Development and Internal Validation of an Interpretable Machine Learning Model to Predict Readmissions in a United States Healthcare System
Abstract
1. Introduction
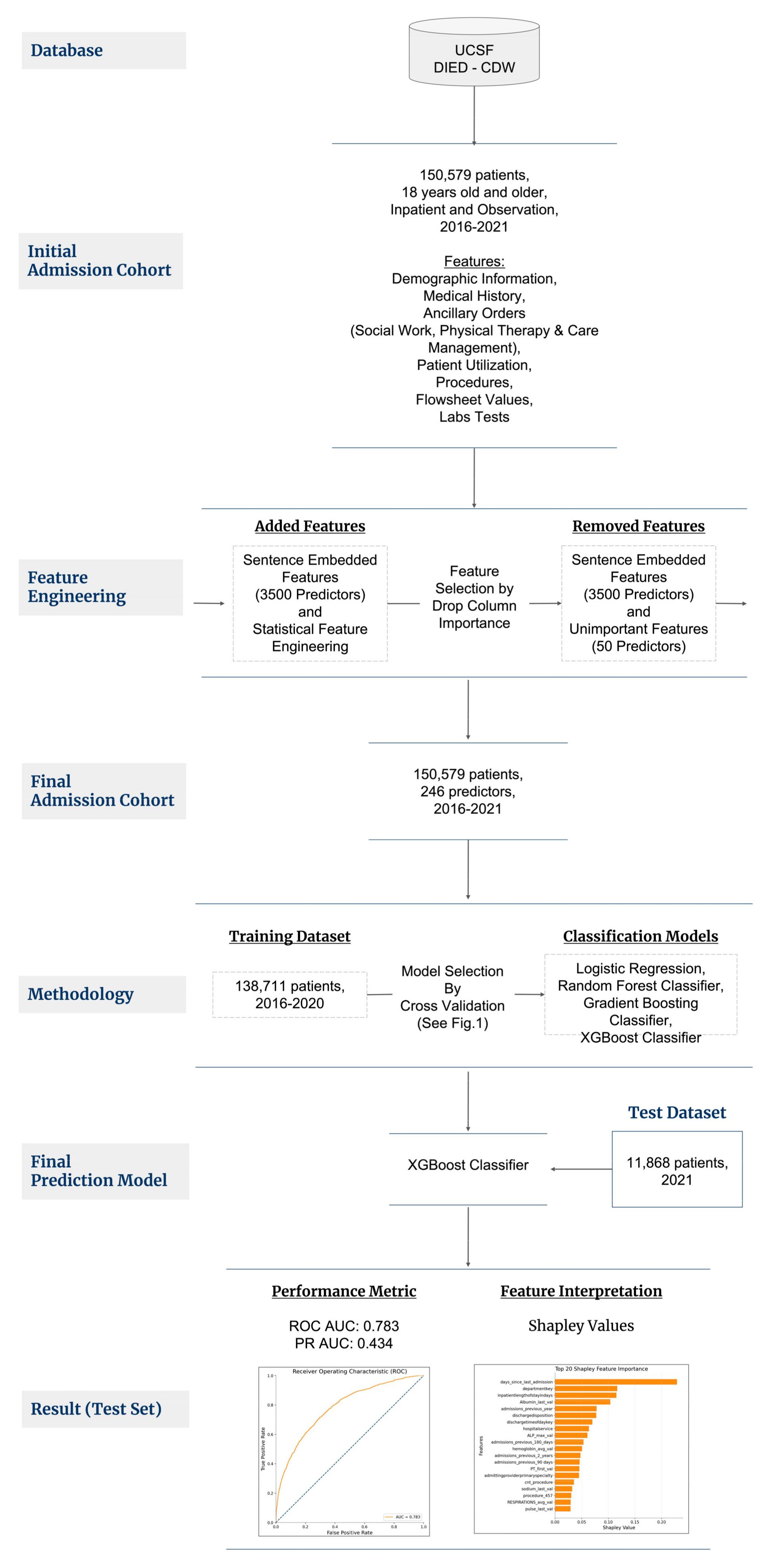
2. Materials and Methods
2.1. Patient Selection
2.2. Outcome Variable
2.3. Feature Engineering, Selection, and Imputation
2.4. Modeling Process
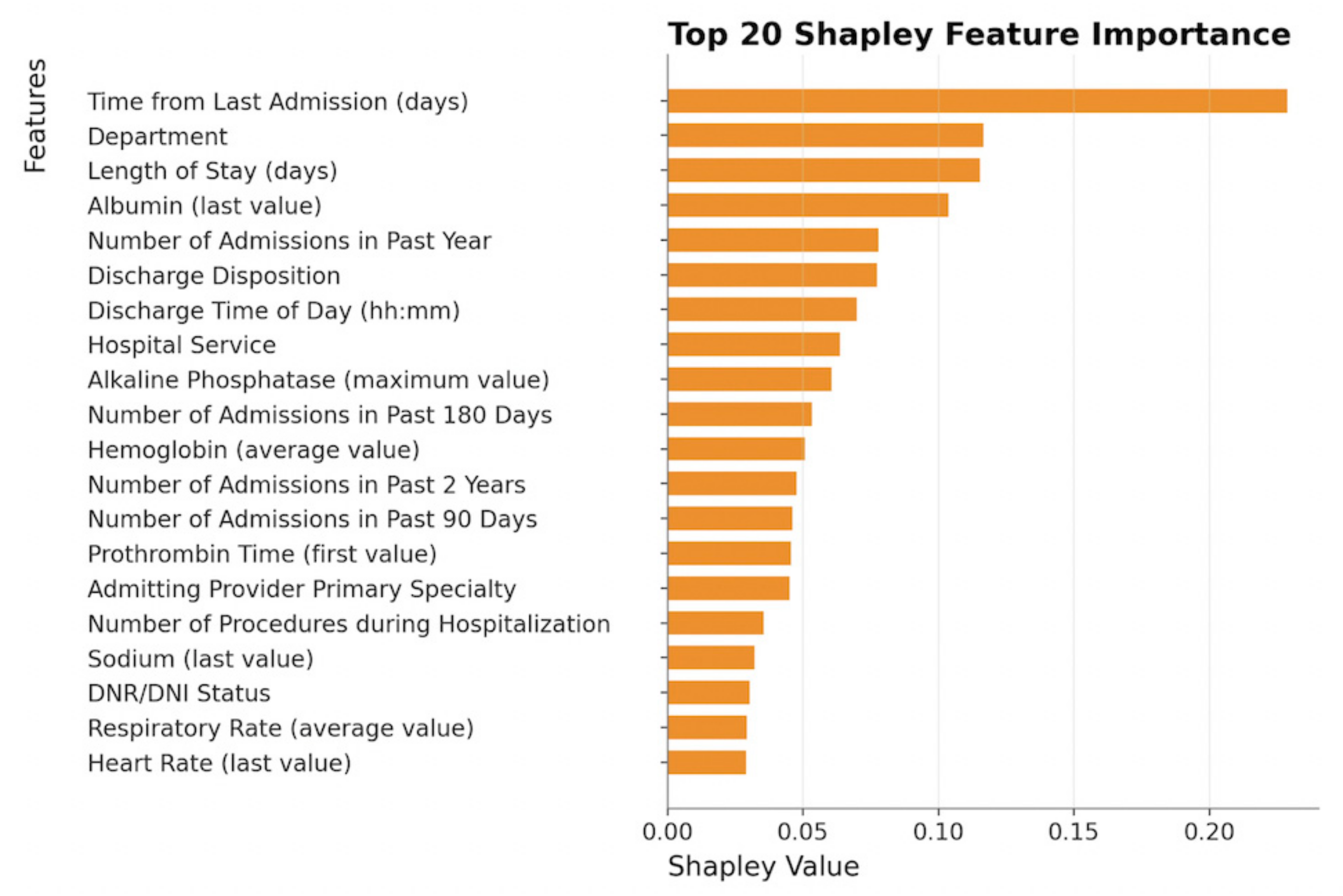
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Features | ||
|---|---|---|
| patientage | PT_first_val | venous_last_val |
| financialclass | PT_last_val | Amphetamine |
| postalcode | PaCO2_min_val | Benzo |
| sex | PaCO2_max_val | SBP_min_val |
| firstrace | PaCO2_avg_val | SBP_max_val |
| ethnicity | PaCO2_first_val | SBP_avg_val |
| maritalstatus | PaCO2_last_val | SBP_first_val |
| preferredlanguage | PaO2_min_val | SBP_last_val |
| smokingstatus | PaO2_max_val | DBP_min_val |
| Readmission | PaO2_avg_val | DBP_max_val |
| admissionsource | PaO2_first_val | DBP_avg_val |
| dischargetimeofdaykey | PaO2_last_val | DBP_first_val |
| admissiontype | PvCO2_min_val | DBP_last_val |
| inpatientlengthofstayindays | PvCO2_max_val | oxygen_amt_min_val |
| dischargedisposition | PvCO2_avg_val | oxygen_amt_max_val |
| departmentkey | PvCO2_first_val | oxygen_amt_avg_val |
| hospitalservice | PvCO2_last_val | oxygen_amt_first_val |
| admittingprovidertype | Urea_min_val | oxygen_amt_last_val |
| admittingproviderprimaryspecialty | Urea_max_val | 5_class_oxygen_device_min_val |
| principalproblemdiagnosisname | Urea_avg_val | 5_class_oxygen_device_avg_val |
| cnt_procedure | Urea_first_val | oxygen_device_min_val |
| days_since_last_admission | Urea_last_val | oxygen_device_max_val |
| admissions_previous_year | WBC_min_val | oxygen_device_avg_val |
| admissions_previous_2_years | WBC_max_val | oxygen_device_first_val |
| admissions_previous_90 days | WBC_avg_val | oxygen_device_last_val |
| admissions_previous_180_days | WBC_first_val | SP_O2_min_val |
| arrivalmethod | WBC_last_val | SP_O2_max_val |
| acuitylevel | arterial_min_val | SP_O2_avg_val |
| primarychiefcomplaintname | arterial_max_val | SP_O2_first_val |
| primaryeddiagnosisname | arterial_avg_val | SP_O2_last_val |
| edvisits_last_year | arterial_first_val | pulse_min_val |
