrane [14] | Naive Bayes (NB) k-nearest neighbor (KNN) | 97.51% for KNN and 96.19% for NB |
| S. H. Kassani, M. J. Wesolowski, and K. A. Schneider [29] | VGG19, MobileNet, and DenseNet | 98.13% |
| F. A. Spanhol, L. S. Oliveira, C. Petitjean, and L. Heutte [31] | Ensemble models | 85.6% |
| Kowal et al. [32] | Deep learning model | 92.4% |
| A. Al Nahid, M. A. Mehrabi, and Y. Kong [36] | CNN, LSTM, K-means clustering, Mean-Shift clustering and SVM | 96.0% |
| Our Proposed Approach | BoDMCF + SVM | 99.92% |
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA. Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA. Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, G.; Shuib, L.; Wahab, A.W.A.; Mujtaba, G.; Mujtaba, G.; Nweke, H.F.; Al-garadi, M.A.; Zulfiqar, F.; Raza, G.; Azmi, N.A. Deep learning-based breast cancer classification through medical imaging modalities: State of the art and research challenges. Artif. Intell. Rev. 2019, 53, 1655–1720. [Google Scholar] [CrossRef]
- Birdwell, R.L.; Ikeda, D.M.; O’Shaughnessy, K.F.; Sickles, E.A. Mammographic characteristics of 115 missed cancers later detected with screening mammography and the potential utility of computer-aided detection. Radiology 2001, 219, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Doi, K. Current status and future potential of computer-aided diagnosis in medical imaging. Br. J. Radiol. 2005, 78, 21–30. [Google Scholar] [CrossRef]
- Salama, M.S.; Eltrass, A.S.; Elkamchouchi, H.M. An Improved Approach for Computer-Aided Diagnosis of Breast Cancer in Digital Mammography. In Proceedings of the 2018 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Rome, Italy, 11–13 June 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Eldredge, J.D.; Hannigan, G.G. Emerging trends in health sciences librarian. Heal. Sci. Librariansh. 2014, 1, 53–84. [Google Scholar] [CrossRef]
- Kasban, H.; El-Bendary, M.A.M.; Salama, D.H. A Comparative Study of Brain Imaging Techniques. Int. J. Inf. Sci. Intell. Syst. 2015, 4, 37–58. [Google Scholar]
- He, L.; Long, L.R.; Antani, S.; Thoma, G.R. Histology image analysis for carcinoma detection and grading. Comput. Methods Programs Biomed. 2012, 107, 538–556. [Google Scholar] [CrossRef]
- Yan, R.; Ren, F.; Wang, Z.; Wang, L.; Zhang, T.; Liu, Y.; Rao, X.; Zheng, C.; Zhang, F. Breast cancer histopathological image classification using a hybrid deep neural network. Methods 2019, 173, 52–60. [Google Scholar] [CrossRef]
- Alarabeyyat, A.; Alhanahnah, M. Breast Cancer Detection Using K-Nearest Neighbor Machine Learning Algorithm. In Proceedings of the 9th International Conference on Developments in eSystems Engineering (DeSE), Liverpool, UK, 31 August–1 September 2016; pp. 35–39. [Google Scholar]
- Prabhakar, S.K.; Rajaguru, H. Performance Analysis of Breast Cancer Classification with Softmax Discriminant Classifier and Linear Discriminant Analysis. In Proceedings of the International Conference on Biomedical and Health Informatics, Thessaloniki, Greece, 18–21 November 2017; Maglaveras, N., Chouvarda, I., de Carvalho, P., Eds.; Springer: Singapore, 2018; Volume 66. [Google Scholar]
- Asri, M.H.; Moatassime, H.A. Using Machine Learning Algorithms for Breast Cancer Risk Prediction and Diagnosis. Procedia Comput. Sci. 2016, 83, 1064–1073. [Google Scholar] [CrossRef]
- Amrane, M.; Oukid, S.; Gagaoua, I.; Ensari, T. Breast cancer classification using machine learning. In Proceedings of the 2018 Electric Electronics, Computer Science, Biomedical Engineerings’ Meeting (EBBT), Istanbul, Turkey, 18–19 April 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–4. [Google Scholar]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Zheng, B.; Yoon, S.W.; Lam, S.S. Breast cancer diagnosis based on feature extraction using a hybrid of K-means and support vector machine algorithms. Expert Syst. Appl. 2014, 41, 1476–1482. [Google Scholar] [CrossRef]
- Yin, F.F.; Giger, M.L.; Doi, K.; Vyborny, C.J.; Schmidt, R.A. Computerized detection of masses in digital mammograms: Automated alignment of breast images and its effect on bilateral-subtraction technique. Med. Phys. 1994, 21, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Eltonsy, N.H.; Tourassi, G.D.; Elmaghraby, A.S. A concentric morphology model for the detection of masses in mammography. IEEE Trans. Med. Imaging 2007, 26, 880–889. [Google Scholar] [CrossRef]
- Barata, C.; Marques, J.S.; Celebi, M.E. Improving dermoscopy image analysis using color constancy. In Proceedings of the 2014 IEEE International Conference on Image Processing (ICIP), Paris, France, 27–30 October 2014; Volume 19, pp. 3527–3531. [Google Scholar] [CrossRef]
- Pinto, A.; Pereira, S.; Correia, H.; Oliveira, J.; Rasteiro, D.M.L.D.; Silva, C.A. Brain Tumour Segmentation based on Extremely Randomized Forest with high-level features. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 3037–3040. [Google Scholar] [CrossRef]
- Tustison, N.J.; Shrinidhi, K.L.; Wintermark, M.; Durst, C.R.; Kandel, B.M.; Gee, J.C.; Grossman, M.C.; Avants, B.B. Optimal Symmetric Multimodal Templates and Concatenated Random Forests for Supervised Brain Tumor Segmentation (Simplified) with ANTsR. Neuroinformatics 2015, 13, 209–225. [Google Scholar] [CrossRef]
