Phenolic Compounds from Irradiated Olive Wastes: Optimization of the Heat-Assisted Extraction Using Response Surface Methodology
Abstract
1. Introduction
2. Materials and Methods
2.1. Standards and Reagents
2.2. Olive Pomace Samples
2.3. Irradiation Experiments
2.4. Heat-Assisted Extraction (HAE)
2.5. Analysed Responses
2.5.1. Extraction Yield
2.5.2. Phenolic Fingerprinting and Quantification
2.6. Extraction Optimization by Response Surface Methodology
2.6.1. Screening Test of Factors and Level Range for Phenolic Compounds Extraction
2.6.2. Experimental Design
2.7. Statistical Analysis
3. Results and Discussion
3.1. Single-Factor Effects for Polyphenolic Extractions
3.1.1. Effect of Extraction Time on Polyphenolic Content and Extractability Yield
3.1.2. Effect of Solvent Concentration on Polyphenolic Content and Extractability Yield
3.2. Optimization of HAE Process in Olive Pomace
3.2.1. Extracting Modelling and Analysis of Variance
3.2.2. Factorial and General Optimization
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Madureira, J.; Dias, M.I.; Pinela, J.; Calhelha, R.C.; Barros, L.; Santos-Buelga, C.; Margaça, F.M.A.; Ferreira, I.C.F.R.; Cabo Verde, S. The use of gamma radiation for extractability improvement of bioactive compounds in olive oil wastes. Sci. Total Environ. 2020, 727, 138706. [Google Scholar] [CrossRef]
- Malapert, A.; Reboul, E.; Loonis, M.; Dangles, O.; Tomao, V. Direct and Rapid Profiling of Biophenols in Olive Pomace by UHPLC-DAD-MS. Food Anal. Methods 2018, 11, 1001–1010. [Google Scholar] [CrossRef]
- Nunes, M.A.; Costa, A.S.G.; Bessada, S.; Santos, J.; Puga, H.; Alves, R.C.; Freitas, V.; Oliveira, M.B.P.P. Olive pomace as a valuable source of bioactive compounds: A study regarding its lipid- and water-soluble components. Sci. Total Environ. 2018, 644, 229–236. [Google Scholar] [CrossRef]
- Faustino, M.; Veiga, M.; Sousa, P.; Costa, E.M.; Silva, S.; Pintado, M. Agro-food byproducts as a new source of natural food additives. Molecules 2019, 24, 1056. [Google Scholar] [CrossRef] [PubMed]
- Galanakis, C.M.; Aldawoud, T.M.S.; Rizou, M.; Rowan, N.J.; Ibrahim, S.A. Food Ingredients and Active Compounds against the Coronavirus Disease (COVID-19) Pandemic: A Comprehensive Review. Foods 2020, 9, 1701. [Google Scholar] [CrossRef] [PubMed]
