Machine Learning Applied to Tree Crop Yield Prediction Using Field Data and Satellite Imagery: A Case Study in a Citrus Orchard
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Region
2.2. Data
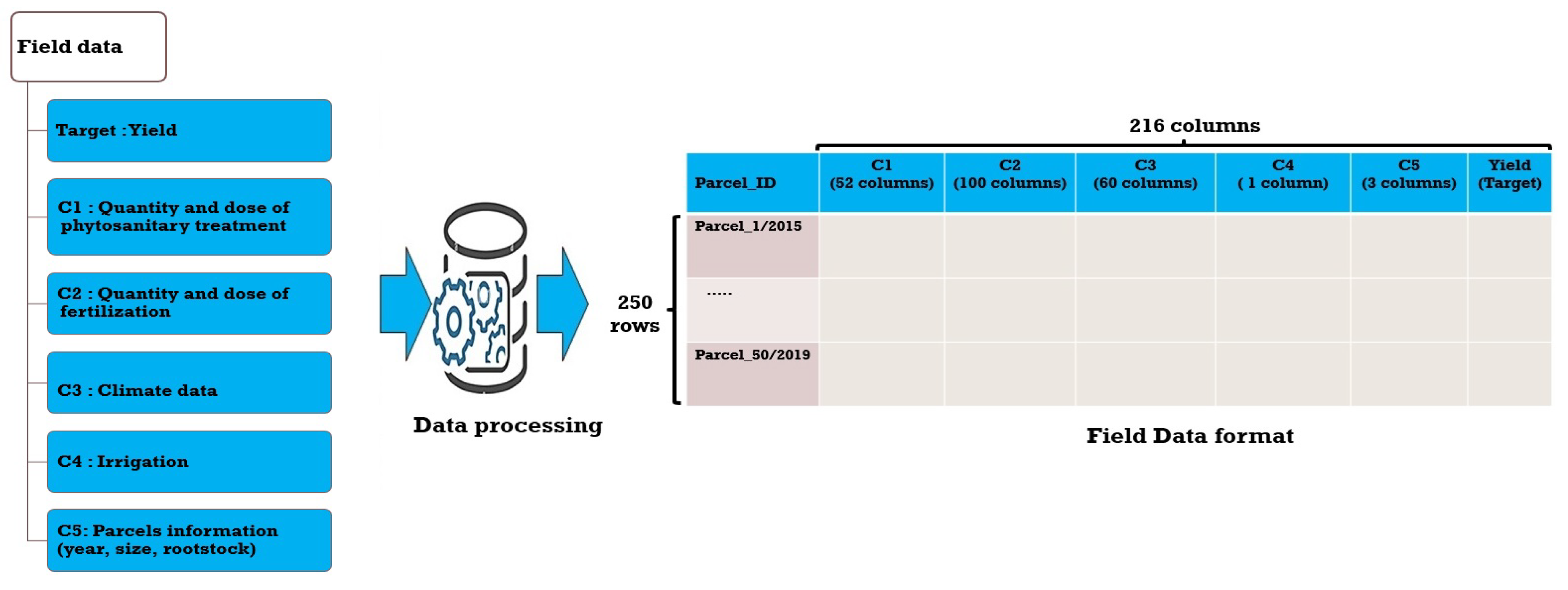
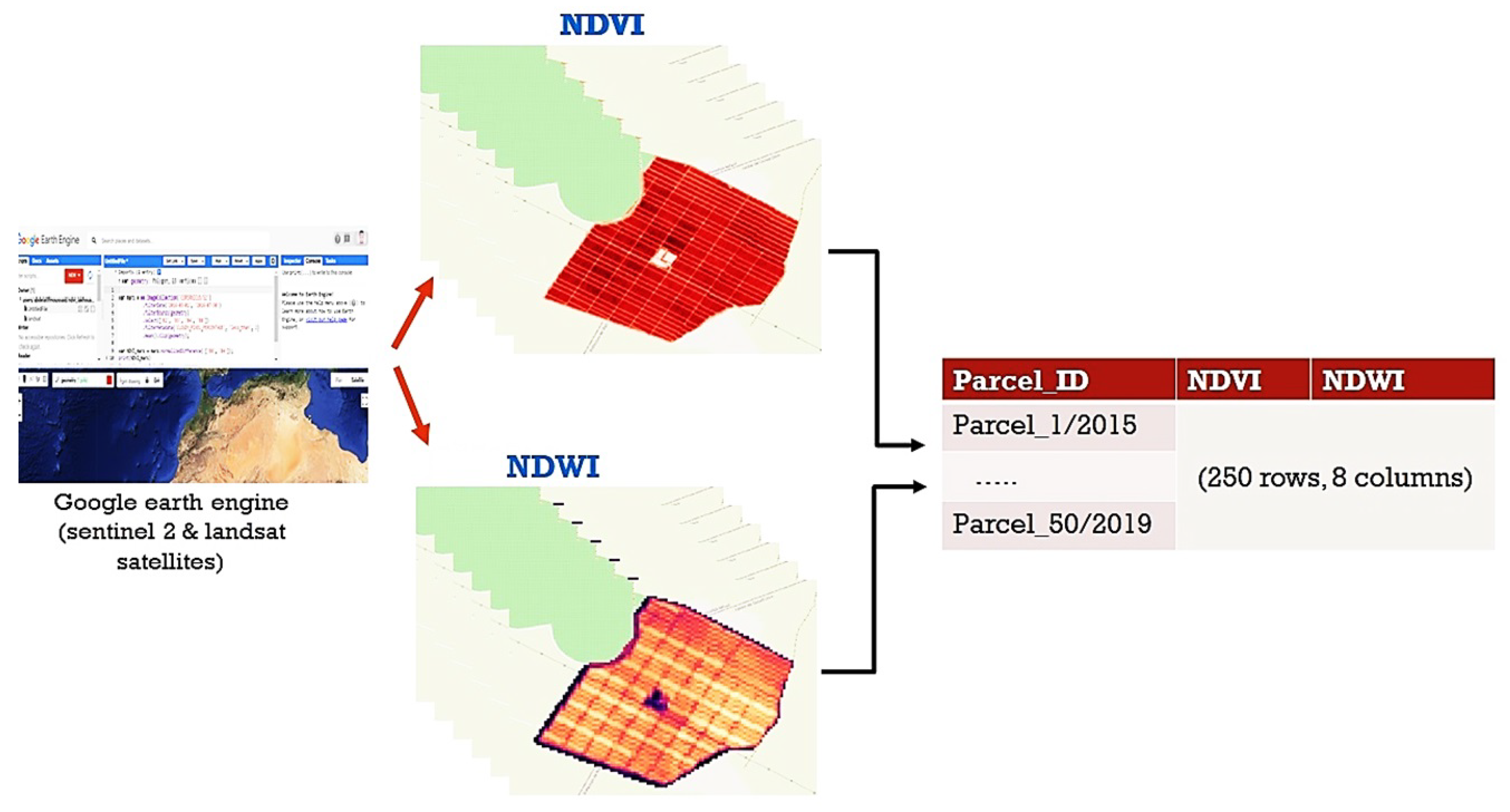
2.2.1. Data Acquisition
2.2.2. Data Processing
2.2.3. Data Exploration
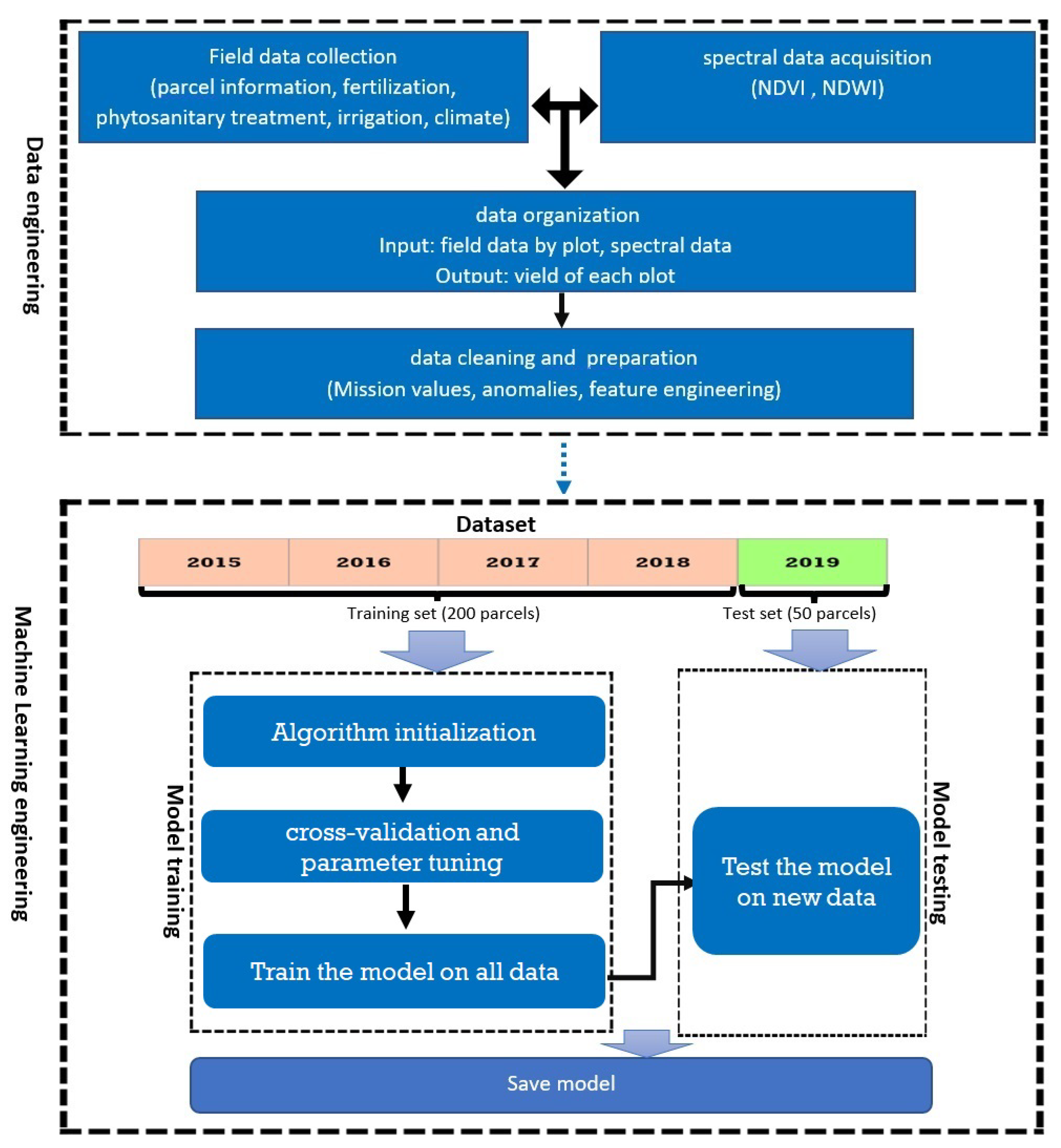
2.3. Our Approach
3. Results and Discussion
3.1. Cross-Validation and Model Selection
| Algorithm 1 Orthogonal Matching Pursuit |
Input: Initialization: Repeat ; match step: ; identify step: ; update step: ; ; Until stop criterion satisfied; output: ; |
3.2. Discussion of Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Patrício, D.I.; Rieder, R. Computer vision and artificial intelligence in precision agriculture for grain crops: A systematic review. Comput. Electron. Agric. 2018, 153, 69–81. [Google Scholar] [CrossRef]
- Linaza, M.T.; Posada, J.; Bund, J.; Eisert, P.; Quartulli, M.; Döllner, J.; Pagani, A.; G Olaizola, I.; Barriguinha, A.; Moysiadis, T.; et al. Data-driven artificial intelligence applications for sustainable precision agriculture. Agronomy 2021, 11, 1227. [Google Scholar] [CrossRef]
- Sharma, A.; Jain, A.; Gupta, P.; Chowdary, V. Machine learning applications for precision agriculture: A comprehensive review. IEEE Access 2020, 9, 4843–4873. [Google Scholar] [CrossRef]
- Lu, Y.; Young, S. A survey of public datasets for computer vision tasks in precision agriculture. Comput. Electron. Agric. 2020, 178, 105760. [Google Scholar] [CrossRef]
- Rashid, M.; Bari, B.S.; Yusup, Y.; Kamaruddin, M.A.; Khan, N. A comprehensive review of crop yield prediction using machine learning approaches with special emphasis on palm oil yield prediction. IEEE Access 2021, 9, 63406–63439. [Google Scholar] [CrossRef]
- Michler, J.D.; Tjernström, E.; Verkaart, S.; Mausch, K. Money matters: The role of yields and profits in agricultural technology adoption. Am. J. Agric. Econ. 2019, 101, 710–731. [Google Scholar] [CrossRef]
- Anderson, N.T.; Walsh, K.B.; Wulfsohn, D. Technologies for forecasting tree fruit load and harvest timing—From ground, sky and time. Agronomy 2021, 11, 1409. [Google Scholar] [CrossRef]
- Yan, B.; Fan, P.; Lei, X.; Liu, Z.; Yang, F. A real-time apple targets detection method for picking robot based on improved YOLOv5. Remote Sens. 2021, 13, 1619. [Google Scholar] [CrossRef]