- Cheng, G.; Li, Z.; Yao, X.; Guo, L.; Wei, Z. Remote sensing image scene classification using bag of convolutional features. IEEE Geosci. Remote Sens. Lett. 2017, 14, 1735–1739. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2016, 63, 1455–1462. [Google Scholar] [CrossRef]
- Vaccaro, F.; Bertini, M.; Uricchio, T.; Del Bimbo, A. Image Retrieval using Multi-scale CNN Features Pooling. In Proceedings of the 2020 International Conference on Multimedia Retrieval, Dublin, Ireland, 8–11 June 2020; pp. 311–315. [Google Scholar]
- Kassani, S.H.; Kassani, P.H.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Classification of histopathological biopsy images using ensemble of deep learning networks. arXiv 2019, arXiv:1909.11870. [Google Scholar]
- Maqsood, S.; Damaševičius, R.; Maskeliūnas, R. TTCNN: A Breast Cancer Detection and Classification towards Computer-Aided Diagnosis Using Digital Mammography in Early Stages. Appl. Sci. 2022, 12, 3273. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast Cancer Histopathological Image Classification using Convolutional Neural Networks. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016. [Google Scholar]
- Kowal, M.; Skobel, M.; Gramacki, A.; Korbicz, J. Breast cancer nuclei segmentation and classification based on a deep learning approach. Int. J. Appl. Math. Comput. Sci. 2021, 31, 85–106. [Google Scholar]
- Shen, R.; Yan, K.; Tian, K.; Jiang, C.; Zhou, K. Breast mass detection from the digitized X-ray mammograms based on the combination of deep active learning and self-paced learning. Futur. Gener. Comput. Syst. 2019, 101, 668–679. [Google Scholar] [CrossRef]
- Byra, M.; Piotrzkowska-Wroblewska, H.; Dobruch-Sobczak, K.; Nowicki, A. Combining Nakagami imaging and convolutional neural network for breast lesion classification. In Proceedings of the 2017 IEEE International Ultrasonics Symposium (IUS), Washington, DC, USA, 6–9 September 2017; pp. 5–8. [Google Scholar] [CrossRef]
- Nejad, E.M.; Affendey, L.S.; Latip, R.B.; Ishak, I.B. Classification of histopathology images of breast into benign and malignant using a single-layer convolutional neural network. ACM Int. Conf. Proc. Ser. 2017, 1313, 50–53. [Google Scholar] [CrossRef]
- Al Nahid, A.; Mehrabi, M.A.; Kong, Y. Histopathological breast cancer image classification by deep neural network techniques guided by local clustering. Biomed Res. Int. 2018, 2018, 2362108. [Google Scholar] [CrossRef]
- Ogundokun, R.O.; Misra, S.; Douglas, M.; Damaševičius, R.; Maskeliūnas, R. Medical Internet-of-Things Based Breast Cancer Diagnosis Using Hyperparameter-Optimized Neural Networks. Future Internet 2022, 14, 153. [Google Scholar] [CrossRef]
- Vogado, L.H.S.; Veras, R.M.S.; Araujo, F.H.D.; Silva, R.R.V.; Aires, K.R.T. Leukemia diagnosis in blood slides using transfer learning in CNNs and SVM for classification. Eng. Appl. Artif. Intell. 2018, 72, 415–422. [Google Scholar] [CrossRef]
- Gandomkar, Z.; Brennan, P.C.; Mello-Thoms, C. MuDeRN: Multi-category classification of breast histopathological image using deep residual networks. Artif. Intell. Med. 2018, 88, 14–24. [Google Scholar] [CrossRef]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast Cancer Multi-classification from Histopathological Images with Structured Deep Learning Model. Sci. Rep. 2017, 7, 4172. [Google Scholar] [CrossRef] [PubMed]
- Wichakam, I.; Vateekul, P. Combining Deep Convolutional Networks for Mass Detection on Digital Mammograms. In Proceedings of the 2016 8th International Conference on Knowledge and Smart Technology (KST), Chiangmai, Thailand, 3–6 February 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 239–244. [Google Scholar]
- Devnath, L.; Luo, S.; Summons, P.; Wang, D. Automated detection of pneumoconiosis with multilevel deep features learned from chest X-ray radiographs. Comput. Biol. Med. 2021, 129, 104125. [Google Scholar] [CrossRef]
- Devnath, L.; Summons, P.; Luo, S.; Wang, D.; Shaukat, K.; Hameed, I.A.; Aljuaid, H. Computer-Aided Diagnosis of Coal Workers’ Pneumoconiosis in Chest X-ray Radiographs Using Machine Learning: A Systematic Literature Review. Int. J. Environ. Res. Public Health 2022, 19, 6439. [Google Scholar] [CrossRef]
- Devnath, L.; Fan, Z.; Luo, S.; Summons, P. Detection and Visualisation of Pneumoconiosis Using an Ensemble of Multi-Dimensional Deep Features Learned from Chest X-rays. Int. J. Environ. Res. Public Health 2022, 19, 11193. [Google Scholar] [CrossRef] [PubMed]
- Huynh, B.Q.; Li, H.; Giger, M.L. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J. Med. Imaging 2016, 3, 034501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Qi, S.; Monkam, P.; Li, C.; Yang, F.; Yao, Y.D.; Qian, W. Ensemble learners of multiple deep CNNs for pulmonary nodules classification using ct images. IEEE Access 2019, 7, 110358–110371. [Google Scholar] [CrossRef]
- Filipczuk, P.; Fevens, T.; Krzyzak, A.; Monczak, R. Computer-aided breast cancer diagnosis based on the analysis of cytological images of fine needle biopsies. IEEE Trans. Med. Imaging 2013, 32, 2169–2178. [Google Scholar] [CrossRef]
- George, Y.M.; Zayed, H.H.; Roushdy, M.I.; Elbagoury, B.M. Remote computer-aided breast cancer detection and diagnosis system based on cytological images. IEEE Syst. J. 2014, 8, 949–964. [Google Scholar] [CrossRef]
- Subhadra, D. Basic Histology: A Color Atlas & Text. Female Reproductive System. Chapter 17. 2016. Available online: https://www.jaypeedigital.com/book/9789352501786/chapter/ch17 (accessed on 25 July 2022).