- De Bruno, A.; Romeo, R.; Fedele, F.L.; Sicari, A.; Piscopo, A.; Poiana, M. Antioxidant activity shown by olive pomace extracts. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2018, 53, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, S.M.; Guyot, S.; Marnet, N.; Lopes-da-Silva, J.A.; Renard, C.M.G.C.; Coimbra, M.A. Characterisation of phenolic extracts from olive pulp and olive pomace by electrospray mass spectrometry. J. Sci. Food Agric. 2005, 85, 21–32. [Google Scholar] [CrossRef]
- Fernández-Bolaños, J.; Rodríguez, G.; Rodríguez, R.; Guillén, R.; Jiménez, A. Extraction of interesting organic compounds from olive oil waste. Grasas Aceites 2006, 57, 95–106. [Google Scholar] [CrossRef]
- Suárez, M.; Romero, M.-P.; Ramo, T.; Macià, A.; Motilva, M.-J. Methods for Preparing Phenolic Extracts from Olive Cake for Potential Application as Food Antioxidants. J. Agric. Food Chem. 2009, 57, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Galanakis, C.M. Recovery of high added-value components from food wastes: Conventional, emerging technologies and commercialized applications. Trends Food Sci. Technol. 2012, 26, 68–87. [Google Scholar] [CrossRef]
- Galanakis, C.M. Emerging technologies for the production of nutraceuticals from agricultural by-products: A viewpoint of opportunities and challenges. Food Bioprod. Process. 2013, 91, 575–579. [Google Scholar] [CrossRef]
- Alu’datt, M.H.; Alli, I.; Ereifej, K.; Alhamad, M.; Al-Tawaha, A.R.; Rababah, T. Optimisation, characterisation and quantification of phenolic compounds in olive cake. Food Chem. 2010, 123, 117–122. [Google Scholar] [CrossRef]
- Vitali Čepo, D.; Albahari, P.; Zovko Končić, M.; Radić, K.; Jurmanović, S.; Jug, M. Solvent Extraction and Chromatographic Determination of Polyphenols in Olive Pomace. Food Health Dis. 2017, 6, 7–14. [Google Scholar]
- Böhmer-Maas, B.W.; Otero, D.M.; Zambiazi, R.C.; Aranha, B.C. Optimization of the extraction of phenolic compounds from olive pomace using response surface methodology. Rev. Ceres 2020, 67, 181–190. [Google Scholar] [CrossRef]
- Zuorro, A. Modelling of polyphenol recovery from olive pomace by response surface methodology. Int. Rev. Model. Simul. 2014, 7, 1023–1028. [Google Scholar] [CrossRef]
- Madureira, J.; Pimenta, A.I.; Popescu, L.; Besleaga, A.; Dias, M.I.; Santos, P.M.P.; Melo, R.; Ferreira, I.C.F.R.; Cabo Verde, S.; Margaça, F.M.A. Effects of gamma radiation on cork wastewater: Antioxidant activity and toxicity. Chemosphere 2017, 169, 139–145. [Google Scholar] [CrossRef]
- Guerreiro, D.; Madureira, J.; Silva, T.; Melo, R.; Santos, P.M.P.; Ferreira, A.; Trigo, M.J.; Falcão, A.N.; Margaça, F.M.A.; Cabo Verde, S. Post-harvest treatment of cherry tomatoes by gamma radiation: Microbial and physicochemical parameters evaluation. Innov. Food Sci. Emerg. Technol. 2016, 36, 1–9. [Google Scholar] [CrossRef]