- Parico, A.I.B.; Ahamed, T. Real time pear fruit detection and counting using YOLOv4 models and deep SORT. Sensors 2021, 21, 4803. [Google Scholar] [CrossRef]
- Wan, S.; Goudos, S. Faster R-CNN for multi-class fruit detection using a robotic vision system. Comput. Netw. 2020, 168, 107036. [Google Scholar] [CrossRef]
- Nawaz, R.; Abbasi, N.A.; Hafiz, I.A.; Khalid, A. Impact of climate variables on growth and development of Kinnow fruit (Citrus nobilis Lour x Citrus deliciosa Tenora) grown at different ecological zones under climate change scenario. Sci. Hortic. 2020, 260, 108868. [Google Scholar] [CrossRef]
- Wan, L.J.; Tian, Y.; He, M.; Zheng, Y.Q.; Lyu, Q.; Xie, R.J.; Ma, Y.Y.; Deng, L.; Yi, S.L. Effects of Chemical Fertilizer Combined with Organic Fertilizer Application on Soil Properties, Citrus Growth Physiology, and Yield. Agriculture 2021, 11, 1207. [Google Scholar] [CrossRef]
- Dutta, S.K.; Gurung, G.; Yadav, A.; Laha, R.; Mishra, V.K. Factors associated with citrus fruit abscission and management strategies developed so far: A review. N. Z. J. Crop Hortic. Sci. 2022, 1–22. [Google Scholar] [CrossRef]
- Vincent, C.; Morillon, R.; Arbona, V.; Gómez-Cadenas, A. Citrus in changing environments. In The Genus Citrus; Elsevier: Amsterdam, The Netherlands, 2020; pp. 271–289. [Google Scholar]
- Moussaid, A.; El Fkihi, S.; Zennayi, Y. Citrus Orchards Monitoring based on Remote Sensing and Artificial Intelligence Techniques: A Review of the Literature. In Proceedings of the 2nd International Conference on Advanced Technologies for Humanity—ICATH, Rabat, Morocco, 20–21 November 2020; IEEE: New York, NY, USA, 2020; pp. 172–178. [Google Scholar]
- Mngadi, M.; Odindi, J.; Mutanga, O. The utility of Sentinel-2 spectral data in quantifying above-ground carbon stock in an urban reforested landscape. Remote Sens. 2021, 13, 4281. [Google Scholar] [CrossRef]
- Galphade, M.; More, N.; Wagh, A.; Nikam, V. Crop Yield Prediction Using Weather Data and NDVI Time Series Data. In Advances in Data Computing, Communication and Security; Springer: Berlin/Heidelberg, Germany, 2022; pp. 261–271. [Google Scholar]
- Moussaid, A.; Fkihi, S.E.; Zennayi, Y. Tree Crowns Segmentation and Classification in Overlapping Orchards Based on Satellite Images and Unsupervised Learning Algorithms. J. Imaging 2021, 7, 241. [Google Scholar] [CrossRef]
- Ye, X.; Sakai, K.; Manago, M.; Asada, S.i.; Sasao, A. Prediction of citrus yield from airborne hyperspectral imagery. Precis. Agric. 2007, 8, 111–125. [Google Scholar] [CrossRef]