| edvisits_last_2_years | arterial_last_val | pulse_max_val |
| edvisits_last_90_days | creatinie_min_val | pulse_avg_val |
| SLP consult | creatinie_max_val | pulse_first_val |
| Nutrition consult | creatinie_avg_val | pulse_last_val |
| SLP plan order | creatinie_first_val | r_number_ppl_assist_min_val |
| Observation status | creatinie_last_val | r_number_ppl_assist_max_val |
| Palliative care consult | eGFRhigh_min_val | r_number_ppl_assist_avg_val |
| 5150 order | eGFRhigh_avg_val | r_number_ppl_assist_first_val |
| Psych consult | eGFRhigh_first_val | r_number_ppl_assist_last_val |
| Social work consult | eGFRhigh_last_val | R ED RISK OF FALL ADULT SCORE_min_val |
| DNR/DNI order | eGFRlow_min_val | R ED RISK OF FALL ADULT SCORE_first_val |
| Home health order | eGFRlow_max_val | R IP STRATIFY MOBILITY SCORE_avg_val |
| Cardiology consult | eGFRlow_avg_val | R IP STRATIFY MOBILITY SCORE_first_val |
| SNF discharge order | eGFRlow_first_val | R IP STRATIFY TOTAL SCORE_max_val |
| Inpatient psychiatry order | eGFRlow_last_val | R IP STRATIFY TOTAL SCORE_avg_val |
| SNF discharge attending contact | glucose_min_val | R IP STRATIFY TOTAL SCORE_first_val |
| ALP_min_val | glucose_max_val | R IP STRATIFY TRANSFER AND MOBILITY SUM_min_val |
| ALP_max_val | glucose_avg_val | R IP STRATIFY TRANSFER AND MOBILITY SUM_avg_val |
| ALP_avg_val | glucose_first_val | R IP STRATIFY TRANSFER AND MOBILITY SUM_first_val |
| ALP_first_val | glucose_last_val | R IP STRATIFY TRANSFER SCORE_min_val |
| ALP_last_val | hemoglobin_min_val | R IP STRATIFY TRANSFER SCORE_max_val |
| ALT_min_val | hemoglobin_max_val | R IP STRATIFY TRANSFER SCORE_avg_val |
| ALT_max_val | hemoglobin_avg_val | R IP STRATIFY TRANSFER SCORE_first_val |
| ALT_avg_val | hemoglobin_first_val | R NU-DESC DISORIENTATION_max_val |
| ALT_first_val | hemoglobin_last_val | R NU-DESC DISORIENTATION_avg_val |
| ALT_last_val | pH_min_val | R NU-DESC DISORIENTATION_first_val |
| AST_min_val | pH_max_val | R NU-DESC DISORIENTATION_last_val |
| AST_max_val | pH_avg_val | R NU-DESC INAPPROPRIATE BEHAVIOR_avg_val |
| AST_avg_val | pH_first_val | R NU-DESC INAPPROPRIATE BEHAVIOR_last_val |
| AST_first_val | pH_last_val | R NU-DESC INAPPROPRIATE COMMUNICATION_max_val |
