Bayesian Integrative Modeling of Genome-Scale Metabolic and Regulatory Networks
Abstract
1. Introduction
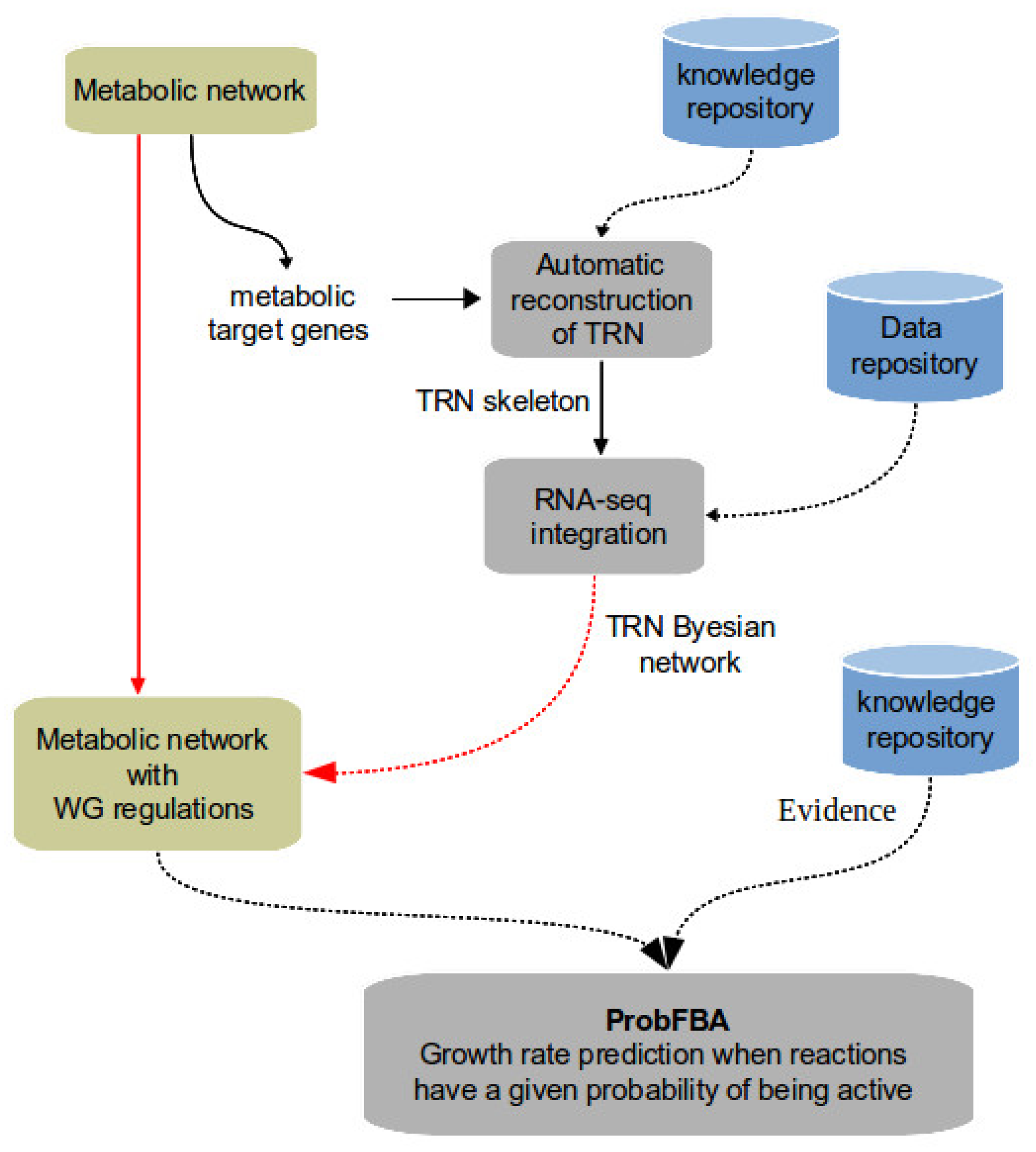
2. Methods
2.1. Constraint-Based Modeling of Metabolic Networks
2.2. Bayesian Modeling of Transcriptional Regulatory Networks
3. Results
3.1. Reconstruction of Regulatory Networks
| Algorithm 1: A SPARQL query on the Pathway Commons endpoint. |
| Input: a gene g identified later by ENTITY Output: List of genes that regulates g PREFIX bp: <http://www.biopax.org/release/biopax-level3.owl#> SELECT ?source_name WHERE { ?regul rdf:type bp:TemplateReactionRegulation . ?regul bp:controller ?source . ?source bp:displayName ?source_name . ?regul bp:controlled ?target . ?target bp:displayName ?target_name . FILTER (regex(?target_name, "ENTITY", "i")) } |
| Algorithm 2: Reconstruction of regulatory network using Pathway Commons SPARQL endpoint. |
| Input: Set of relevant metabolic target genes = Output: List of TF-genes interactions Initialization: GenesToBeExplored:=, Interactions:= ∅ while GenesToBeExplored do let g in GenesToBeExplored remove g from GenesToBeExplored TF=GetTFcontrollers(g) /* perform the above SPARQL query */ for all r in TF do add to Interactions if r is not in GenesToBeExplored add r to GenesToBeExplored end if end for end while |
3.2. Probabilistic Transcriptional Regulatory Network
4. Case Study: Hepatocellular Carcinoma (HCC): Results and Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rigden, D.J.; Fernández-Suárez, X.M.; Galperin, M.Y. The 2016 database issue of Nucleic Acids Research and an updated molecular biology database collection. Nucl. Acids Res. 2016, 44, 1–6. [Google Scholar] [CrossRef]
- Thiele, I.; Palsson, B.Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93. [Google Scholar] [CrossRef] [PubMed]
- Gama-Castro, S.; Salgado, H.; Santos-Zavaleta, A.; Ledezma-Tejeida, D.; Muniz-Rascado, L.; García-Sotelo, J.S.; Alquicira-Hernández, K.; Martínez-Flores, I.; Pannier, L.; Castro-Mondragón, J.A.; et al. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucl. Acids Res. 2015, 44, D133–D143. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, M.B.; Kundaje, A.; Hariharan, M.; Landt, S.G.; Yan, K.K.; Cheng, C.; Mu, X.J.; Khurana, E.; Rozowsky, J.; Alexander, R.; et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012, 489, 91. [Google Scholar] [CrossRef] [PubMed]
- Ihmels, J.; Levy, R.; Barkai, N. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat. Biotechnol. 2003, 22, 86. [Google Scholar] [CrossRef]