- De Ollas, C.; Morillón, R.; Fotopoulos, V.; Puértolas, J.; Ollitrault, P.; Gómez-Cadenas, A.; Arbona, V. Facing climate change: Biotechnology of iconic Mediterranean woody crops. Front. Plant Sci. 2019, 10, 427. [Google Scholar] [CrossRef]
- Vogel, E.; Donat, M.G.; Alexander, L.V.; Meinshausen, M.; Ray, D.K.; Karoly, D.; Meinshausen, N.; Frieler, K. The effects of climate extremes on global agricultural yields. Environ. Res. Lett. 2019, 14, 054010. [Google Scholar] [CrossRef]
- Nagaz, K.; El Mokh, F.; Ben Hassen, N.; Masmoudi, M.; Ben Mechlia, N.; Baba Sy, M.; Belkheiri, O.; Ghiglieri, G. Impact of deficit irrigation on yield and fruit quality of orange Trees (Citrus sinensis, l. Osbeck, cv. Meski maltaise) in southern Tunisia. Irrig. Drain. 2020, 69, 186–193. [Google Scholar] [CrossRef]
- Cai, A.; Xu, M.; Wang, B.; Zhang, W.; Liang, G.; Hou, E.; Luo, Y. Manure acts as a better fertilizer for increasing crop yields than synthetic fertilizer does by improving soil fertility. Soil Tillage Res. 2019, 189, 168–175. [Google Scholar] [CrossRef]
- Morugán-Coronado, A.; Linares, C.; Gómez-López, M.D.; Faz, Á.; Zornoza, R. The impact of intercropping, tillage and fertilizer type on soil and crop yield in fruit orchards under Mediterranean conditions: A meta-analysis of field studies. Agric. Syst. 2020, 178, 102736. [Google Scholar] [CrossRef]
- Buczko, U.; van Laak, M.; Eichler-Löbermann, B.; Gans, W.; Merbach, I.; Panten, K.; Peiter, E.; Reitz, T.; Spiegel, H.; von Tucher, S. Re-evaluation of the yield response to phosphorus fertilization based on meta-analyses of long-term field experiments. Ambio 2018, 47, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, R.; Xia, S.; Wang, L.; Liu, C.; Zhang, R.; Fan, Z.; Chen, F.; Liu, Y. Interactions between N, P and K fertilizers affect the environment and the yield and quality of satsumas. Glob. Ecol. Conserv. 2019, 19, e00663. [Google Scholar] [CrossRef]
- Coble, K.H.; Mishra, A.K.; Ferrell, S.; Griffin, T. Big data in agriculture: A challenge for the future. Appl. Econ. Perspect. Policy 2018, 40, 79–96. [Google Scholar] [CrossRef]
- Cravero, A.; Sepúlveda, S. Use and adaptations of machine learning in big data—Applications in real cases in agriculture. Electronics 2021, 10, 552. [Google Scholar] [CrossRef]