| AST_last_val | platelets_min_val | R NU-DESC INAPPROPRIATE COMMUNICATION_avg_val |
| Albumin_min_val | platelets_max_val | R NU-DESC PSYCHOMOTOR RETARDATION_avg_val |
| Albumin_max_val | platelets_avg_val | R NU-DESC PSYCHOMOTOR RETARDATION_first_val |
| Albumin_avg_val | platelets_first_val | R NU-DESC SCORE V2_max_val |
| Albumin_first_val | platelets_last_val | R NU-DESC SCORE V2_avg_val |
| Albumin_last_val | potassium_min_val | R NU-DESC SCORE V2_first_val |
| BNP_min_val | potassium_max_val | R NU-DESC SCORE V2_last_val |
| BNP_max_val | potassium_avg_val | RESPIRATIONS_min_val |
| BNP_avg_val | potassium_first_val | RESPIRATIONS_max_val |
| BNP_first_val | potassium_last_val | RESPIRATIONS_avg_val |
| Bicarb_min_val | sodium_min_val | RESPIRATIONS_first_val |
| Bicarb_max_val | sodium_max_val | RESPIRATIONS_last_val |
| Bicarb_avg_val | sodium_avg_val | TEMPERATURE_min_val |
| Bicarb_first_val | sodium_first_val | TEMPERATURE_max_val |
| Bicarb_last_val | sodium_last_val | TEMPERATURE_avg_val |
| Bilirubin_min_val | troponin_min_val | TEMPERATURE_first_val |
| Bilirubin_max_val | troponin_max_val | TEMPERATURE_last_val |
| Bilirubin_avg_val | troponin_avg_val | year_discharge_date |
| Bilirubin_first_val | troponin_first_val | |
| Bilirubin_last_val | venous_min_val | |
| PT_min_val | venous_max_val | |
| PT_max_val | venous_avg_val | |
| PT_avg_val | venous_first_val | |
Appendix B
- -
- Learning objective: ‘binary:logistic’
- -
- Learning rate: 0.1
- -
- Maximum depth: 5
- -
- Number of trees: 100
- -
- Scale_pos_weight: 6.08
- -
- Evaluation Metric: AUC-PR
- -
- Minimum sample leafs: 98
- -
- Maximum features: 0.152
- -
- Maximum depth: 8
- -
- Number of trees: 100
- -
- Learning rate: 0.1
- -
- n_estimators: 250
- -
- min_samples_leaf: 98
- -
- max_features: 0.152
- -
- max_depth: 8
- -
- default parameters from sklearn library, LogisticRegression module.
References
- Hospital Readmissions Reduction Program (HRRP)|CMS. Available online: https://www.cms.gov/Medicare/Medicare- (accessed on 20 July 2022).