- Schlitt, T.; Brazma, A. Current approaches to gene regulatory network modelling. BMC Bioinform. 2007, 8, S9. [Google Scholar] [CrossRef]
- Davidson, E.H.; Rast, J.P.; Oliveri, P.; Ransick, A.; Calestani, C.; Yuh, C.H.; Minokawa, T.; Amore, G.; Hinman, V.; Arenas-Mena, C.; et al. A genomic regulatory network for development. Science 2002, 295, 1669–1678. [Google Scholar] [CrossRef]
- Covert, M.W.; Schilling, C.H.; Palsson, B. Regulation of gene expression in flux balance models of metabolism. J. Theor. Biol. 2001, 213, 73–88. [Google Scholar] [CrossRef]
- Shlomi, T.; Eisenberg, Y.; Sharan, R.; Ruppin, E. A genome-scale computational study of the interplay between transcriptional regulation and metabolism. Mol. Syst. Biol. 2007, 3, 101. [Google Scholar] [CrossRef]
- Covert, M.W.; Knight, E.M.; Reed, J.L.; Herrgard, M.J.; Palsson, B.O. Integrating high-throughput and computational data elucidates bacterial networks. Nature 2004, 429, 92. [Google Scholar] [CrossRef]
- Machado, D.; Herrgård, M.J. Systematic evaluation of methods for integration of transcriptomic data into constraint-based models of metabolism. PLoS Comput. Biol. 2014, 10, e1003580. [Google Scholar] [CrossRef] [PubMed]
- Folger, O.; Jerby, L.; Frezza, C.; Gottlieb, E.; Ruppin, E.; Shlomi, T. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 2011, 7, 1. [Google Scholar] [CrossRef]
- Chandrasekaran, S.; Price, N.D. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 17845–17850. [Google Scholar] [CrossRef] [PubMed]
- Colijn, C.; Brandes, A.; Zucker, J.; Lun, D.S.; Weiner, B.; Farhat, M.R.; Cheng, T.Y.; Moody, D.B.; Murray, M.; Galagan, J.E. Interpreting expression data with metabolic flux models: predicting Mycobacterium tuberculosis mycolic acid production. PLoS Comput. Biol. 2009, 5, e1000489. [Google Scholar] [CrossRef] [PubMed]
- Covert, M.W.; Xiao, N.; Chen, T.J.; Karr, J.R. Integrating metabolic, transcriptional regulatory and signal transduction models in Escherichia coli. Bioinformatics 2008, 24, 2044–2050. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.A.; Palsson, B.O. Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 2008, 4, e1000082. [Google Scholar] [CrossRef]
- Agren, R.; Bordel, S.; Mardinoglu, A.; Pornputtapong, N.; Nookaew, I.; Nielsen, J. Reconstruction of genome-Scale Active Metabolic Networks for 69 Human Cell Types and 16 Cancer Types Using INIT. PLoS Comput. Biol. 2012, 8, e1002518. [Google Scholar] [CrossRef]
- Marmiesse, L.; Peyraud, R.; Cottret, L. FlexFlux: combining metabolic flux and regulatory network analyses. BMC Syst. Biol. 2015, 9, 93. [Google Scholar] [CrossRef]