- Xu, B.; Wang, N.; Chen, T.; Li, M. Empirical Evaluation of Rectified Activations in Convolutional Network. arXiv 2015, arXiv:1505.00853. [Google Scholar]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the ICLR 2015 3rd International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Brown, M.J.; Hutchinson, L.A.; Rainbow, M.J.; Deluzio, K.J.; de Asha, A.R. A comparison of self-selected walking speeds and walking speed variability when data are collected during repeated discrete trials and during continuous walking. J. Appl. Biomech. 2017, 33, 384–387. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 248–255. [Google Scholar]
- Szegedy, P.S.C.; Liu, W.; Jia, Y. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 163–182. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–1 July 2015; Volume 1, pp. 448–456. [Google Scholar]
- Xiao, Y.; Decencière, E.; Velasco-Forero, S.; Burdin, H.; Bornschlögl, T.; Bernerd, F.; Warrick, E.; Baldeweck, T. A new color augmentation method for deep learning segmentation of histological images. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 886–890. [Google Scholar]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Zerouaoui, H.; Idri, A. Deep hybrid architectures for binary classification of medical breast cancer images. Biomed. Signal Process. Control 2022, 71, 103226. [Google Scholar] [CrossRef]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Singh, J.; Thakur, D.; Ali, F.; Gera, T.; Kwak, K.S. Deep feature extraction and classification of android malware images. Sensors 2020, 20, 7013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar]
- Aljuaid, H.; Alturki, N.; Alsubaie, N.; Cavallaro, L.; Liotta, A. Computer-aided diagnosis for breast cancer classification using deep neural networks and transfer learning. Comput. Methods Programs Biomed. 2022, 223, 106951. [Google Scholar] [CrossRef] [PubMed]
- Nemenyi, P.B. Distribution-Free Multiple Comparisons. Ph.D. Thesis, Princeton University, Princeton, NJ, USA, 1963. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]





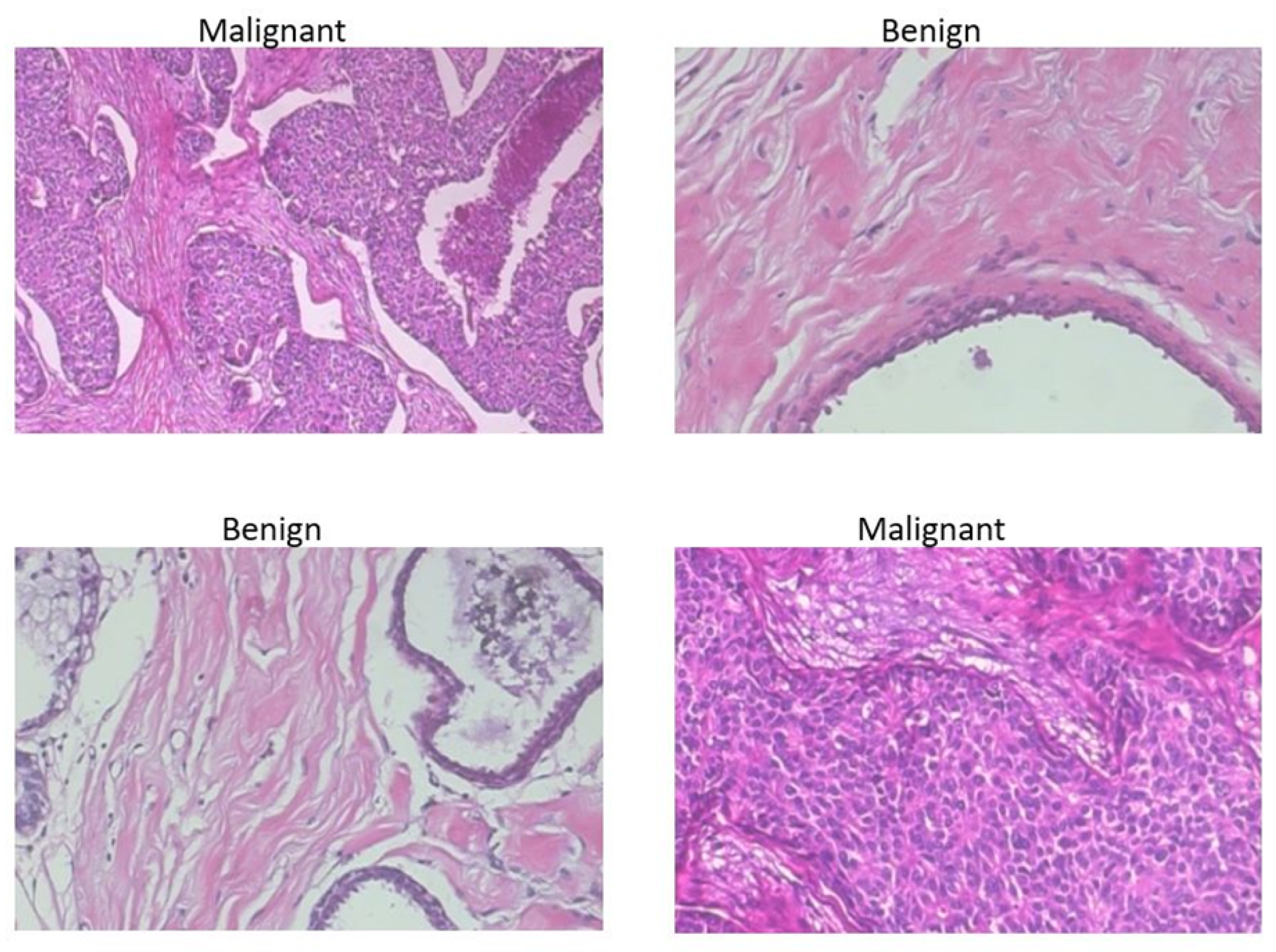

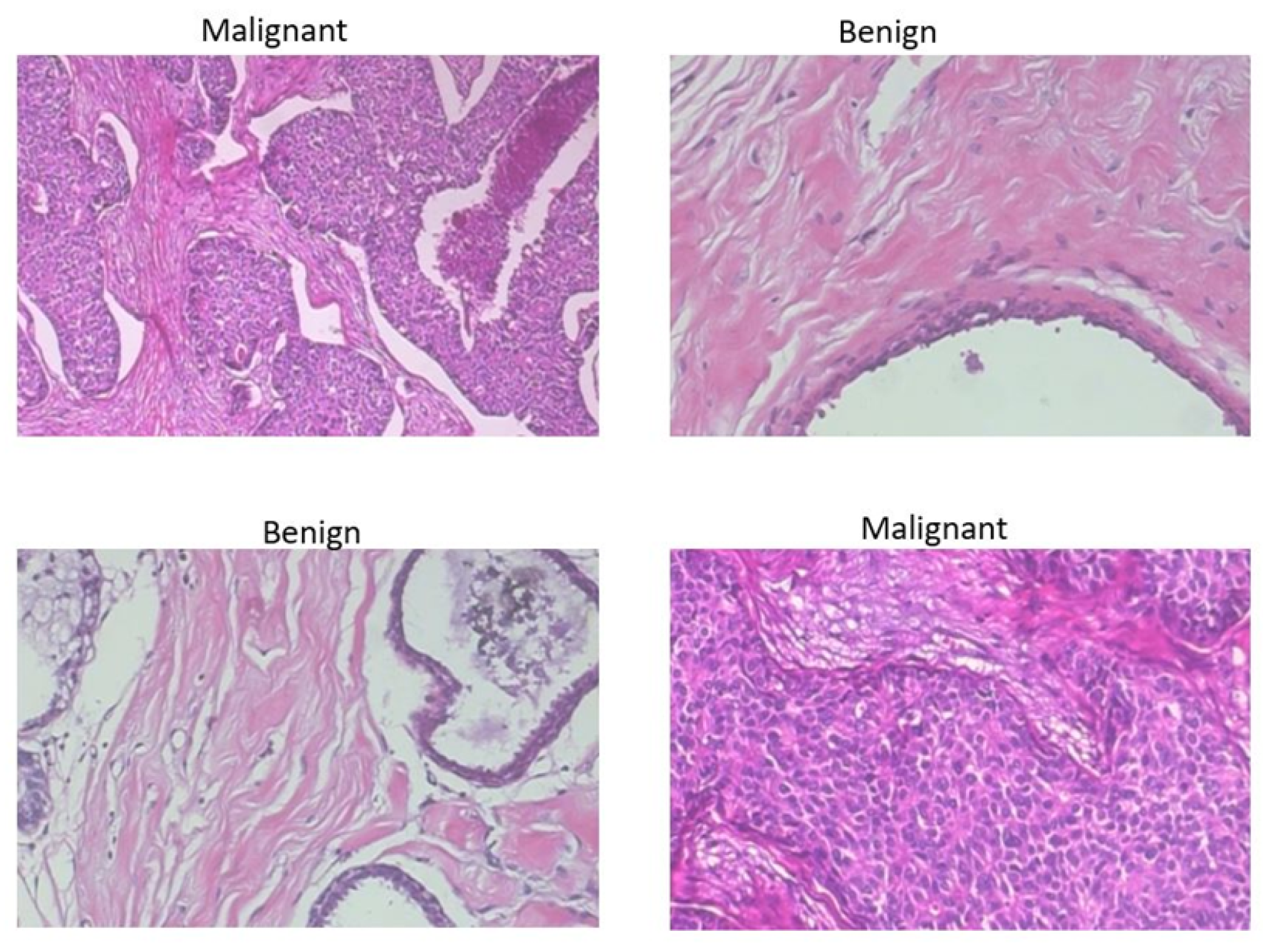



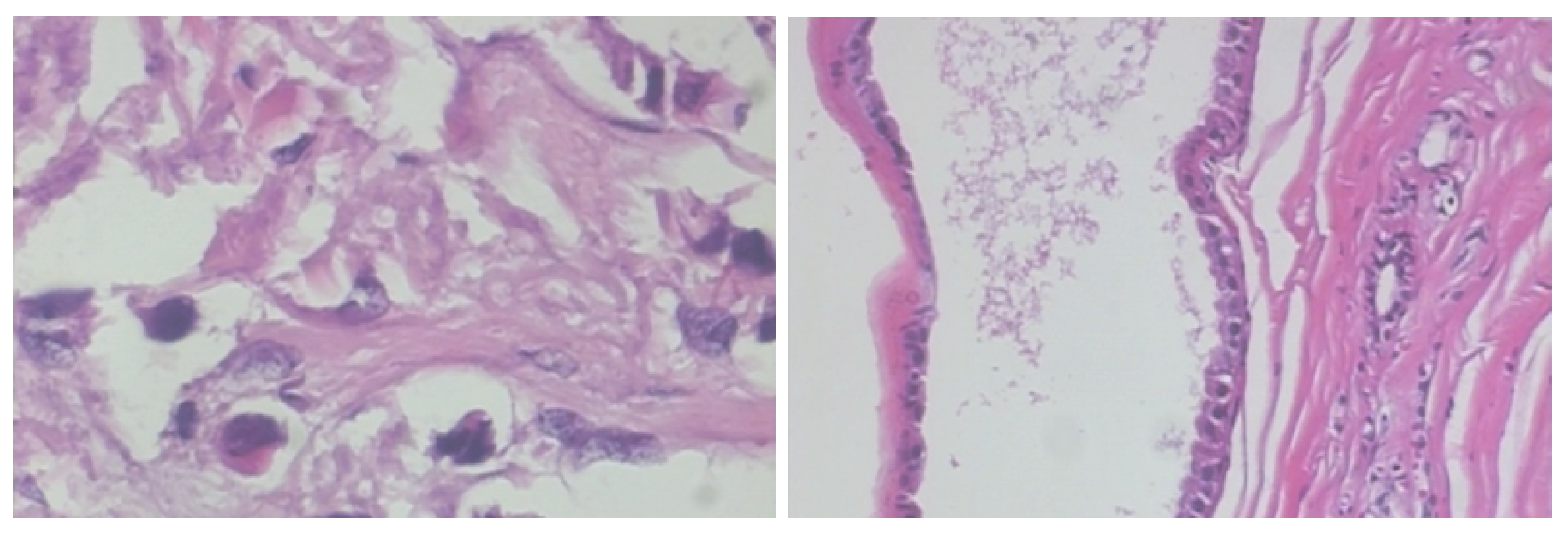
| Authors | Models | ML Problem | Summary of Approach | Accuracy |
|---|---|---|---|---|
| R. Yan et al. [10] | CNN and RNN | Four-class classification into malignant and benign subtypes | A CNN was used to extract image patches. Then an RNN was used to fuse the patch features and make the final image classification. | 91.3% |