- Auerbach, A.D.; Kripalani, S.; Vasilevskis, E.E.; Sehgal, N.; Lindenauer, P.K.; Metlay, J.P.; Fletcher, G.; Ruhnke, G.W.; Flanders, S.A.; Kim, C.; et al. Preventability and causes of readmissions in a national cohort of general medicine patients. JAMA Intern. Med. 2016, 176, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Zumbrunn, S.; Beck, K.; Vincent, A.; Loretz, N.; Müller, J.; Amacher, S.A.; Schaefert, R.; Hunziker, S. Interventions to Improve Communication at Hospital Discharge and Rates of Readmission: A Systematic Review and Meta-analysis. JAMA Netw. Open 2021, 4, e2119346. [Google Scholar] [CrossRef] [PubMed]
- Kripalani, S.; Theobald, C.N.; Anctil, B.; Vasilevskis, E.E. Reducing hospital readmission rates: Current strategies and future directions. Annu. Rev. Med. 2014, 65, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.-T.; Liao, J.C.; Chen, M.-H.; Chang, C.-M.; Li, C.-T. Predictive modeling for 14-day unplanned hospital readmission risk by using machine learning algorithms. BMC Med. Inf. Decis. Mak. 2021, 21, 288. [Google Scholar] [CrossRef]
- Li, Q.; Yao, X.; Échevin, D. How Good Is Machine Learning in Predicting All-Cause 30-Day Hospital Readmission? Evidence From Administrative Data. Value Health 2020, 23, 1307–1315. [Google Scholar] [CrossRef]
- Allam, A.; Nagy, M.; Thoma, G.; Krauthammer, M. Neural networks versus Logistic regression for 30 days all-cause readmission prediction. Sci. Rep. 2019, 9, 9277. [Google Scholar] [CrossRef]
- Mišić, V.V.; Gabel, E.; Hofer, I.; Rajaram, K.; Mahajan, A. Machine learning prediction of postoperative emergency department hospital readmission. Anesthesiology 2020, 132, 968–980. [Google Scholar] [CrossRef]
- AHA Guide. Available online: https://guide.prod.iam.aha.org/guide/hospitalProfile/6930043 (accessed on 14 November 2022).
- Chen, Q.; Peng, Y.; Lu, Z. BioSentVec: Creating sentence embeddings for biomedical texts. In Proceedings of the 2019 IEEE International Conference on Healthcare Informatics (ICHI), Xi’an, China, 10–13 June 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Q.; Yang, Z.; Lin, H.; Lu, Z. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Sci. Data 2019, 6, 52. [Google Scholar] [CrossRef]
- Parr, T.; Turgutlu, K.; Csiszar, C.; Howard, J. Beware Default Random Forest Importances. Available online: https://explained.ai/rf-importance/ (accessed on 27 October 2022).
