Label-Free Biosensing Method for the Detection of a Pancreatic Cancer Biomarker Based on Dielectrophoresis Spectroscopy
Abstract
1. Introduction
2. DEP Theory
3. Materials and Methods
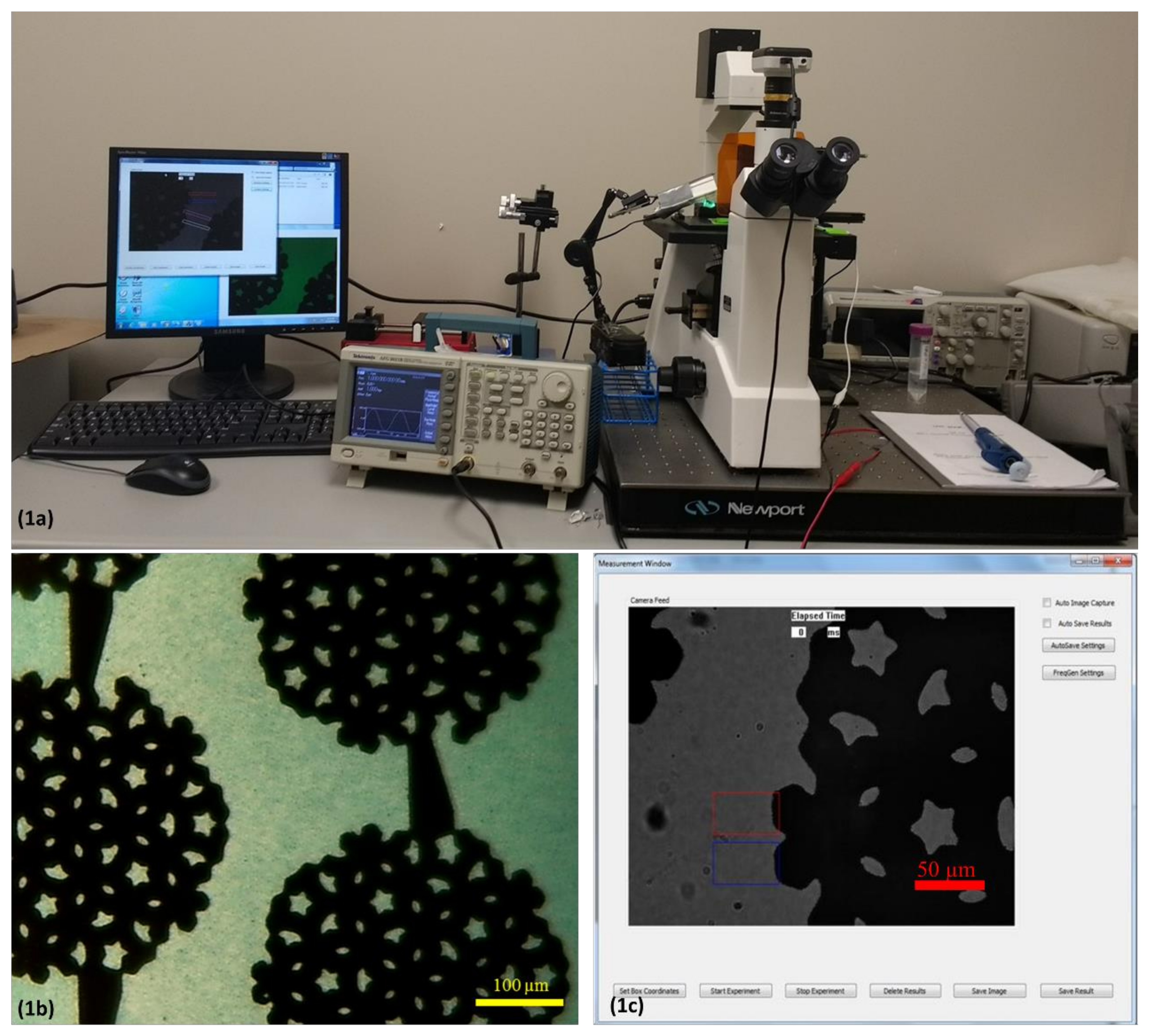
3.1. DEP Spectroscopy Application
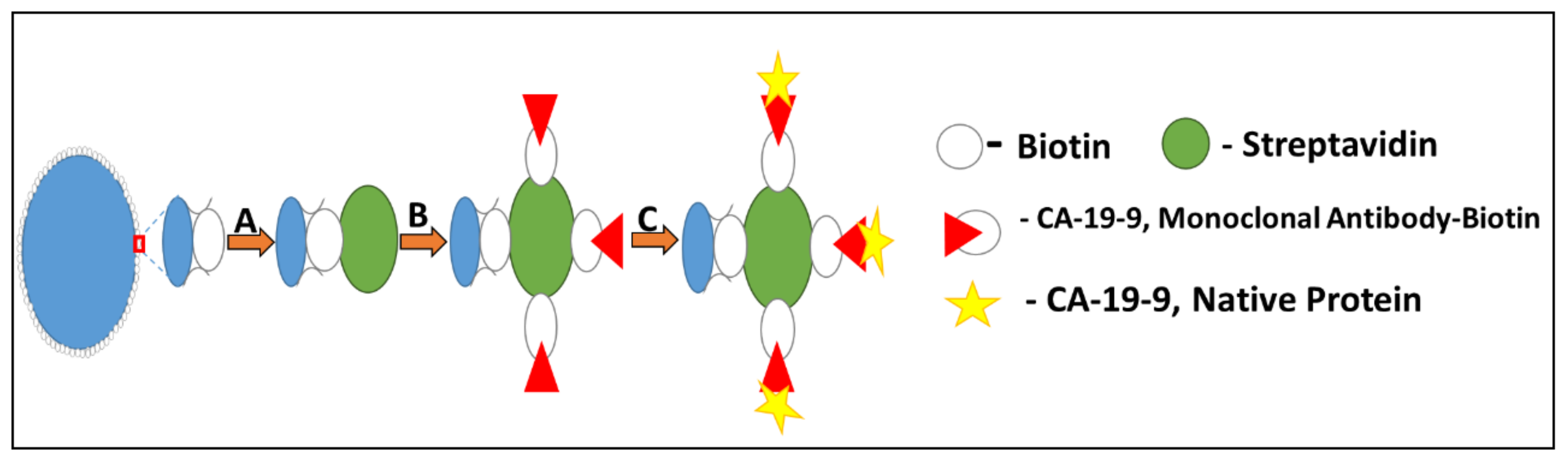
3.2. Sampe Preparation