| M. Amrane [14] | Naive Bayes (NB) and k-nearest neighbor (KNN) | Binary classification (malignant or benign) | For NB, data was split into blocks of 2 classes and 2 sets of features T and classes D and statistical analysis were performed. K-nearest neighbor, pick an instance from the testing sets and calculate its distance with the training set. | 97.51% for KNN and 96.19% for NB |
| S. H. Kassani, M. J. Wesolowski, and K. A. Schneider [29] | VGG19, MobileNet, and DenseNet. | Binary classification (malignant or benign) | Ensemble model was used for the feature extraction. Then classification was done using a Multi-Layer Perceptron (MLP) classifier | 98.13% |
| F. A. Spanhol, L. S. Oliveira, C. Petitjean, and L. Heutte [31] | Ensemble models | Binary classification (malignant or benign) | Various CNNs were using a fusion rule for breast cancer classification | 85.6% |
| Kowal et al. [32] | Deep learning model | Binary classification (malignant or benign) | Segmentation, feature extraction and classification were performed on individual cell nuclei of cytological images. | 92.4% |
| A. Al Nahid, M. A. Mehrabi, and Y. Kong [36] | CNN, LSTM, K-means clustering, Mean-Shift clustering and SVM | Binary classification (malignant or benign) | A set of biomedical breast cancer images were classified using novel DNN models guided by an unsupervised clustering method | 96.0% |
| Z. Gandomkar, P. C. Brennan, and C. Mello-Thoms [39] | Deep residual network (ResNet) | Multi class classification into Subtypes of malignant and benign | Approach consisted of two stages. In the first stage, ResNet layers classified patches from the images as benign or malignant. In the second statge, images were classified into subtypes of malignant and benign | 98.52%, 97.90%, 98.33%, and 97.66% in 40×, 100×, 200× and 400× magnification factors respectively |
| Magnification | Malignant | Benign | Total |
|---|---|---|---|
| 40× | 1370 (68.67%) | 652 (32.68%) | 1995 |
| 100× | 1437 (69.05%) | 644 (30.95%) | 2081 |
| 200× | 1390 (69.05%) | 623 (30.94%) | 2013 |
| 400× | 1232 (67.69%) | 588 (32.31%) | 1820 |
| 5429 (68.64%) | 2480 (31.14%) | 7909 |
| Iteration | Activation Stength | Pyramid Level |
|---|---|---|
| 1 | 0.35 | 1 |
| 2 | 0.31 | 1 |
| 3 | 0.59 | 1 |
| 4 | 1.19 | 1 |
| 5 | 1.87 | 1 |
| 6 | 2.56 | 1 |
| 7 | 3.12 | 1 |
| 8 | 3.56 | 1 |
| 9 | 3.87 | 1 |
| 10 | 4.15 | 1 |
| Iteration | Activation Strength | Pyramid Level |
|---|---|---|
| 1 | 0.32 | 1 |