- Cabo Verde, S.; Trigo, M.J.; Sousa, M.B.; Ferreira, A.; Ramos, A.C.; Nunes, I.; Junqueira, C.; Melo, R.; Santos, P.M.P.; Botelho, M.L. Effects of gamma radiation on raspberries: Safety and quality issues. J. Toxicol. Environ. Health A 2013, 76, 291–303. [Google Scholar] [CrossRef]
- Elias, M.I.; Madureira, J.; Santos, P.M.P.; Carolino, M.M.; Margaça, F.M.A.; Verde, S.C. Preservation treatment of fresh raspberries by e-beam irradiation. Innov. Food Sci. Emerg. Technol. 2020, 66, 102487. [Google Scholar] [CrossRef]
- Barkaoui, S.; Madureira, J.; Santos, P.M.P.; Margaça, F.M.A.; Miloud, N.B.; Mankai, M.; Boudhrioua, N.M.; Cabo Verde, S. Effect of Ionizing Radiation and Refrigeration on the Antioxidants of Strawberries. Food Bioprocess Technol. 2020, 13, 1516–1527. [Google Scholar] [CrossRef]
- Pereira, E.; Barros, L.; Antonio, A.L.; Cabo Verde, S.; Santos-Buelga, C.; Ferreira, I.C.F.R.; Rodrigues, P. Is gamma radiation suitable to preserve phenolic compounds and to decontaminate mycotoxins in aromatic plants? A case-study with aloysia citrodora paláu. Molecules 2017, 22, 347. [Google Scholar] [CrossRef]
- Box, G.E.P.; Wilson, K.B. On the Experimental Attainment of Optimum Conditions. J. R. Stat. Soc. 1951, 13, 1–45. [Google Scholar] [CrossRef]
- Whittaker, B.; Watts, M.F. The influence of dose rate, ambient temperature and time on the radiation response of Harwell PMMA dosimeters. Radiat. Phys. Chem. 2001, 60, 101–110. [Google Scholar] [CrossRef]
- Pinela, J.; Prieto, M.A.; Pereira, E.; Jabeur, I.; Barreiro, M.F.; Barros, L.; Ferreira, I.C.F.R. Optimization of heat- and ultrasound-assisted extraction of anthocyanins from Hibiscus sabdariffa calyces for natural food colorants. Food Chem. 2019, 275, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Primo da Silva, L.; Pereira, E.; Prieto, M.A.; Simal-Gandara, J.; Pires, T.C.S.P.; Alves, M.J.; Calhelha, R.; Barros, L.; Ferreira, I.C.F.R. Rubus ulmifolius Schott as a Novel Source of Food Colorant: Extraction optimization of coloring pigments and incorporation in a bakery product. Molecules 2019, 24, 2181. [Google Scholar] [CrossRef] [PubMed]
- Melgar, B.; Dias, M.I.; Ciric, A.; Sokovic, M.; Garcia-Castello, E.M.; Rodriguez-Lopez, A.D.; Barros, L.; Ferreira, I.C.R.F. Bioactive characterization of Persea americana Mill. by-products: A rich source of inherent antioxidants. Ind. Crops Prod. 2018, 111, 212–218. [Google Scholar] [CrossRef]