- Ihbach, F.Z.; Kchikach, A.; Jaffal, M.; El Azzab, D.; Chalikakis, K.; Mazzili, N.; Guerin, R.; Jourani, E.S. Study of an Aquifer in a Semi-arid Area Using MRS, FDEM, TDEM and ERT Methods (Youssoufia and Khouribga, Morocco). In Proceedings of the Conference of the Arabian Journal of Geosciences, Hammamet, Tunisia, 12–15 November 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 73–76. [Google Scholar]
- Roh, Y.; Heo, G.; Whang, S.E. A survey on data collection for machine learning: A big data-ai integration perspective. IEEE Trans. Knowl. Data Eng. 2019, 33, 1328–1347. [Google Scholar] [CrossRef]
- Segarra, J.; Buchaillot, M.L.; Araus, J.L.; Kefauver, S.C. Remote sensing for precision agriculture: Sentinel-2 improved features and applications. Agronomy 2020, 10, 641. [Google Scholar] [CrossRef]
- Leslie, C.R.; Servina, L.O.; Miller, H.M. Landsat and Agriculture: Case Studies on the Uses and Benefits of Landsat Imagery in Agricultural Monitoring and Production; US Department of the Interior, US Geological Survey: Reston, VA, USA, 2017.
- Shanmugapriya, P.; Rathika, S.; Ramesh, T.; Janaki, P. Applications of remote sensing in agriculture-A Review. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2270–2283. [Google Scholar] [CrossRef]
- Giovos, R.; Tassopoulos, D.; Kalivas, D.; Lougkos, N.; Priovolou, A. Remote sensing vegetation indices in viticulture: A critical review. Agriculture 2021, 11, 457. [Google Scholar] [CrossRef]
- Sishodia, R.P.; Ray, R.L.; Singh, S.K. Applications of remote sensing in precision agriculture: A review. Remote Sens. 2020, 12, 3136. [Google Scholar] [CrossRef]
- Rickman, J.; Balasubramanian, G.; Marvel, C.; Chan, H.; Burton, M.T. Machine learning strategies for high-entropy alloys. J. Appl. Phys. 2020, 128, 221101. [Google Scholar] [CrossRef]
- Khfif, K.; Mokrini, F.; Sbaghi, M. Population monitoring of males steriles of Mediterranean fruit fly (Ceratitis capitata Wiedemann, 1824) in citrus orchards of the Moulouya region. Afr. Mediterr. Agric. J. 2022, 135, 123–135. [Google Scholar]
- Otero, A.; Goni, C.; Jifon, J.; Syvertsen, J. High temperature effects on citrus orange leaf gas exchange, flowering, fruit quality and yield. In Proceedings of the IX International Symposium on Integrating Canopy, Rootstock and Environmental Physiology in Orchard Systems 903, Geneva, NY, USA, 4–8 August 2008; pp. 1069–1075. [Google Scholar]