3.3. Pearl-Shaped Interdigitated Electrode
3.4. Illumination Setup
3.5. Frequency Sweep and Image Processing
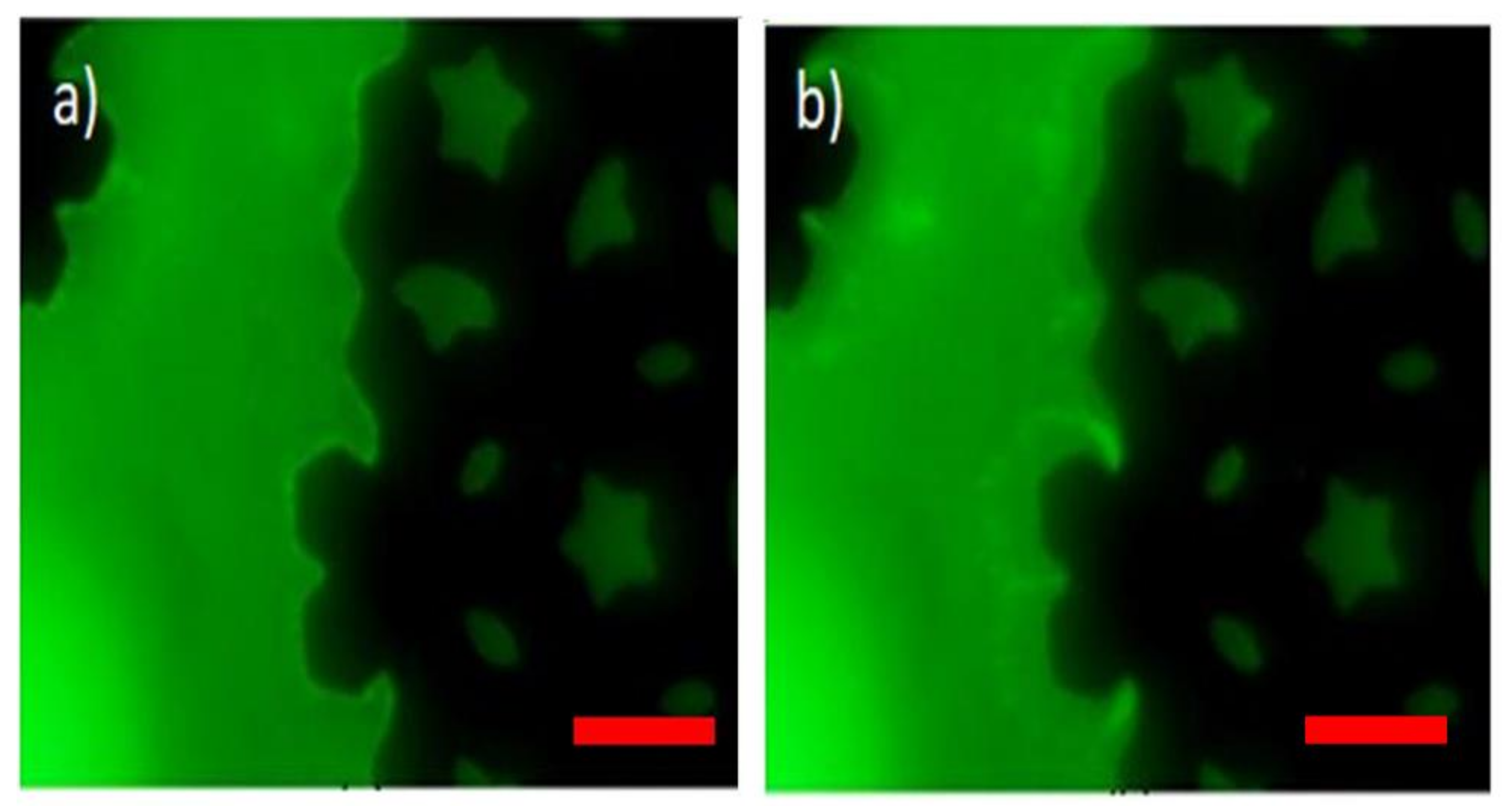
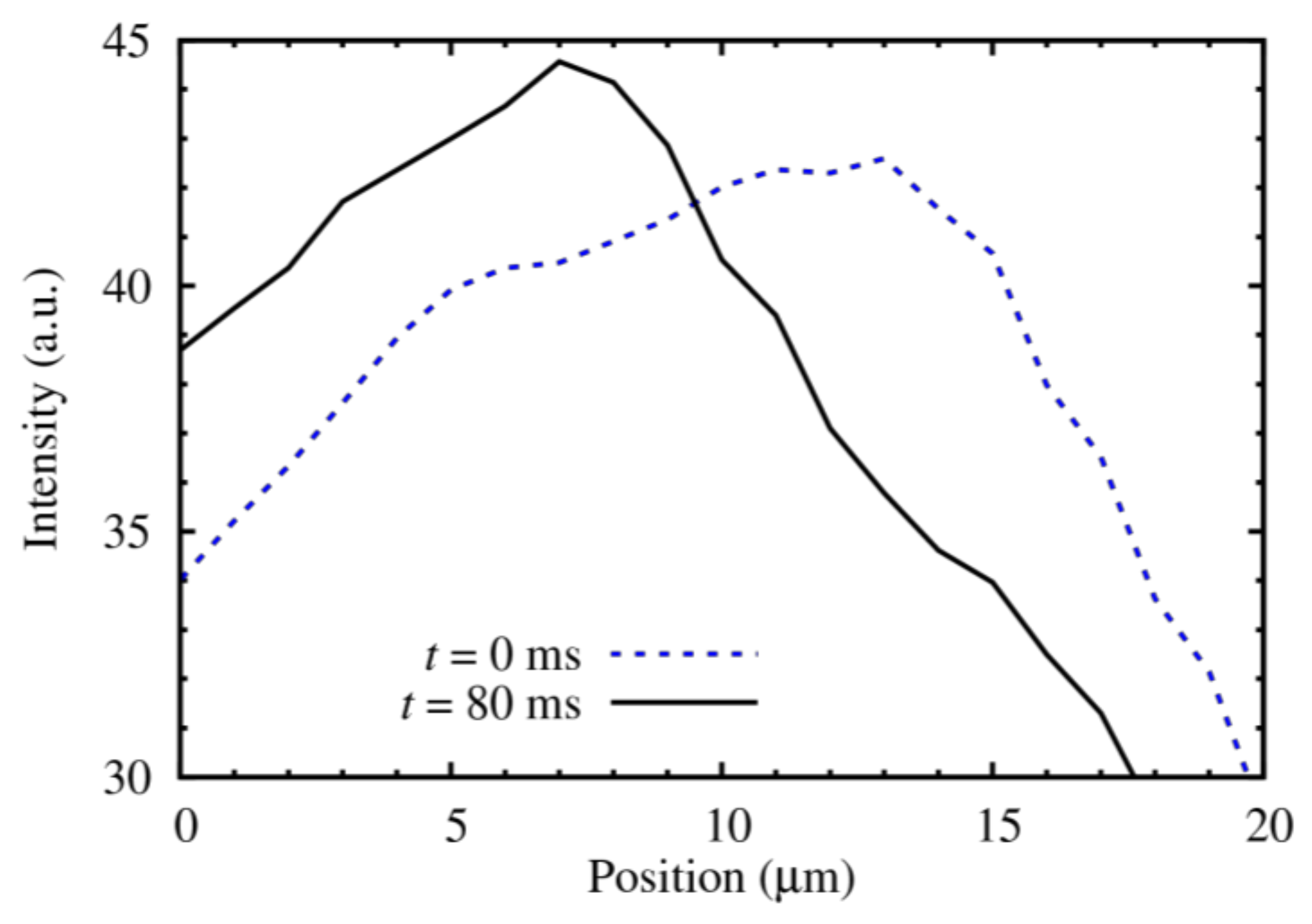
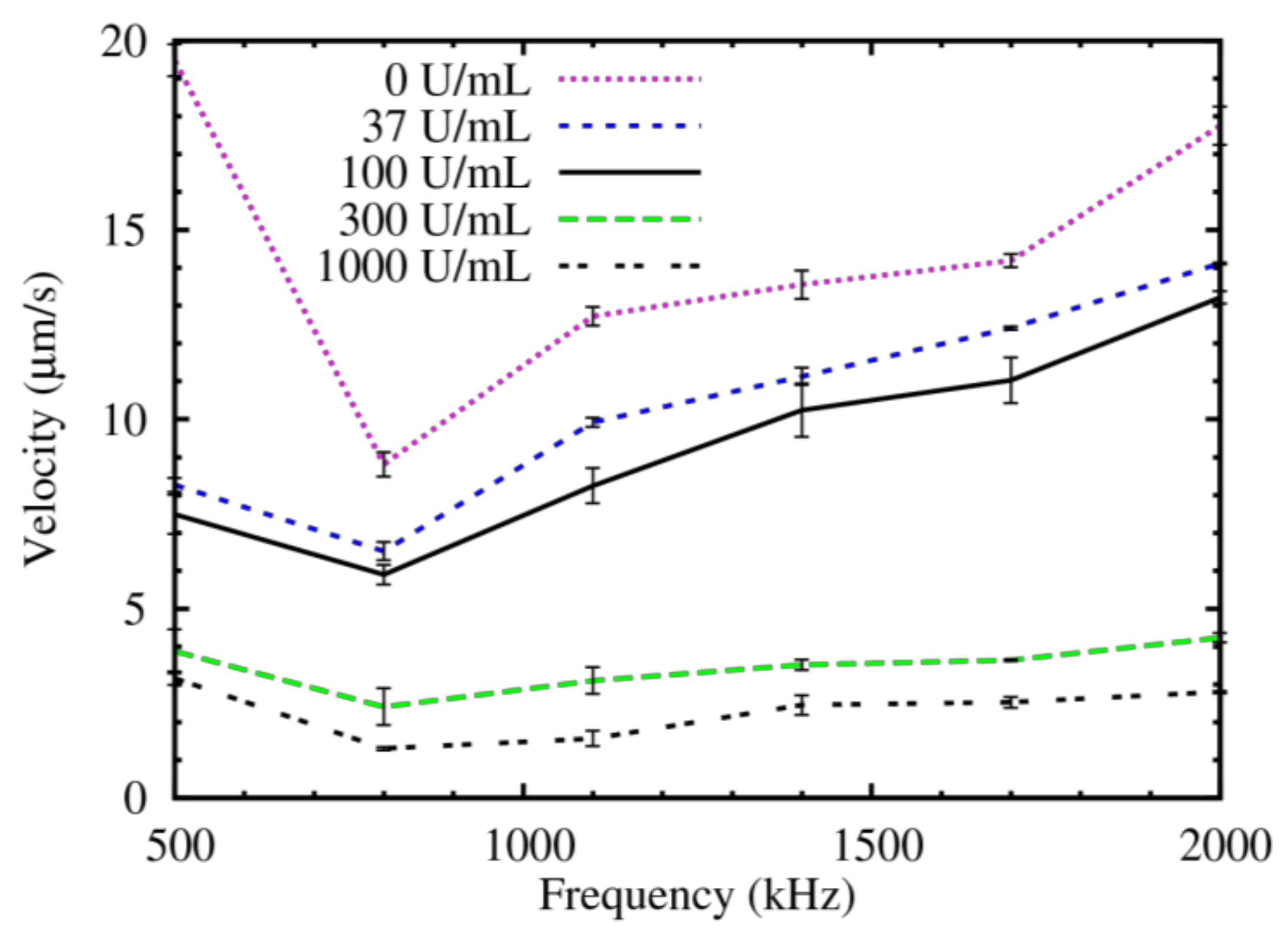
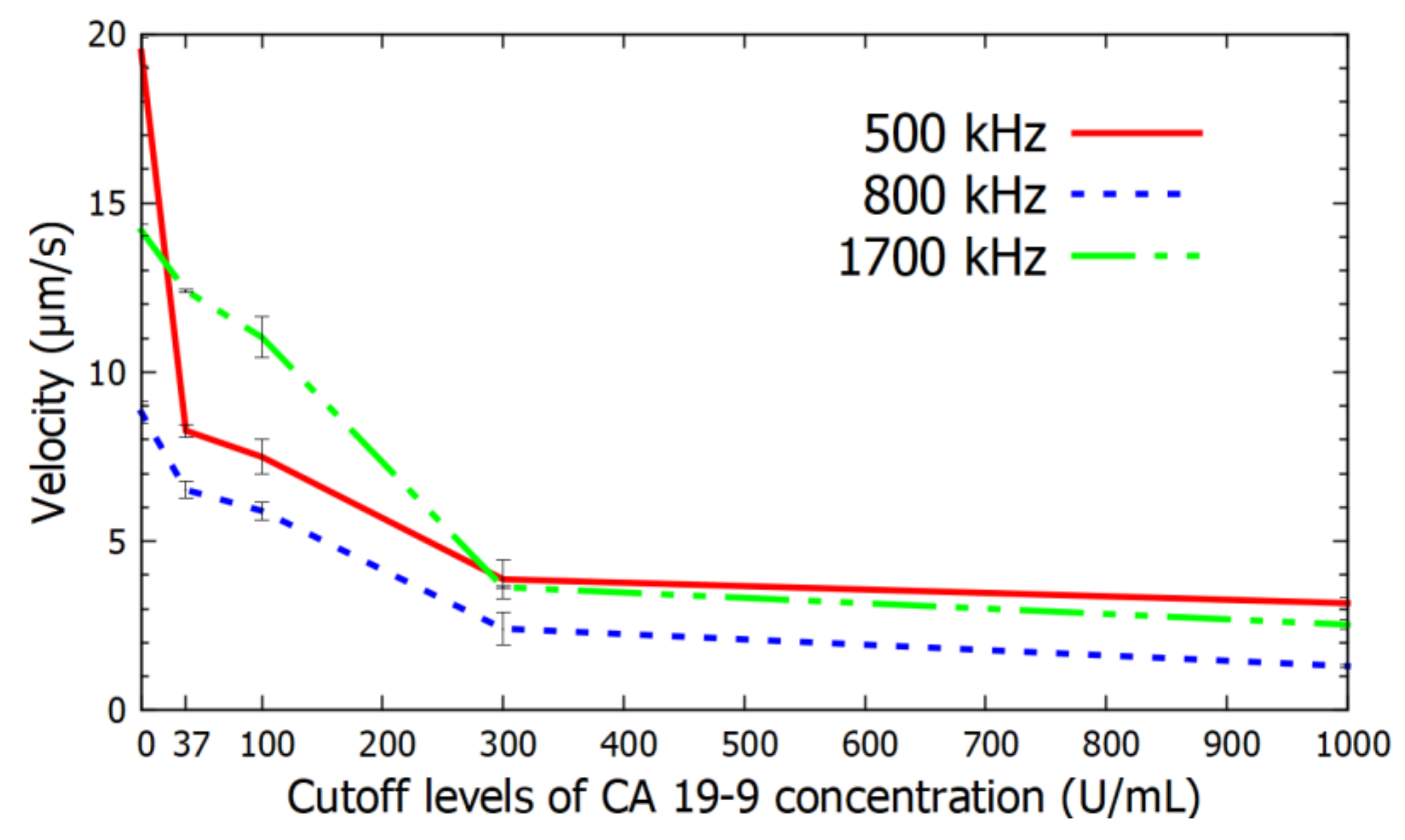