- Jensen, P.A.; Papin, J.A. Functional integration of a metabolic network model and expression data without arbitrary thresholding. Bioinformatics 2010, 27, 541–547. [Google Scholar] [CrossRef]
- Orth, J.; Thiele, I.; Palsson, B. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245. [Google Scholar] [CrossRef]
- Roy, S.; Ernst, J.; Kharchenko, P.; Kheradpour, P.; Negre, N.; Eaton, M.; Olin, J.; Bristow, C.; Ma, L.; Lin, M. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 2010, 330, 1787–1797. [Google Scholar] [PubMed]
- Edwards, J.S.; Palsson, B.O. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. USA 2000, 97, 5528–5533. [Google Scholar] [CrossRef]
- Bordbar, A.; Monk, J.M.; King, Z.A.; Palsson, B.O. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genetics 2014, 15, 107. [Google Scholar] [CrossRef] [PubMed]
- Karlebach, G.; Shamir, R. Modelling and analysis of gene regulatory networks. Nat. Rev. Mol. Cell Biol. 2008, 9, 770. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Smith, V.A.; Wang, P.P.; Hartemink, A.J.; Jarvis, E.D. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics 2004, 20, 3594–3603. [Google Scholar] [CrossRef] [PubMed]
- Vijesh, N.; Chakrabarti, S.K.; Sreekumar, J. Modeling of gene regulatory networks: A review. J. Biomed. Sci. Eng. 2013, 6, 223. [Google Scholar] [CrossRef]
- Chickering, D.M. Learning Bayesian Networks is NP-Complete. In Learning from Data—Fifth International Workshop on Artificial Intelligence and Statistics, AISTATS 1995, Key West, FL, USA, January 1995; Fisher, D., Lenz, H., Eds.; Springer: Berlin, Germany, 1995; pp. 121–130. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef]
- Han, H.; Shim, H.; Shin, D.; Shim, J.E.; Ko, Y.; Shin, J.; Kim, H.; Cho, A.; Kim, E.; Lee, T.; et al. TRRUST: A reference database of human transcriptional regulatory interactions. Sci. Rep. 2015, 5, 11432. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2017, 46, D380–D386. [Google Scholar] [CrossRef]
- Regulation and signalization graph assembly through Linked Open Data. Available online: https://hal.archives-ouvertes.fr/hal-01768420/document (accessed on 1 July 2017).
- Gatto, F.; Miess, H.; Schulze, A.; Nielsen, J. Flux balance analysis predicts essential genes in clear cell renal cell carcinoma metabolism. Sci. Rep. 2015, 5, 10738. [Google Scholar] [CrossRef] [PubMed]
- SPARQL Query Language for RDF. Available online: https://drops.dagstuhl.de/opus/volltexte/2015/4986/pdf/13.pdf (accessed on 13 November 2019).