| 2 | 0.35 | 1 |
| 3 | 0.58 | 1 |
| 4 | 0.98 | 1 |
| 5 | 1.51 | 1 |
| 6 | 1.94 | 1 |
| 7 | 2.37 | 1 |
| 8 | 2.73 | 1 |
| 9 | 3.09 | 1 |
| 10 | 3.29 | 1 |
| Iteration | Activation Strength | Pyramid Level |
|---|---|---|
| 1 | 0.94 | 1 |
| 2 | 1.18 | 1 |
| 3 | 2.92 | 1 |
| 4 | 5.34 | 1 |
| 5 | 7.22 | 1 |
| 6 | 8.50 | 1 |
| 7 | 9.41 | 1 |
| 8 | 10.04 | 1 |
| 9 | 10.60 | 1 |
| 10 | 10.88 | 1 |
| Hyperparameter | Value |
|---|---|
| Train-Test ratio | 70:30 |
| optimization algorithm | stochastic gradient descent |
| activation function | ReLu |
| Mini Batch Size | 20 |
| Max Epochs | 30 |
| Initial Learn Rate | 0.00125 |
| Learn-Rate Drop Factor | 0.1 |
| Learn-Rate Drop Period | 20 |
| Model | Accuracy | Precision | F1-Score | Recall | AUC | Kappa | MCC |
|---|---|---|---|---|---|---|---|
| 40× Renent18 | 0.9064 | 0.8047 | 0.8607 | 0.9251 | 0.9115 | 0.8667 | 0.7950 |
| 40× InceptionResnetV2 | 0.7843 | 0.6021 | 0.7261 | 0.9144 | 0.8197 | 0.6924 | 0.5937 |
| 40× InceptionV3 | 0.8679 | 0.8418 | 0.7710 | 0.7112 | 0.8252 | 0.8247 | 0.6839 |
| 40× Densenet201 | 0.8963 | 0.9433 | 0.8110 | 0.7112 | 0.8459 | 0.8629 | 0.7555 |
| 40× Resnet50 | 0.9381 | 0.9261 | 0.8981 | 0.8717 | 0.9200 | 0.9135 | 0.8545 |
| 40× Efficientnetbo | 0.8311 | 0.7389 | 0.7248 | 0.7112 | 0.7984 | 0.7749 | 0.6033 |
| 40× Squeezenet | 0.9281 | 0.8429 | 0.8917 | 0.9465 | 0.9331 | 0.8970 | 0.8413 |
| 40× Shufflenet | 0.8428 | 0.8252 | 0.7152 | 0.6310 | 0.7851 | 0.7972 | 0.6197 |
| 100× Renent18 | 0.9327 | 0.8995 | 0.8901 | 0.8808 | 0.9184 | 0.9060 | 0.8417 |
| 100× InceptionResnetV2 | 0.8173 | 0.7582 | 0.6705 | 0.6010 | 0.7576 | 0.7669 | 0.5535 |
| 100× InceptionV3 | 0.9022 | 0.9342 | 0.8232 | 0.7358 | 0.8563 | 0.8701 | 0.7673 |
| 100× Densenet201 | 0.9183 | 0.9329 | 0.8571 | 0.7927 | 0.8836 | 0.8892 | 0.8056 |
| 100× Resnet50 | 0.9119 | 0.8670 | 0.8556 | 0.8446 | 0.8933 | 0.8783 | 0.7924 |
| 100× Efficientnetbo | 0.8526 | 0.8301 | 0.7341 | 0.6580 | 0.7989 | 0.8087 | 0.6422 |
| 100× Squeezenet | 0.9455 | 0.8630 | 0.9175 | 0.9793 | 0.9548 | 0.9216 | 0.8809 |
| 100× Shufflenet | 0.8606 | 0.7454 | 0.7873 | 0.8342 | 0.8533 | 0.8075 | 0.6865 |
| 200× Renent18 | 0.8791 | 0.9831 | 0.7607 | 0.6203 | 0.8078 | 0.8460 | 0.7178 |
| 200× InceptionResnetV2 | 0.8692 | 0.9030 | 0.7539 | 0.6471 | 0.8079 | 0.8313 | 0.6853 |
| 200× Inceptionv3 | 0.8957 | 0.9079 | 0.8142 | 0.7380 | 0.8522 | 0.8611 | 0.7504 |
| 200× Densenet201 | 0.8891 | 0.9000 | 0.8012 | 0.7219 | 0.8430 | 0.8531 | 0.7340 |