- Emmert-Streib, F.; Yli-Harja, O.; Dehmer, M. Explainable artificial intelligence and machine learning: A reality rooted perspective. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2020, 10, e1368. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A review of machine learning interpretability methods. Entropy 2020, 23, 18. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Mou, S.; Lian, J.; Ren, W. Solving a system of linear equations: From centralized to distributed algorithms. Annu. Rev. Control. 2019, 47, 306–322. [Google Scholar] [CrossRef]
- Jiao, S.; Song, J.; Liu, B. A Review of Decision Tree Classification Algorithms for Continuous Variables. In Proceedings of the Journal of Physics: Conference Series, The 2020 second International Conference on Artificial Intelligence Technologies and Application (ICAITA), Dalian, China, 21–23 August 2020; Volume 1651, p. 012083. [Google Scholar]
- Huettmann, F. Boosting, Bagging and Ensembles in the Real World: An Overview, some Explanations and a Practical Synthesis for Holistic Global Wildlife Conservation Applications Based on Machine Learning with Decision Trees. In Machine Learning for Ecology and Sustainable Natural Resource Management; Springer: Berlin/Heidelberg, Germany, 2018; pp. 63–83. [Google Scholar]
- Azmi, S.S.; Baliga, S. An Overview of Boosting Decision Tree Algorithms utilizing AdaBoost and XGBoost Boosting strategies. Int. Res. J. Eng. Technol. 2020, 7, 6867–6870. [Google Scholar]
- Hancock, J.T.; Khoshgoftaar, T.M. CatBoost for big data: An interdisciplinary review. J. Big Data 2020, 7, 1–45. [Google Scholar] [CrossRef]
- Kadiyala, A.; Kumar, A. Applications of python to evaluate the performance of decision tree-based boosting algorithms. Environ. Prog. Sustain. Energy 2018, 37, 618–623. [Google Scholar] [CrossRef]
- Siedhoff, N.E.; Schwaneberg, U.; Davari, M.D. Machine learning-assisted enzyme engineering. Methods Enzymol. 2020, 643, 281–315. [Google Scholar]
- Ding, J.; Chen, L.; Gu, Y. Perturbation analysis of orthogonal matching pursuit. IEEE Trans. Signal Process. 2013, 61, 398–410. [Google Scholar] [CrossRef]
- Khosravy, M.; Gupta, N.; Patel, N.; Duque, C.A. Recovery in compressive sensing: A review. Compressive Sens. Healthc. 2020, 25–42. [Google Scholar] [CrossRef]
- Koc-San, D.; Selim, S.; Aslan, N.; San, B.T. Automatic citrus tree extraction from UAV images and digital surface models using circular Hough transform. Comput. Electron. Agric. 2018, 150, 289–301. [Google Scholar] [CrossRef]
- Csillik, O.; Cherbini, J.; Johnson, R.; Lyons, A.; Kelly, M. Identification of citrus trees from unmanned aerial vehicle imagery using convolutional neural networks. Drones 2018, 2, 39. [Google Scholar] [CrossRef]