- Chanioti, S.; Tzia, C. Optimization of ultrasound-assisted extraction of oil from olive pomace using response surface technology: Oil recovery, unsaponifiable matter, total phenol content and antioxidant activity. LWT Food Sci. Technol. 2017, 79, 178–189. [Google Scholar] [CrossRef]
- Garcia-Castello, E.M.; Rodriguez-Lopez, A.D.; Mayor, L.; Ballesteros, R.; Conidi, C.; Cassano, A. Optimization of conventional and ultrasound assisted extraction of flavonoids from grapefruit (Citrus paradisi L.) solid wastes. LWT Food Sci. Technol. 2015, 64, 1114–1122. [Google Scholar] [CrossRef]
- Santos Felix, A.C.; Novaes, C.G.; Pires Rocha, M.; Barreto, G.E.; do Nascimento, B.B.; Giraldez Alvarez, L.D. Mixture Design and Doehlert Matrix for the Optimization of the Extraction of Phenolic Compounds from Spondias mombin L Apple Bagasse Agroindustrial Residues. Front. Chem. 2018, 5, 1–8. [Google Scholar] [CrossRef]
- Shi, J.; Yu, J.; Pohorly, J.; Young, J.C.; Bryan, M.; Wu, Y. Optimization of the extraction of polyphenols from grape seed meal by aqueous ethanol solution. J. Food Agric. Environ. 2003, 1, 42–47. [Google Scholar]
- Belwal, T.; Chemat, F.; Venskutonis, P.R.; Cravotto, G.; Jaiswal, D.K.; Bhatt, I.D.; Devkota, H.P.; Luo, Z. Recent advances in scaling-up of non-conventional extraction techniques: Learning from successes and failures. TrAC Trends Anal. Chem. 2020, 127, 115895. [Google Scholar] [CrossRef]
- Goldsmith, C.D.; Vuong, Q.V.; Stathopoulos, C.E.; Roach, P.D.; Scarlett, C.J. Ultrasound increases the aqueous extraction of phenolic compounds with high antioxidant activity from olive pomace. LWT Food Sci. Technol. 2018, 89, 284–290. [Google Scholar] [CrossRef]
- Hadidi, M.; Amoli, P.I.; Jelyani, A.Z.; Kasiri, Z.; Rouhafza, A.; Ibarz, A.; Khaksar, F.B.; Tabrizi, S.T. Polysaccharides from pineapple core as a canning by-product: Extraction optimization, chemical structure, antioxidant and functional properties. Int. J. Biol. Macromol. 2020, 163, 2357–2364. [Google Scholar] [CrossRef] [PubMed]
- Pavez, I.C.; Lozano-Sánchez, J.; Borrás-Linares, I.; Nuñez, H.; Robert, P.; Segura-Carretero, A. Obtaining an Extract Rich in Phenolic Compounds from Olive Pomace by Pressurized Liquid Extraction. Molecules 2019, 24, 3108. [Google Scholar] [CrossRef] [PubMed]
- Irakli, M.; Chatzopoulou, P.; Ekateriniadou, L. Optimization of ultrasound-assisted extraction of phenolic compounds: Oleuropein, phenolic acids, phenolic alcohols and flavonoids from olive leaves and evaluation of its antioxidant activities. Ind. Crops Prod. 2018, 124, 382–388. [Google Scholar] [CrossRef]