| 200× Resnet50 | 0.8642 | 0.7561 | 0.7908 | 0.8289 | 0.8545 | 0.8128 | 0.6922 |
| 200× Efficientnetbo | 0.8245 | 0.7396 | 0.7022 | 0.6684 | 0.7815 | 0.7704 | 0.5798 |
| 200× Squeezenet | 0.9205 | 0.9017 | 0.8667 | 0.8342 | 0.8967 | 0.8907 | 0.8114 |
| 200× Shufflenet | 0.8526 | 0.8451 | 0.7295 | 0.6417 | 0.7945 | 0.8099 | 0.6421 |
| 400× Renent18 | 0.8681 | 0.9127 | 0.7616 | 0.6534 | 0.8118 | 0.8273 | 0.6918 |
| 400× InceptionResnetV2 | 0.8168 | 0.7262 | 0.7093 | 0.6932 | 0.7844 | 0.7545 | 0.5760 |
| 400× InceptionV3 | 0.7253 | 0.5422 | 0.6901 | 0.9489 | 0.7839 | 0.5988 | 0.5351 |
| 400× Densenet201 | 0.8626 | 0.8686 | 0.7604 | 0.6761 | 0.8137 | 0.8183 | 0.6765 |
| 400× Resnet50 | 0.8846 | 0.9185 | 0.7974 | 0.7045 | 0.8374 | 0.8460 | 0.7311 |
| 400× Efficientnetbo | 0.8278 | 0.6971 | 0.7552 | 0.8239 | 0.8268 | 0.7585 | 0.6290 |
| 400× Squeezenet | 0.9231 | 0.9524 | 0.7947 | 0.6818 | 0.8328 | 0.8499 | 0.7383 |
| 400× Shufflenet | 0.8736 | 0.9350 | 0.7692 | 0.6534 | 0.8159 | 0.8346 | 0.7068 |
| Model | Accuracy | Precision | F1-Score | Recall | AUC | Kappa | MCC |
|---|---|---|---|---|---|---|---|
| 40× Resnet18 | 0.9064 | 0.8619 | 0.8478 | 0.8342 | 0.8867 | 0.8706 | 0.7804 |
| 40× InceptionResnetV2 | 0.9214 | 0.8723 | 0.8746 | 0.8770 | 0.9093 | 0.8899 | 0.8174 |
| 40× InceptionV3 | 0.9348 | 0.9353 | 0.8908 | 0.8503 | 0.9118 | 0.9095 | 0.8464 |
| 40× Densenet201 | 0.9615 | 0.9409 | 0.9383 | 0.9358 | 0.9545 | 0.9451 | 0.9104 |
| 40× Resnet50 | 0.9548 | 0.9546 | 0.9257 | 0.8984 | 0.9395 | 0.9364 | 0.8941 |
| 40× Efficientnetbo | 0.9448 | 0.9326 | 0.9096 | 0.8877 | 0.9293 | 0.9225 | 0.8705 |
| 40× Squeezenet | 0.8395 | 0.7301 | 0.7496 | 0.7701 | 0.8205 | 0.7817 | 0.6324 |
| 40× Shufflenet | 0.8746 | 0.8146 | 0.7945 | 0.7754 | 0.8476 | 0.8297 | 0.7048 |
| 100× Renent18 | 0.9199 | 0.8743 | 0.8698 | 0.8653 | 0.9048 | 0.8887 | 0.8119 |
| 100× InceptionResnetV2 | 0.9006 | 0.8466 | 0.8377 | 0.8290 | 0.8809 | 0.8634 | 0.7662 |
| 100× InceptionV3 | 0.9022 | 0.8743 | 0.8338 | 0.7969 | 0.8729 | 0.8671 | 0.7663 |
| 100× Densenet201 | 0.9151 | 0.8763 | 0.8602 | 0.8446 | 0.8956 | 0.8828 | 0.7995 |
| 100× Resnet50 | 0.9391 | 0.8894 | 0.9031 | 0.9171 | 0.9330 | 0.9140 | 0.8589 |
| 100× Efficientnetb0 | 0.9295 | 0.8821 | 0.8866 | 0.8912 | 0.9189 | 0.9012 | 0.8355 |
| 100× Squeezenet | 0.8542 | 0.7713 | 0.7612 | 0.7513 | 0.8258 | 0.8041 | 0.6563 |
| 100× Shufflenet | 0.9022 | 0.8511 | 0.8399 | 0.8290 | 0.8820 | 0.8657 | 0.7697 |
| 200× Renent18 | 0.8940 | 0.8090 | 0.8342 | 0.8610 | 0.8849 | 0.8527 | 0.7572 |