- Osco, L.P.; De Arruda, M.d.S.; Junior, J.M.; Da Silva, N.B.; Ramos, A.P.M.; Moryia, É.A.S.; Imai, N.N.; Pereira, D.R.; Creste, J.E.; Matsubara, E.T.; et al. A convolutional neural network approach for counting and geolocating citrus-trees in UAV multispectral imagery. Isprs J. Photogramm. Remote Sens. 2020, 160, 97–106. [Google Scholar] [CrossRef]






| Fertilization products | - N (nitrogen) - P(phosphorus) - K (Potassium) - NITRIC ACID - PHOSPHORIC ACID - SULFURIC ACID -ACTICAL - ACTIRAIL - ENABLE - AMMONITRATE - Aggis TE Fe6 - BERELEX (Lozenge) - BIOCURE -BORONIA POWDER - BUDMAX - CALICO Ca-Mg - CALCIUM - DEVSOL - FENGIB - FERTILOG 20 - FITOSOL - LIME BLOSSOM - FOSFITAL - MANURE - GOËMAR BM 86 E - GREENSTIM - KRISTAFEED 46N - KYLATE -Kelpak - LIQUID MAP -MAXIM - MICROQUEL - MOLYBDATE - NACAR - CALCIUM NITRATE - POTASH NITRATE - NITROPLUS - NITROPLUS 9+B GA - SEQUESTREN - SIAPTON - SOLQUEL - COPPER SULFATE - IRON SULFATE - MANGANESE SULPHATE - SOLUBLE SULPHATE OF POTASH - ZINC SULPHATE - SUPRALEX -Samfer - Stop it - TENSOTEC - UREA - UREA 46% -VIGOMAX |
| Phytosanitary treatment products | Product against Mites - Product against Whitefly - Product against Ceratitis - Product against Snails - Product against California rental - Product against aphids - growth regulator - Product against Thrips |
| Model | MAE | MSE | |
|---|---|---|---|
| ridge | Ridge Regression | 0.3225 | 0.1676 |
| catboost | CatBoost Regressor | 0.2631 | 0.1174 |
| gbr | Gradient Boosting Regressor | 0.2608 | 0.1107 |
| rf | Random Forest Regressor | 0.2677 | 0.1175 |
| en | Elastic Net | 0.2963 | 0.1435 |
| ada | AdaBoost Regressor | 0.2618 | 0.1158 |
| br | Bayesian Ridge | 0.2932 | 0.1392 |
| lasso | Lasso Regression | 0.2918 | 0.1380 |
| et | Extra Trees Regressor | 0.2767 | 0.1303 |
| lightgbm | Light Gradient Boosting Machine | 0.2829 | 0.1311 |
| xgboost | Extreme Gradient Boosting | 0.3192 | 0.1586 |
| huber | Huber Regressor | 0.3506 | 0.2661 |
| omp | Orthogonal Matching Pursuit | 0.2585 | 0.1092 |
| llar | Lasso Least Angle Regression | 0.3025 | 0.1667 |
| dt | Decision Tree Regressor | 0.3468 | 0.2114 |
| knn | K Neighbors Regressor | 0.2705 | 0.1204 |
| lr | Linear Regression | 0.6027 | 0.6159 |
| Model | Field Dat | Field + Spectral Data | |||
|---|---|---|---|---|---|
| MAE | MSE | MAE | MSE | ||
| ridge | Ridge Regression | 0.2680 | 0.1140 | 0.2525 | 0.1034 |