- Fu, X.; Belwal, T.; Cravotto, G.; Luo, Z. Sono-physical and sono-chemical effects of ultrasound: Primary applications in extraction and freezing operations and influence on food components. Ultrason. Sonochem. 2020, 60, 104726. [Google Scholar] [CrossRef]
- Balzan, S.; Taticchi, A.; Cardazzo, B.; Urbani, S.; Servili, M.; Di Lecce, G.; Zabalza, I.B.; Rodriguez-Estrada, M.T.; Novelli, E.; Fasolato, L. Effect of phenols extracted from a by-product of the oil mill on the shelf-life of raw and cooked fresh pork sausages in the absence of chemical additives. LWT Food Sci. Technol. 2017, 85, 89–95. [Google Scholar] [CrossRef]
- Chaves-López, C.; Serio, A.; Mazzarrino, G.; Martuscelli, M.; Scarpone, E.; Paparella, A. Control of household mycoflora in fermented sausages using phenolic fractions from olive mill wastewaters. Int. J. Food Microbiol. 2015, 207, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Elama, C.; Tarawa, M.; Al-Rimawi, F. Oleuropein from Olive Leaf Extract as Natural Antioxidant of Frozen Hamburger. J. Food Sci. Eng. 2017, 7, 406–412. [Google Scholar] [CrossRef][Green Version]
- Guglielmotti, M.; Passaghe, P.; Buiatti, S. Use of olive (Olea europaea L.) leaves as beer ingredient, and their influence on beer chemical composition and antioxidant activity. J. Food Sci. 2020, 85, 2278–2285. [Google Scholar] [CrossRef]
- Cedola, A.; Cardinali, A.; D’Antuono, I.; Conte, A.; Del Nobile, M.A. Cereal foods fortified with by-products from the olive oil industry. Food Biosci. 2020, 33, 100490. [Google Scholar] [CrossRef]
- Cedola, A.; Palermo, C.; Centonze, D.; Del Nobile, M.A.; Conte, A. Characterization and bio-accessibility evaluation of olive leaf extract-enriched “Taralli”. Foods 2020, 9, 1268. [Google Scholar] [CrossRef] [PubMed]
- Galanakis, C.M.; Tsatalas, P.; Charalambous, Z.; Galanakis, I.M. Control of microbial growth in bakery products fortified with polyphenols recovered from olive mill wastewater. Environ. Technol. Innov. 2018, 10, 1–15. [Google Scholar] [CrossRef]
- Georgakouli, K.; Mpesios, A.; Kouretas, D.; Petrotos, K.; Mitsagga, C.; Giavasis, I.; Jamurtas, A.Z. The effects of an olive fruit polyphenol-enriched yogurt on body composition, blood redox status, physiological and metabolic parameters and yogurt microflora. Nutrients 2016, 8, 344. [Google Scholar] [CrossRef] [PubMed]
- Palmeri, R.; Parafati, L.; Trippa, D.; Siracusa, L.; Arena, E.; Restuccia, C.; Fallico, B. Addition of olive leaf extract (OLE) for producing fortified fresh pasteurized milk with an extended shelf life. Antioxidants 2019, 8, 255. [Google Scholar] [CrossRef] [PubMed]