| 200× InceptionResnetV2 | 0.9421 | 0.9000 | 0.9072 | 0.9144 | 0.9344 | 0.9182 | 0.8651 |
| 200× InceptionV3 | 0.8990 | 0.8663 | 0.8301 | 0.7968 | 0.8708 | 0.8627 | 0.7598 |
| 200× Densenet201 | 0.9400 | 0.8989 | 0.8767 | 0.8556 | 0.9062 | 0.8968 | 0.8239 |
| 200× Resnet50 | 0.9338 | 0.9016 | 0.8919 | 0.8824 | 0.9196 | 0.9075 | 0.8443 |
| 200× Efficientnetbo | 0.9437 | 0.9048 | 0.9096 | 0.9144 | 0.9356 | 0.9206 | 0.8687 |
| 200× Squeezenet | 0.9056 | 0.8385 | 0.8496 | 0.8610 | 0.8933 | 0.8689 | 0.7810 |
| 200× Shufflenet | 0.9040 | 0.8817 | 0.8371 | 0.7968 | 0.8744 | 0.8695 | 0.7712 |
| 400× Renent18 | 0.8516 | 0.7811 | 0.7652 | 0.7500 | 0.8250 | 0.7977 | 0.6571 |
| 400× InceptionResnetV2 | 0.8663 | 0.8160 | 0.7847 | 0.7557 | 0.8373 | 0.8176 | 0.6890 |
| 400× InceptionV3 | 0.9011 | 0.8466 | 0.8466 | 0.8466 | 0.8868 | 0.8609 | 0.7736 |
| 400× Densenet201 | 0.8900 | 0.8650 | 0.8319 | 0.8011 | 0.8708 | 0.8555 | 0.7575 |
| 400× Resnet50 | 0.9176 | 0.8659 | 0.8732 | 0.8807 | 0.9079 | 0.8828 | 0.8123 |
| 400× Efficientnetbo | 0.9176 | 0.8503 | 0.9034 | 0.8760 | 0.9139 | 0.8819 | 0.8152 |
| 400× Squeezenet | 0.8626 | 0.7919 | 0.7851 | 0.7784 | 0.8406 | 0.8109 | 0.6842 |
| 400× Shufflenet | 0.8828 | 0.8111 | 0.8202 | 0.8295 | 0.8688 | 0.8359 | 0.7334 |
| Model | Accuracy | Precision | F1-Score | Recall | AUC | Kappa | MCC |
|---|---|---|---|---|---|---|---|
| Renent18 | 0.8896 | 0.9382 | 0.7975 | 0.6935 | 0.8363 | 0.8548 | 0.7396 |
| InceptionResnetV2 | 0.8609 | 0.8405 | 0.7559 | 0.6868 | 0.8136 | 0.8169 | 0.6666 |
| InceptionV3 | 0.8845 | 0.9211 | 0.7896 | 0.6909 | 0.8319 | 0.8482 | 0.7262 |
| Densenet201 | 0.9166 | 0.9535 | 0.8529 | 0.7715 | 0.8772 | 0.8872 | 0.8043 |
| Resnet50 | 0.9048 | 0.8960 | 0.8383 | 0.7876 | 0.8729 | 0.8704 | 0.7745 |
| Efficientnetb0 | 0.8281 | 0.8590 | 0.6634 | 0.5403 | 0.7499 | 0.7857 | 0.5827 |
| Squeezenet | 0.9460 | 0.9195 | 0.9201 | 0.9207 | 0.9419 | 0.9287 | 0.8835 |
| Shufflenet | 0.8643 | 0.8879 | 0.7500 | 0.6492 | 0.8059 | 0.8241 | 0.6752 |
| Model | Accuracy | Sensitivity | AUC | Fscore | TPR | FPR | MCC | Kappa | Prec | Spec |
|---|---|---|---|---|---|---|---|---|---|---|
| Densenet201 | 0.9815 | 0.9677 | 0.9777 | 0.9704 | 0.9677 | 0.0123 | 0.9569 | 0.8458 | 0.9730 | 0.9877 |
| Resnet50 | 0.9899 | 0.9892 | 0.9897 | 0.9840 | 0.9892 | 0.0098 | 0.9766 | 0.8410 | 0.9787 | 0.9902 |
| Efficientnetb0 | 0.9836 | 0.9718 | 0.9804 | 0.9737 | 0.9718 | 0.0110 | 0.9618 | 0.8447 | 0.9757 | 0.9890 |