| catboost | CatBoost Regressor | 0.2477 | 0.1057 | 0.2477 | 0.1057 |
| gbr | Gradient Boosting Regressor | 0.2649 | 0.1281 | 0.2475 | 0.1024 |
| rf | Random Forest Regressor | 0.2689 | 0.1287 | 0.2497 | 0.1046 |
| en | Elastic Net | 0.2760 | 0.1287 | 0.2536 | 0.1049 |
| ada | AdaBoost Regressor | 0.2682 | 0.1333 | 0.2484 | 0.1085 |
| br | Bayesian Ridge | 0.2758 | 0.1299 | 0.2585 | 0.1093 |
| lasso | Lasso Regression | 0.2759 | 0.1265 | 0.2592 | 0.1099 |
| et | Extra Trees Regressor | 0.2594 | 0.1187 | 0.2452 | 0.1092 |
| lightgbm | Light Gradient Boosting Machine | 0.2593 | 0.1153 | 0.2610 | 0.1138 |
| xgboost | Extreme Gradient Boosting | 0.2926 | 0.1703 | 0.2645 | 0.1165 |
| huber | Huber Regressor | 0.3816 | 0.2752 | 0.2825 | 0.1305 |
| omp | Orthogonal Matching Pursuit | 0.2489 | 0.0843 | 0.2315 | 0.0748 |
| llar | Lasso Least Angle Regression | 0.3036 | 0.1666 | 0.2982 | 0.1665 |
| dt | Decision Tree Regressor | 0.3187 | 0.1828 | 0.3270 | 0.1678 |
| knn | K Neighbors Regressor | 0.3166 | 0.1789 | 0.3327 | 0.2036 |
| lr | Linear Regression | 0.5705 | 0.5696 | 0.4129 | 0.3327 |
| Field Data | Field + Spectral Data | |||
|---|---|---|---|---|
| Parcel_ID | Parcel Information and Climate | Parcel Information, Climate and Phytosanitary Treatment | Parcel Information, Climate, Phytosanitary Treatment and Fertilization | Parcel Information, Climate, Phytosanitary Treatment Fertilization and Spectral Data |
| 0 | 0.3789 | 0.2853 | 0.2738 | 0.2731 |
| 1 | 0.2309 | 0.1667 | 0.1564 | 0.0929 |
| 2 | 0.1739 | 0.1697 | 0.1477 | 0.1424 |
| 3 | 0.3097 | 0.2397 | 0.0847 | 0.0421 |
| 4 | 0.5509 | 0.4903 | 0.23 | 0.1822 |
| 5 | 0.3201 | 0.2697 | 0.2642 | 0.2174 |
| 6 | 0.7632 | 0.2803 | 0.2366 | 0.1774 |
| 7 | 1.259 | 0.531 | 0.3538 | 0.2465 |
| 8 | 0.8286 | 0.373 | 0.2851 | 0.2353 |
| 9 | 0.8662 | 0.6127 | 0.3483 | 0.2976 |
| 10 | 0.6043 | 0.3091 | 0.1267 | 0.0846 |
| 11 | 0.2791 | 0.2722 | 0.2698 | 0.2319 |
| 12 | 0.2436 | 0.2252 | 0.1425 | 0.0966 |
| 13 | 0.2943 | 0.0306 | 0.1619 | 0.1175 |
| 14 | 0.4927 | 0.2881 | 0.2317 | 0.1954 |
| 15 | 0.3473 | 0.2983 | 0.1745 | 0.1516 |
| 16 | 0.2703 | 0.2523 | 0.2121 | 0.1651 |
| 17 | 0.2965 | 0.2625 | 0.2411 | 0.193 |
| 18 | 0.3455 | 0.2803 | 0.0972 | 0.0937 |
| 19 | 0.3957 | 0.3198 | 0.2981 | 0.2291 |
| 20 | 0.4877 | 0.396 | 0.1861 | 0.1401 |
| 21 | 0.7791 | 0.1548 | 0.0373 | 0.0191 |
| 22 | 0.3168 | 0.2838 | 0.2621 | 0.2523 |
| 23 | 0.3878 | 0.3806 | 0.3188 | 0.2805 |
| 24 | 0.3236 | 0.2997 | 0.2981 | 0.2918 |