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liu, X.; Anstey, J.; Li, R.; Sarabu, C.; Sono, R.; Butte, A.J. Rethinking PICO in the Machine Learning Era: ML-PICO. Appl. Clin. Inform. 2021, 12, 407–416. [Google Scholar] [CrossRef]
- Hyndman, R.J.; Athanasopoulos, G. Forecasting: Principles and Practice, 2nd ed. Available online: https://otexts.com/fpp2/ (accessed on 27 October 2022).
- Omphalos. Uber’s Parallel and Language-Extensible Time Series Backtesting Tool|Uber Blog. Available online: https://www.uber.com/blog/omphalos/ (accessed on 27 October 2022).
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. Adv. Neural Inf. Process. Syst. 2017. Available online: https://papers.nips.cc/paper/2017/hash/8a20a8621978632d76c43dfd28b67767-Abstract.html (accessed on 20 July 2022).
- Slack, D.; Hilgard, S.; Jia, E.; Singh, S.; Lakkaraju, H. Fooling LIME and SHAP: Adversarial Attacks on Post hoc Explanation Methods. arXiv 2019, arXiv:1911.02508. [Google Scholar] [CrossRef]
- Shapley, L.S. A value for n-person games. In The Shapley Value: Essays in Honor of Lloyd S. Shapley; Roth, A.E., Ed.; Cambridge University Press: Cambridge, UK, 1988; pp. 31–40. [Google Scholar] [CrossRef]
- Sundararajan, M.; Najmi, A. The many Shapley values for model explanation. arXiv 2019, arXiv:1908.08474. [Google Scholar] [CrossRef]
- Štrumbelj, E.; Kononenko, I. Explaining prediction models and individual predictions with feature contributions. Knowl. Inf. Syst. 2014, 41, 647–665. [Google Scholar] [CrossRef]
- Huang, Y.; Talwar, A.; Chatterjee, S.; Aparasu, R.R. Application of machine learning in predicting hospital readmissions: A scoping review of the literature. BMC Med. Res. Methodol. 2021, 21, 96. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.F.; Irie, S.M.; Russo, C.A.; Pav, V.; O’Connor, S.L.; Wensky, S.G. Lessons Learned in Providing Claims-Based Data to Participants in Health Care Innovation Models. Am. J. Med. Qual. 2019, 34, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.; Nieves-Robbins, N.; Spieker, E.; Louwers, T.; Hazel, D.; Marquardt, J.; Solveson, K.; Zahid, A.; Ahmad, M.; Barnhill, R.; et al. Development and Prospective Validation of a Machine Learning-Based Risk of Readmission Model in a Large Military Hospital. Appl. Clin. Inform. 2019, 10, 316–325. [Google Scholar] [CrossRef]
- Ko, M.; Chen, E.; Agrawal, A.; Rajpurkar, P.; Avati, A.; Ng, A.; Basu, S.; Shah, N.H. Improving hospital readmission prediction using individualized utility analysis. J. Biomed. Inform. 2021, 119, 103826. [Google Scholar] [CrossRef]
- Schiltz, N.K.; Dolansky, M.A.; Warner, D.F.; Stange, K.C.; Gravenstein, S.; Koroukian, S.M. Impact of instrumental activities of daily living limitations on hospital readmission: An observational study using machine learning. J. Gen. Intern. Med. 2020, 35, 2865–2872. [Google Scholar] [CrossRef]
- Papanicolas, I.; Riley, K.; Abiona, O.; Arvin, M.; Atsma, F.; Bernal-Delgado, E.; Bowden, N.; Blankart, C.R.; Deeny, S.; Estupiñán-Romero, F.; et al. Differences in health outcomes for high-need high-cost patients across high-income countries. Health Serv. Res. 2021, 56, 1347–1357. [Google Scholar] [CrossRef]