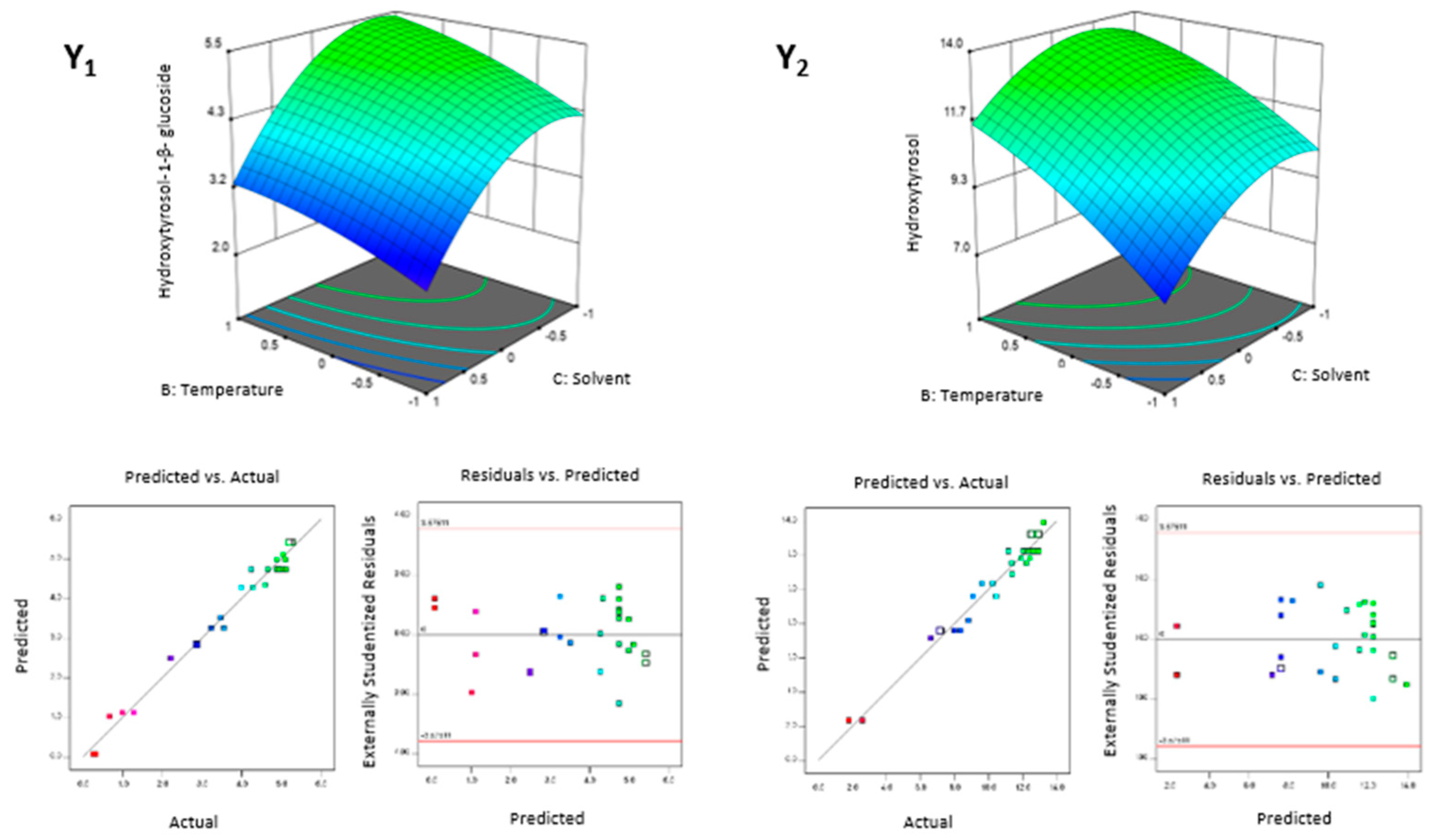

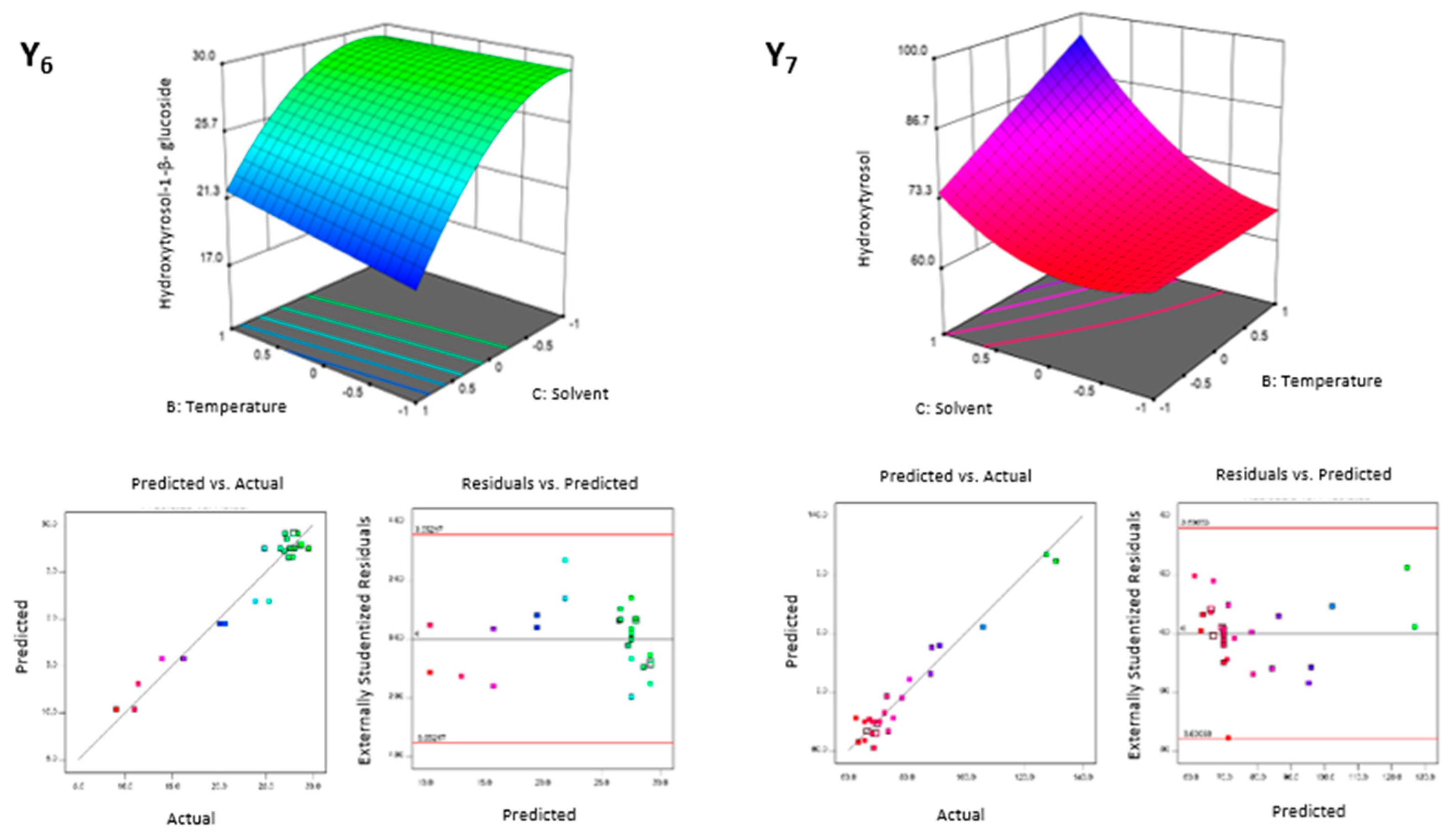
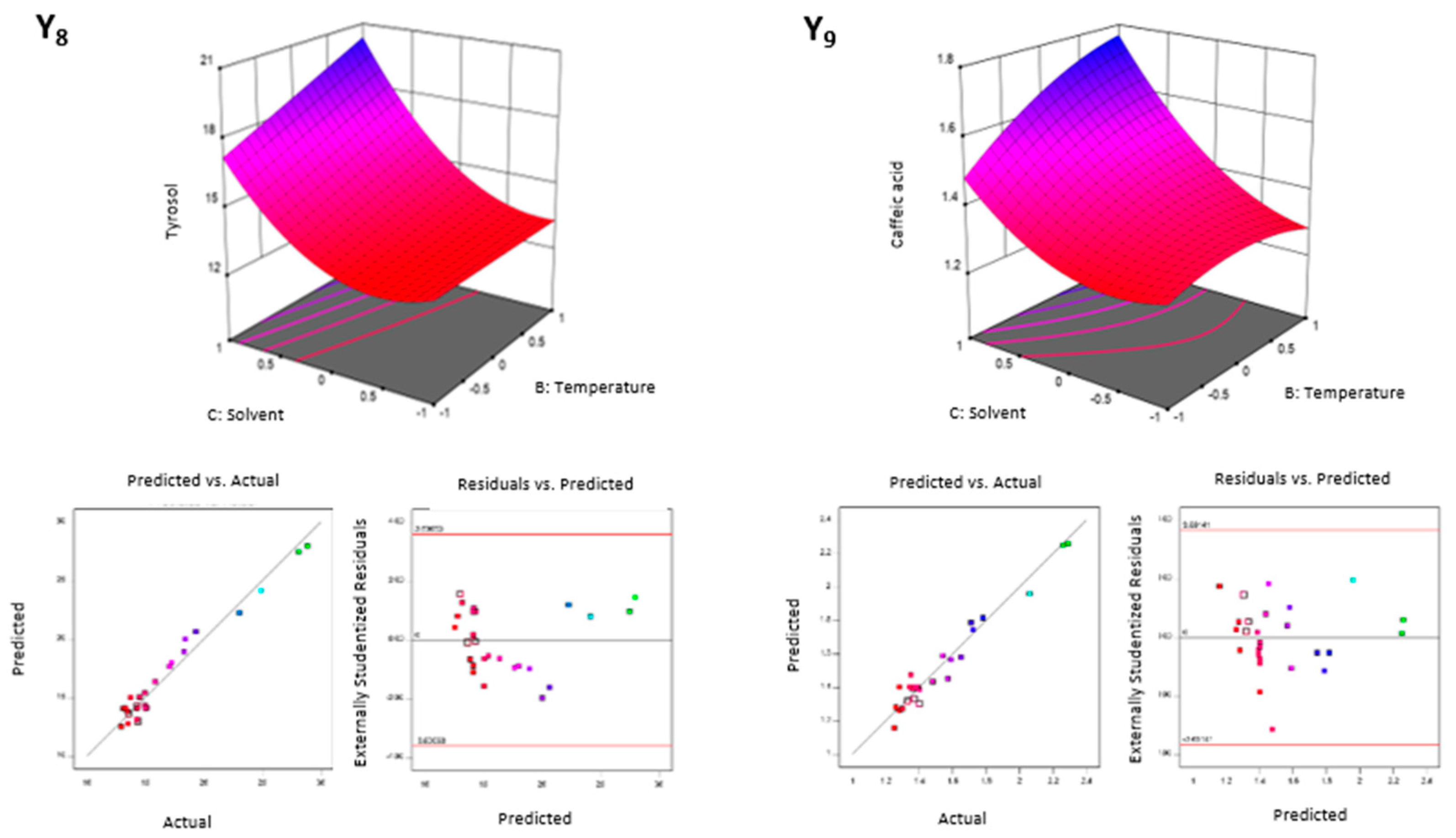

| Factors | Responses | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Run | X1 | X2 | X3 | X1:t min | X2: T °C | X3: S % | Y1 | Y2 | Y3 | Y4 | Y5 | Y6 | Y7 | Y8 | Y9 | Y10 | Y11 |
| 1 | −1 | −1 | −1 | 40.3 | 37.2 | 20.3 | 4.28 | 10.23 | 2.08 | 0.21 | 16.79 | 27.01 | 63.36 | 12.92 | 1.28 | 104.57 | 16.12 |
| 2 | −1 | −1 | 1 | 40.3 | 37.2 | 79.7 | 2.19 | 7.25 | 1.72 | 0.16 | 11.31 | 20.08 | 75.23 | 15.81 | 1.57 | 112.70 | 10.90 |
| 3 | −1 | 1 | −1 | 40.3 | 72.8 | 20.3 | 5.26 | 12.93 | 2.55 | 0.24 | 20.98 | 28.05 | 69.41 | 14.33 | 1.40 | 113.18 | 16.98 |
| 4 | −1 | 1 | 1 | 40.3 | 72.8 | 79.7 | 3.23 | 11.35 | 2.37 | 0.23 | 17.18 | 23.84 | 88.49 | 18.41 | 1.71 | 132.44 | 13.23 |
| 5 | 1 | −1 | −1 | 99.7 | 37.2 | 20.3 | 3.98 | 9.61 | 2.09 | 0.19 | 15.87 | 28.34 | 69.60 | 15.13 | 1.40 | 114.47 | 13.81 |
| 6 | 1 | −1 | 1 | 99.7 | 37.2 | 79.7 | 2.21 | 8.35 | 1.84 | 0.16 | 12.55 | 20.66 | 78.26 | 17.24 | 1.65 | 117.81 | 10.67 |
| 7 | 1 | 1 | −1 | 99.7 | 72.8 | 20.3 | 5.19 | 12.50 | 2.62 | 0.26 | 20.57 | 27.91 | 66.37 | 13.56 | 1.37 | 109.20 | 18.83 |
| 8 | 1 | 1 | 1 | 99.7 | 72.8 | 79.7 | 3.55 | 12.22 | 2.59 | 0.24 | 18.61 | 25.30 | 91.16 | 19.30 | 1.78 | 137.55 | 13.67 |
| 9 | 1.68 | 0 | 0 | 120 | 55 | 50 | 4.87 | 12.55 | 2.64 | 0.26 | 20.33 | 29.51 | 73.64 | 14.24 | 1.48 | 118.87 | 17.80 |
| 10 | −1.68 | 0 | 0 | 20 | 55 | 50 | 4.23 | 11.17 | 2.38 | 0.25 | 18.02 | 28.03 | 72.34 | 14.50 | 1.59 | 116.46 | 14.59 |
| 11 | 0 | −1.68 | 0 | 70 | 25 | 50 | 3.46 | 8.81 | 1.79 | 0.16 | 14.22 | 27.38 | 68.68 | 13.51 | 1.25 | 110.82 | 12.63 |
| 12 | 0 | 1.68 | 0 | 70 | 85 | 50 | 5.04 | 13.22 | 2.91 | 0.25 | 21.42 | 27.23 | 73.01 | 14.94 | 1.37 | 116.55 | 18.49 |
| 13 | 0 | 0 | −1.68 | 70 | 55 | 0 | 4.58 | 11.38 | 2.32 | 0.23 | 18.51 | 26.95 | 67.10 | 13.74 | 1.36 | 109.15 | 16.99 |
| 14 | 0 | 0 | 1.68 | 70 | 55 | 100 | 0.66 | 6.59 | 1.51 | 0.13 | 8.89 | 11.37 | 106.03 | 24.85 | 2.06 | 144.31 | 5.83 |
| 15 | −1.68 | −1.68 | −1.68 | 20 | 25 | 0 | 2.86 | 7.13 | 1.42 | 0.14 | 11.54 | 28.68 | 69.86 | 14.28 | 1.33 | 114.15 | 9.96 |
| 16 | −1.68 | −1.68 | 1.68 | 20 | 25 | 100 | 0.26 | 1.78 | 0.52 | 0.04 | 2.60 | 9.04 | 62.59 | 18.30 | 1.35 | 91.28 | 2.89 |
| 17 | −1.68 | 1.68 | −1.68 | 20 | 85 | 0 | 5.09 | 12.43 | 2.46 | 0.23 | 20.21 | 27.84 | 68.06 | 13.44 | 1.26 | 110.60 | 18.28 |