- Cerami, E.G.; Gross, B.E.; Demir, E.; Rodchenkov, I.; Babur, Ö.; Anwar, N.; Schultz, N.; Bader, G.D.; Sander, C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011, 39, 685–690. [Google Scholar] [CrossRef]
- Haibe-Kains, B.; Olsen, C.; Djebbari, A.; Bontempi, G.; Correll, M.; Bouton, C.; Quackenbush, J. Predictionet: Inference for Predictive Networks Designed for (but not limited to) Genomic Data. Nucleic Acids Res. 2011, 40, D866–D875. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yuan, C.; Druzdzel, M.J. Importance sampling algorithms for Bayesian networks: Principles and performance. Math. Comput. Modell. 2006, 43, 1189–1207. [Google Scholar] [CrossRef]
- Ankur, A.; Abinash, P. Pgmpy: Probabilistic Graphical Models using Python. In Proceedings of the 14th Python in Science Conference, Austin, TX, USA, 6–12 July 2015; pp. 6–11. [Google Scholar]
- Covert, M.W.; Famili, I.; Palsson, B.O. Identifying constraints that govern cell behavior: A key to converting conceptual to computational models in biology? Biotechnol. Bioeng. 2003, 84, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.P.; Sabatini, D.M. Cancer cell metabolism: Warburg and beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Nwosu, Z.C.; Megger, D.A.; Hammad, S.; Sitek, B.; Roessler, S.; Ebert, M.P.; Meyer, C.; Dooley, S. Identification of the Consistently Altered Metabolic Targets in Human Hepatocellular Carcinoma. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 303–323. [Google Scholar] [CrossRef]
- Agren, R.; Mardinoglu, A.; Asplund, A.; Kampf, C.; Uhlen, M.; Nielsen, J. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol. Syst. Biol. 2014, 10, 3. [Google Scholar] [CrossRef]
- Pornputtapong, N.; Nookaew, I.; Nielsen, J. Human metabolic atlas: An online resource for human metabolism. Database 2015. [Google Scholar] [CrossRef]
- Brunk, E.; Sahoo, S.; Zielinski, D.C.; Altunkaya, A.; Drager, A.; Mih, N.; Gatto, F.; Nilsson, A.; Preciat Gonzalez, G.A.; Aurich, M.K.; et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 2018, 36, 272. [Google Scholar] [CrossRef]
- Love, M.; Anders, S.; Huber, W. Beginner’s guide to using the DESeq2 package. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar] [PubMed]
- Stefanini, M. Enzymes, isozymes, and enzyme variants in the diagnosis of cancer. A short review. Cancer 1985, 55, 1931–1936. [Google Scholar] [CrossRef]






| Approaches | Functionality | Model of GRN | Transcriptomic Data | Large-Scale Models | |||
|---|---|---|---|---|---|---|---|
| Building Models | Flux Prediction | Both | Boolean | Non Boolean | |||
| PROM [13] | X | X | X | ||||
| E-flux [14] | X | X | X | ||||
| rFBA [10] SR-FBA [9] iFBA [15] | X | X | X | ||||
| GIMME [16] | X | X | X | ||||
| INIT [17] | X | X | X | X | |||
| FlexFlux [18] | X | X | X | ||||
| MADE [19] | X | X | X | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mhamdi, H.; Bourdon, J.; Larhlimi, A.; Elloumi, M. Bayesian Integrative Modeling of Genome-Scale Metabolic and Regulatory Networks. Informatics 2020, 7, 1. https://doi.org/10.3390/informatics7010001
Mhamdi H, Bourdon J, Larhlimi A, Elloumi M. Bayesian Integrative Modeling of Genome-Scale Metabolic and Regulatory Networks. Informatics. 2020; 7(1):1. https://doi.org/10.3390/informatics7010001
Chicago/Turabian StyleMhamdi, Hanen, Jérémie Bourdon, Abdelhalim Larhlimi, and Mourad Elloumi. 2020. "Bayesian Integrative Modeling of Genome-Scale Metabolic and Regulatory Networks" Informatics 7, no. 1: 1. https://doi.org/10.3390/informatics7010001
APA StyleMhamdi, H., Bourdon, J., Larhlimi, A., & Elloumi, M. (2020). Bayesian Integrative Modeling of Genome-Scale Metabolic and Regulatory Networks. Informatics, 7(1), 1. https://doi.org/10.3390/informatics7010001