| InceptionResnetV2 | 0.9823 | 0.9866 | 0.9835 | 0.9722 | 0.9866 | 0.0196 | 0.9594 | 0.8439 | 0.9582 | 0.9804 |
| InceptionV3 | 0.9836 | 0.9798 | 0.9826 | 0.9739 | 0.9798 | 0.0147 | 0.9620 | 0.8440 | 0.9681 | 0.9853 |
| Renent18 | 0.9777 | 0.9798 | 0.9783 | 0.9649 | 0.9798 | 0.0233 | 0.9488 | 0.8461 | 0.9505 | 0.9767 |
| Shufflenet | 0.9794 | 0.9691 | 0.9766 | 0.9671 | 0.9691 | 0.0160 | 0.9521 | 0.8464 | 0.9652 | 0.9840 |
| Squeezenet | 0.9659 | 0.9664 | 0.9660 | 0.9467 | 0.9664 | 0.0344 | 0.9220 | 0.8512 | 0.9277 | 0.9656 |
| ML Classifier | Acc. | Sens. | Spec. | AUC | F1 | TPR | FPR | MCC | Kappa | Prec. |
|---|---|---|---|---|---|---|---|---|---|---|
| Binary Decision Classifier | 0.9660 | 0.9449 | 0.9761 | 0.9605 | 0.9462 | 0.9449 | 0.0239 | 0.9216 | 0.8529 | 0.9474 |
| Linear Discriminant Analysis (LDA) | 0.9900 | 0.9892 | 0.9902 | 0.9897 | 0.9840 | 0.9892 | 0.0098 | 0.9766 | 0.8410 | 0.9787 |
| Generalized Additive Model | 0.9660 | 0.9489 | 0.9742 | 0.9616 | 0.9464 | 0.9489 | 0.0258 | 0.9218 | 0.8526 | 0.9439 |
| Gradient Boosted Machines (GBM) | 0.9870 | 0.9812 | 0.9890 | 0.9851 | 0.9786 | 0.9812 | 0.0110 | 0.9687 | 0.8429 | 0.9759 |
| K-Nearest Neighbor (KNN) | 0.9800 | 0.9960 | 0.9724 | 0.9842 | 0.9686 | 0.9960 | 0.0276 | 0.9545 | 0.8440 | 0.9427 |
| Naive Bayes (NB) | 0.9730 | 0.9919 | 0.9638 | 0.9779 | 0.9578 | 0.9919 | 0.0362 | 0.9388 | 0.8468 | 0.9260 |
| Support Vector Machines (SVM) | 0.9992 | 0.9987 | 0.9797 | 0.9990 | 0.9987 | 0.9987 | 0.0006 | 0.9980 | 0.8368 | 0.9987 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clement, D.; Agu, E.; Obayemi, J.; Adeshina, S.; Soboyejo, W. Breast Cancer Tumor Classification Using a Bag of Deep Multi-Resolution Convolutional Features. Informatics 2022, 9, 91. https://doi.org/10.3390/informatics9040091
Clement D, Agu E, Obayemi J, Adeshina S, Soboyejo W. Breast Cancer Tumor Classification Using a Bag of Deep Multi-Resolution Convolutional Features. Informatics. 2022; 9(4):91. https://doi.org/10.3390/informatics9040091
Chicago/Turabian StyleClement, David, Emmanuel Agu, John Obayemi, Steve Adeshina, and Wole Soboyejo. 2022. "Breast Cancer Tumor Classification Using a Bag of Deep Multi-Resolution Convolutional Features" Informatics 9, no. 4: 91. https://doi.org/10.3390/informatics9040091
APA StyleClement, D., Agu, E., Obayemi, J., Adeshina, S., & Soboyejo, W. (2022). Breast Cancer Tumor Classification Using a Bag of Deep Multi-Resolution Convolutional Features. Informatics, 9(4), 91. https://doi.org/10.3390/informatics9040091


_Bryant.png)