| 25 | 0.1691 | 0.1638 | 0.1683 | 0.1625 |
| 26 | 0.3772 | 0.2373 | 0.0867 | 0.0489 |
| 27 | 0.3646 | 0.3188 | 0.0777 | 0.0701 |
| 28 | 0.2822 | 0.2492 | 0.1351 | 0.1484 |
| 29 | 0.44 | 0.3521 | 0.328 | 0.2698 |
| 30 | 0.0893 | 0.0589 | 0.0229 | 0.0155 |
| 31 | 0.2962 | 0.2553 | 0.0319 | 0.0037 |
| 32 | 0.3527 | 0.3522 | 0.2478 | 0.2183 |
| 33 | 0.2291 | 0.2221 | 0.2203 | 0.196 |
| 34 | 1.5524 | 0.3627 | 0.2954 | 0.0204 |
| 35 | 0.2556 | 0.2474 | 0.2297 | 0.2169 |
| 36 | 0.7512 | 0.5378 | 0.2104 | 0.1904 |
| 37 | 0.3316 | 0.3034 | 0.1884 | 0.1302 |
| 38 | 0.2945 | 0.294 | 0.2239 | 0.1986 |
| 39 | 0.4909 | 0.3789 | 0.1237 | 0.1027 |
| 40 | 0.6537 | 0.4716 | 0.2405 | 0.2244 |
| 41 | 0.1862 | 0.1775 | 0.1761 | 0.1563 |
| 42 | 0.4248 | 0.3129 | 0.279 | 0.2025 |
| 43 | 0.9766 | 0.8478 | 0.3093 | 0.2596 |
| 44 | 0.2824 | 0.0474 | 0.0357 | 0.0353 |
| 45 | 0.3354 | 0.318 | 0.3145 | 0.2666 |
| 46 | 0.208 | 0.1935 | 0.1888 | 0.1326 |
| 47 | 0.3784 | 0.3301 | 0.247 | 0.1955 |
| 48 | 0.3636 | 0.3032 | 0.1225 | 0.0637 |
| 49 | 0.3317 | 0.2708 | 0.219 | 0.168 |
| Average | 0.4392 | 0.3015 | 0.2032 | 0.1629 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moussaid, A.; El Fkihi, S.; Zennayi, Y.; Lahlou, O.; Kassou, I.; Bourzeix, F.; El Mansouri, L.; Imani, Y. Machine Learning Applied to Tree Crop Yield Prediction Using Field Data and Satellite Imagery: A Case Study in a Citrus Orchard. Informatics 2022, 9, 80. https://doi.org/10.3390/informatics9040080
Moussaid A, El Fkihi S, Zennayi Y, Lahlou O, Kassou I, Bourzeix F, El Mansouri L, Imani Y. Machine Learning Applied to Tree Crop Yield Prediction Using Field Data and Satellite Imagery: A Case Study in a Citrus Orchard. Informatics. 2022; 9(4):80. https://doi.org/10.3390/informatics9040080
Chicago/Turabian StyleMoussaid, Abdellatif, Sanaa El Fkihi, Yahya Zennayi, Ouiam Lahlou, Ismail Kassou, François Bourzeix, Loubna El Mansouri, and Yasmina Imani. 2022. "Machine Learning Applied to Tree Crop Yield Prediction Using Field Data and Satellite Imagery: A Case Study in a Citrus Orchard" Informatics 9, no. 4: 80. https://doi.org/10.3390/informatics9040080
APA StyleMoussaid, A., El Fkihi, S., Zennayi, Y., Lahlou, O., Kassou, I., Bourzeix, F., El Mansouri, L., & Imani, Y. (2022). Machine Learning Applied to Tree Crop Yield Prediction Using Field Data and Satellite Imagery: A Case Study in a Citrus Orchard. Informatics, 9(4), 80. https://doi.org/10.3390/informatics9040080