- Shah, A.A.; Devana, S.K.; Lee, C.; Bugarin, A.; Lord, E.L.; Shamie, A.N.; Park, D.Y.; van der Schaar, M.; SooHoo, N.F. Prediction of Major Complications and Readmission After Lumbar Spinal Fusion: A Machine Learning-Driven Approach. World Neurosurg. 2021, 152, e227–e234. [Google Scholar] [CrossRef]
- Hassan, A.M.; Lu, S.-C.; Asaad, M.; Liu, J.; Offodile, A.C.; Sidey-Gibbons, C.; Butler, C.E. Novel Machine Learning Approach for the Prediction of Hernia Recurrence, Surgical Complication, and 30-Day Readmission after Abdominal Wall Reconstruction. J. Am. Coll. Surg. 2022, 234, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, L.; Lu, L.; Zhu, T. Machine learning prediction of postoperative unplanned 30-day hospital readmission in older adult. Front. Mol. Biosci. 2022, 9, 910688. [Google Scholar] [CrossRef]
- Darabi, N.; Hosseinichimeh, N.; Noto, A.; Zand, R.; Abedi, V. Machine Learning-Enabled 30-Day Readmission Model for Stroke Patients. Front. Neurol. 2021, 12, 638267. [Google Scholar] [CrossRef]
- Lineback, C.M.; Garg, R.; Oh, E.; Naidech, A.M.; Holl, J.L.; Prabhakaran, S. Prediction of 30-Day Readmission After Stroke Using Machine Learning and Natural Language Processing. Front. Neurol. 2021, 12, 649521. [Google Scholar] [CrossRef]
- Hoffman, M.K.; Ma, N.; Roberts, A. A machine learning algorithm for predicting maternal readmission for hypertensive disorders of pregnancy. Am. J. Obstet. Gynecol. MFM 2021, 3, 100250. [Google Scholar] [CrossRef] [PubMed]
- Frizzell, J.D.; Liang, L.; Schulte, P.J.; Yancy, C.W.; Heidenreich, P.A.; Hernandez, A.F.; Bhatt, D.L.; Fonarow, G.C.; Laskey, W.K. Prediction of 30-Day All-Cause Readmissions in Patients Hospitalized for Heart Failure: Comparison of Machine Learning and Other Statistical Approaches. JAMA Cardiol. 2017, 2, 204–209. [Google Scholar] [CrossRef]
- Mortazavi, B.J.; Downing, N.S.; Bucholz, E.M.; Dharmarajan, K.; Manhapra, A.; Li, S.X.; Negahban, S.N.; Krumholz, H.M. Analysis of machine learning techniques for heart failure readmissions. Circ. Cardiovasc. Qual. Outcomes 2016, 9, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-K.; Lan, C.-C.; Tzeng, I.-S.; Wu, C.-W. The COPD-readmission (CORE) score: A novel prediction model for one-year chronic obstructive pulmonary disease readmissions. J. Formos. Med. Assoc. 2021, 120, 1005–1013. [Google Scholar] [CrossRef]
- Goto, T.; Jo, T.; Matsui, H.; Fushimi, K.; Hayashi, H.; Yasunaga, H. Machine Learning-Based Prediction Models for 30-Day Readmission after Hospitalization for Chronic Obstructive Pulmonary Disease. COPD J. Chronic Obstr. Pulm. Dis. 2019, 16, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Burkov, A. The Hundred-Page Machine Learning Book; Burkov, A., Ed.; Anton Burkov: Québec, QC, Canada, 2019; p. 160. [Google Scholar]
- Breiman, L. Arcing the Edge. Ann. Prob. 1998, 26, 1683–1702. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Statist. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Mason, L.; Baxter, J.; Bartlett, P.; Frean, M. Boosting Algorithms as Gradient Descent; MIT Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning, 2nd ed.; Springer: New York, NY, USA, 2009; pp. 106–119. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Gryczynski, J.; Nordeck, C.D.; Welsh, C.; Mitchell, S.G.; O’Grady, K.E.; Schwartz, R.P. Preventing hospital readmission for patients with comorbid substance use disorder: A randomized trial. Ann. Intern. Med. 2021, 174, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Kaya, S.; Sain Guven, G.; Aydan, S.; Toka, O. Predictors of hospital readmissions in internal medicine patients: Application of Andersen’s Model. Int. J. Health Plann. Manag. 2019, 34, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Cruz, P.L.M.; Soares, B.L.d.M.; da Silva, J.E.; Lima, E.; Silva, R.R.d. Clinical and nutritional predictors of hospital readmission within 30 days. Eur. J. Clin. Nutr. 2022, 76, 244–250. [Google Scholar] [CrossRef]
- Arnaud, É.; Elbattah, M.; Gignon, M.; Dequen, G. Deep Learning to Predict Hospitalization at Triage: Integration of Structured Data and Unstructured Text. In Proceedings of the 2020 IEEE International Conference on Big Data (Big Data), Atlanta, GA, USA, 10–13 December 2020; pp. 4836–4841. [Google Scholar] [CrossRef]

| Characteristics | Total Cohort | Patients Readmitted | Patients Not Readmitted |
|---|---|---|---|
| Age | |||
| Mean (SD) | 54.02 (18.56) | 53.41 (18.73) | 54.12 (18.53) |
| Gender | |||
| Male | 66,482 (45.12%) | 10,405 (50.15%) | 56,077 (44.29%) |
| Female | 80,827 (54.85%) | 10,332 (49.80%) | 70,495 (55.68%) |
| Nonbinary | 28 (0.02%) | 7 (0.03%) | 21 (0.02%) |
| Unknown | 21 (0.01%) | 3 (0.01%) | 18 (0.01%) |
| Ethnicity | |||
| Hispanic or Latino | 23,487 (15.94%) | 3980 (19.18%) | 19,507 (15.41%) |
| Not Hispanic or Latino | 120,426 (81.72%) | 16,505 (79.55%) | 103,921 (82.08%) |
| Unknown | 3445 (2.34%) | 262 (1.26%) | 3183 (2.51%) |
| Race | |||
| American Indian or Alaska Native | 1390 (0.94%) | 230 (1.11%) | 1160 (0.92%) |
| Asian | 23,871 (16.20%) | 3528 (17.00%) | 20,343 (16.07%) |
| Black or African American | 13,128 (8.91%) | 2278 (10.98%) | 10,850 (8.57%) |
| Native Hawaiian | 65 (0.04%) | 13 (0.06%) | 52 (0.04%) |
| White or Caucasian | 79,816 (54.17%) | 10,330 (49.79%) | 69,486 (54.89%) |
| Other Pacific Islander | 1605 (1.09%) | 212 (1.02%) | 1393 (1.10%) |
| Other | 27,469 (18.64%) | 4156 (20.03%) | 23,313 (18.42%) |
| Admission Type | |||
| Emergency/Urgent | 90,564 (61.88%) | 14,278 (68.80%) | 76,286 (60.73%) |
| Routine/Elective | 54,241 (37.06%) | 6270 (30.22%) | 47,971 (38.19%) |
| Other | 1553 (1.06%) | 203 (0.98%) | 1350 (1.08%) |
| Insurance | |||
| Commercial | 51,388 (34.82%) | 6031 (29.05%) | 45,357 (35.77%) |
| Medi-Cal | 38,464 (26.06%) | 6646 (32.01%) | 31,818 (25.09%) |
| Medicare | 56,235 (38.11%) | 7922 (38.16%) | 48,313 (38.1%) |
| Other | 1488 (1.01%) | 163 (0.79%) | 1325 (1.04%) |
| Length of Stay | |||
| Mean (SD) | 6.13 (9.45) | 7.53 (9.94) | 5.89 (9.35) |
| Type of Feature | Examples of Features Created for Model(s) |
|---|---|
| Patient utilization | Binary target variable: readmission status within 30 days (0 = No, 1 = Yes);Number of days since last admission; Number of admissions in the past 90 days, 180 days, 1 year, and 2 years; Number of emergency visits in the past 90 days, 180 days, 1 year, and 2 years; |
| Demographic information | Age; sex; race; ethnicity; marital status; preferred language; financial class; postal code; smoking status; BMI |