| 18 | −1.68 | 1.68 | 1.68 | 20 | 85 | 100 | 0.99 | 9.08 | 2.00 | 0.16 | 12.23 | 13.93 | 127.65 | 28.10 | 2.29 | 171.97 | 7.11 |
| 19 | 1.68 | −1.68 | −1.68 | 120 | 25 | 0 | 2.87 | 7.99 | 1.74 | 0.15 | 12.75 | 28.76 | 80.72 | 17.06 | 1.54 | 128.08 | 11.01 |
| 20 | 1.68 | −1.68 | 1.68 | 120 | 25 | 100 | 0.32 | 2.57 | 0.67 | 0.05 | 3.61 | 10.97 | 88.03 | 23.02 | 1.72 | 123.74 | 2.93 |
| 21 | 1.68 | 1.68 | −1.68 | 120 | 85 | 0 | 4.88 | 11.91 | 2.54 | 0.22 | 19.55 | 27.42 | 65.49 | 14.95 | 1.30 | 109.16 | 17.22 |
| 22 | 1.68 | 1.68 | 1.68 | 120 | 85 | 100 | 1.28 | 10.44 | 2.35 | 0.18 | 14.26 | 16.21 | 130.81 | 28.85 | 2.26 | 178.12 | 8.30 |
| 23 | 0 | 0 | 0 | 70 | 55 | 50 | 5.02 | 12.73 | 2.60 | 0.25 | 20.61 | 26.54 | 68.09 | 13.12 | 1.35 | 109.09 | 18.72 |
| 24 | 0 | 0 | 0 | 70 | 55 | 50 | 4.66 | 12.03 | 2.36 | 0.23 | 19.28 | 27.53 | 70.12 | 15.04 | 1.39 | 114.08 | 17.17 |
| 25 | 0 | 0 | 0 | 70 | 55 | 50 | 4.92 | 12.59 | 2.65 | 0.26 | 20.42 | 27.79 | 70.03 | 14.23 | 1.39 | 113.44 | 17.97 |
| 26 | 0 | 0 | 0 | 70 | 55 | 50 | 4.94 | 12.58 | 2.55 | 0.25 | 20.32 | 24.84 | 65.56 | 13.31 | 1.28 | 104.99 | 19.31 |
| 27 | 0 | 0 | 0 | 70 | 55 | 50 | 5.11 | 12.95 | 2.62 | 0.25 | 20.92 | 27.81 | 70.57 | 14.98 | 1.38 | 114.74 | 18.35 |
| 28 | 0 | 0 | 0 | 70 | 55 | 50 | 4.92 | 12.30 | 2.44 | 0.24 | 19.90 | 27.51 | 68.82 | 14.28 | 1.34 | 111.96 | 17.88 |
| ANOVA Estimated Coefficients | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Term | Y1 | Y2 | Y3 | Y4 | Y5 | Y6 | Y7 | Y8 | Y9 | Y10 | Y11 | |||||||||
| Intercept | 4.75 | 12.28 | 2.53 | 19.81 | 19.56 | 27.29 | 69.50 | 14.02 | 1.40 | 112.22 | 17.56 | |||||||||
| A-time | −0.02 | 0.08 | 0.04 | 0.10 | 0.09 | 0.20 | 1.90 | 0.57 | 0.04 | 2.70 | −0.10 | |||||||||
| B-Temp | 0.47 | 1.71 | 0.35 | 2.57 | 2.53 | 0.61 | 5.21 | 0.78 | 0.07 | 6.67 | 1.70 | |||||||||
| C-Solvent | −0.99 | −1.11 | −0.18 | −2.30 | −2.28 | −4.22 | 9.38 | 2.71 | 0.17 | 8.04 | −2.63 | |||||||||
| AB | 0.01 | −0.03 | 0.00 | −0.02 | −0.02 | −0.01 | −1.54 | −0.26 | −0.02 | −1.83 | 0.03 | |||||||||
| AC | 0.03 | 0.11 | 0.01 | 0.15 | 0.15 | 0.19 | 0.83 | 0.06 | 0.00 | 1.09 | 0.06 | |||||||||
| BC | −0.10 | 0.27 | 0.06 | 0.23 | 0.23 | 0.59 | 5.24 | 0.80 | 0.07 | 6.70 | −0.20 | |||||||||
| A2 | −0.03 | −0.09 | −0.01 | −0.12 | −0.13 | 0.50 | 0.70 | 0.10 | 0.04 | 1.35 | −0.32 | Not significant | Significant | |||||||
| B2 | −0.14 | −0.39 | −0.06 | −0.60 | −0.59 | −0.02 | −0.06 | 0.05 | −0.04 | −0.07 | −0.54 | |||||||||