| Procedure information | Number of procedures performed during encounter; Binary indicator for if any procedure was performed (0 = No, 1 = Yes) |
| Lab tests | Mean, minimum, maximum, the first and last value of each unique type of lab test (e.g., creatinine, hemoglobin) that was resulted during the encounter; Binary indicators for if amphetamine, barbiturates, benzo, cocaine, opiates, THC, and Utox was ordered (0 = No, 1 = Yes); Binary indicators for if amphetamine, barbiturates, benzo, cocaine, opiates, THC, and Utox were positive (0 = No, 1 = Yes); |
| Flowsheet values | Mean, minimum, maximum, the first and last value of each unique type of flowsheet value (e.g., heart rate, respiratory rate, nursing mobility scores) that was recorded during the encounter |
| Ancillary orders | Binary indicators for if each given ancillary order (e.g., palliative care consult, DNR/DNI order, social work) was placed (0 = No, 1 = Yes) |
| Textual information such as diagnosis and primary chief complaints | Sentence-embedded vectors generated from textual columns, categorical features that treat unique diagnoses as their own categories |
| Other features that have remained the same as in EHR | Admission source; admission type; inpatient length of stay in days; discharge disposition; department; hospital service; admitting provider type; admitting provider primary specialty; arrival method; acuity level |
| Machine Learning Model | Average Area under the Precision-Recall Curve | Average Training Time (Seconds) |
|---|---|---|
| Logistic Regression | 0.2403 | 81.522 |
| Random Forest | 0.4116 | 106.875 |
| Gradient Boosting | 0.4435 | 489.752 |
| XGBoost | 0.434 | 305.884 |
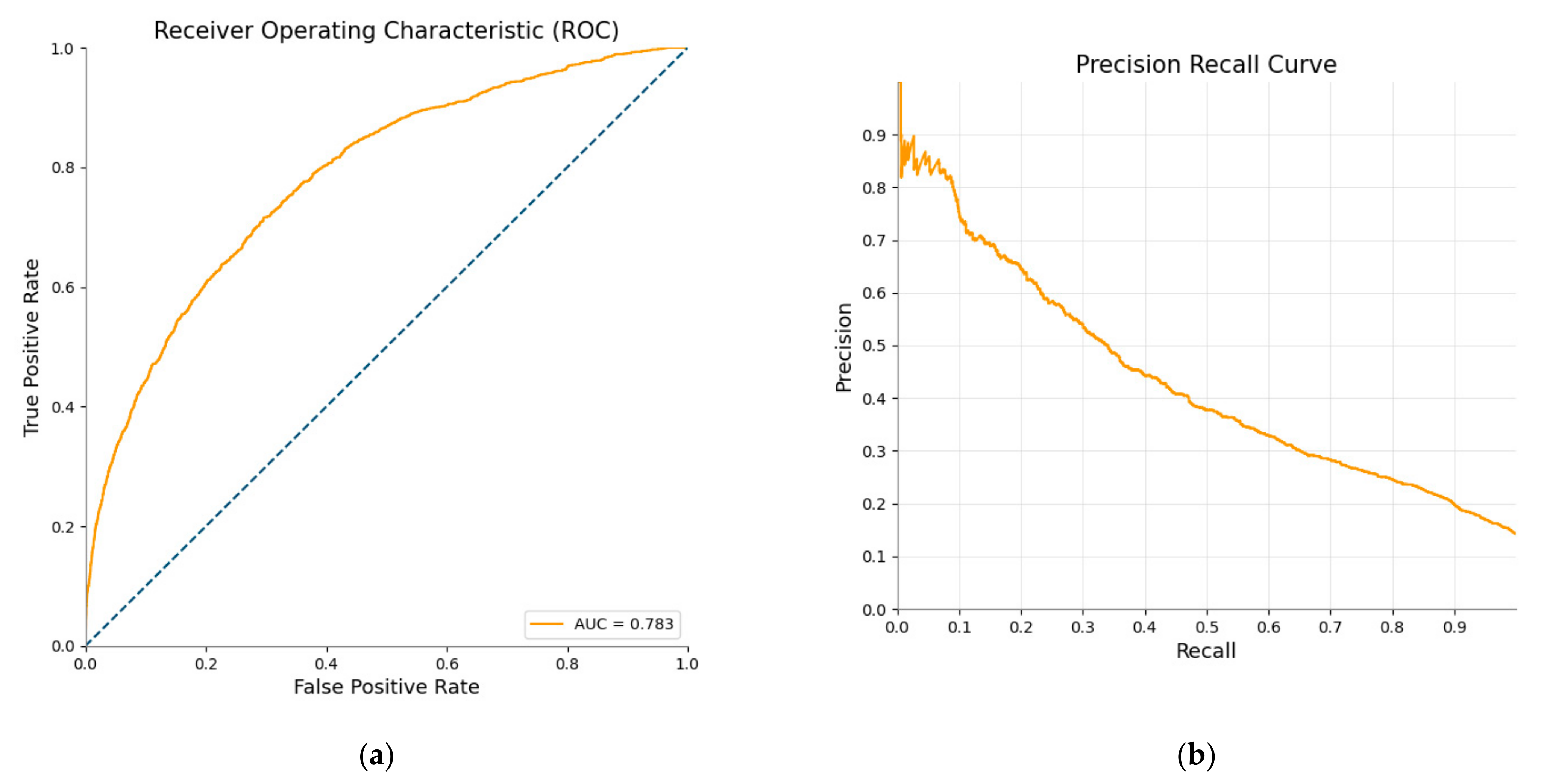
| Test Characteristic | Value |
|---|---|
| AUC | 0.783 |
| AUC-PR | 0.434 |
| Accuracy | 0.713 |
| Precision | 0.283 |
| Recall | 0.701 |
| F1 | 0.403 |
| Threshold | 0.486 |
| True positives | 903 |
| True negatives | 5738 |
| False positives | 2286 |
| False negatives | 384 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, A.L.; Ravi, A.; Arvisais-Anhalt, S.; Muniyappa, A.N.; Liu, X.; Wang, S. Development and Internal Validation of an Interpretable Machine Learning Model to Predict Readmissions in a United States Healthcare System. Informatics 2023, 10, 33. https://doi.org/10.3390/informatics10020033
Luo AL, Ravi A, Arvisais-Anhalt S, Muniyappa AN, Liu X, Wang S. Development and Internal Validation of an Interpretable Machine Learning Model to Predict Readmissions in a United States Healthcare System. Informatics. 2023; 10(2):33. https://doi.org/10.3390/informatics10020033
Chicago/Turabian StyleLuo, Amanda L., Akshay Ravi, Simone Arvisais-Anhalt, Anoop N. Muniyappa, Xinran Liu, and Shan Wang. 2023. "Development and Internal Validation of an Interpretable Machine Learning Model to Predict Readmissions in a United States Healthcare System" Informatics 10, no. 2: 33. https://doi.org/10.3390/informatics10020033
APA StyleLuo, A. L., Ravi, A., Arvisais-Anhalt, S., Muniyappa, A. N., Liu, X., & Wang, S. (2023). Development and Internal Validation of an Interpretable Machine Learning Model to Predict Readmissions in a United States Healthcare System. Informatics, 10(2), 33. https://doi.org/10.3390/informatics10020033