| C2 | −0.72 | −1.11 | −0.22 | −2.06 | −2.04 | −2.90 | 5.51 | 1.85 | 0.10 | 4.56 | −2.01 | |||||||||
| R2 | 0.98 | 0.97 | 0.97 | 0.98 | 0.98 | 0.96 | 0.96 | 0.97 | 0.95 | 0.94 | 0.96 | |||||||||
| Lack of fit | 0.10 | 0.06 | 0.38 | 0.49 | 0.09 | 0.25 | 0.02 | 0.35 | 0.08 | 0.14 | 0.12 | |||||||||
| Decoded Optimization | ||||||||||||||||||||
| Factor | Y1 | Y2 | Y3 | Y4 | Y5 | Y6 | Y7 | Y8 | Y9 | Y10 | Y11 | Global | ||||||||
| Time (min) | 53 | 68 | 120 | 120 | 68 | 120 | 120 | 120 | 120 | 120 | 66 | 120 | ||||||||
| Temperature (°C) | 84.94 | 84.94 | 84.94 | 72.55 | 84.94 | 81.19 | 84.94 | 84.94 | 84.87 | 84.87 | 84.94 | 84.94 | ||||||||
| Solvent (%) | 26 | 41 | 45 | 40 | 36 | 35 | 100 | 100 | 100 | 100 | 28 | 76 | −1.6 | 0 | 1.6 | |||||
| Single Opt Value | 5.63 | 14.16 | 2.99 | 0.28 | 22.86 | 30.65 | 127.46 | 28.23 | 2.27 | 174.86 | 20 | |||||||||
| Global Opt Value (mg/g dry weight or mg/g extract) | 3.50 ± 0.43 | 12.55 ± 0.99 | 2.75 ± 0.21 | 0.24 ± 0.02 | 19.04 ± 1.50 | 25.09 ± 2.45 | 100.77 ± 7.19 | 21.19 ± 1.61 | 1.83 ± 0.12 | 148.88 ± 8.73 | 13.70 ± 1.90 | |||||||||
| Desirability | 0.64 | 0.88 | 0.92 | 0.89 | 0.83 | 0.78 | 0.57 | 0.53 | 0.60 | 0.66 | 0.66 | 0.71 | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madureira, J.; Melgar, B.; Santos-Buelga, C.; Margaça, F.M.A.; Ferreira, I.C.F.R.; Barros, L.; Cabo Verde, S. Phenolic Compounds from Irradiated Olive Wastes: Optimization of the Heat-Assisted Extraction Using Response Surface Methodology. Chemosensors 2021, 9, 231. https://doi.org/10.3390/chemosensors9080231
Madureira J, Melgar B, Santos-Buelga C, Margaça FMA, Ferreira ICFR, Barros L, Cabo Verde S. Phenolic Compounds from Irradiated Olive Wastes: Optimization of the Heat-Assisted Extraction Using Response Surface Methodology. Chemosensors. 2021; 9(8):231. https://doi.org/10.3390/chemosensors9080231
Chicago/Turabian StyleMadureira, Joana, Bruno Melgar, Celestino Santos-Buelga, Fernanda M. A. Margaça, Isabel C. F. R. Ferreira, Lillian Barros, and Sandra Cabo Verde. 2021. "Phenolic Compounds from Irradiated Olive Wastes: Optimization of the Heat-Assisted Extraction Using Response Surface Methodology" Chemosensors 9, no. 8: 231. https://doi.org/10.3390/chemosensors9080231
APA StyleMadureira, J., Melgar, B., Santos-Buelga, C., Margaça, F. M. A., Ferreira, I. C. F. R., Barros, L., & Cabo Verde, S. (2021). Phenolic Compounds from Irradiated Olive Wastes: Optimization of the Heat-Assisted Extraction Using Response Surface Methodology. Chemosensors, 9(8), 231. https://doi.org/10.3390/chemosensors9080231