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Muniraj, T.; Jamidar, P.A.; Aslanian, H.R. Pancreatic cancer: A comprehensive review and update. Dis. Mon. 2013, 59, 368–402. [Google Scholar] [CrossRef] [PubMed]
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Del Villano, B.C.; Brennan, S.; Brock, P.; Bucher, C.; Liu, V.; McClure, M.; Rake, B.; Space, S.; Westrick, B.; Schoemaker, H.; et al. Radioimmunometric assay for a monoclonal antibody-defined tumor marker, CA 19-9. Clin. Chem. 1983, 29, 549–552. [Google Scholar] [PubMed]
- Chang, J.C.; Kundranda, M. Novel Diagnostic and Predictive Biomarkers in Pancreatic Adenocarcinoma. Int. J. Mol. Sci. 2017, 18, 667. [Google Scholar] [CrossRef] [PubMed]
- Distler, M.; Pilarsky, E.; Kersting, S.; Grützmann, R. Preoperative CEA and CA 19-9 are prognostic markers for survival after curative resection for ductal adenocarcinoma of the pancreas—A retrospective tumor marker prognostic study. Int. J. Surg. 2018, 11, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Sturgeon, C.; Lamerz, R.; Haglund, C.; Holubec, V.L.; Klapdor, R.; Nicolini, A.; Topolcan, O.; Heinemann, V. Tumor markers in pancreatic cancer: A European Group on Tumor Markers (EGTM) status report. Ann. Oncol. 2009, 21, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Poruk, K.E.; Gay, D.Z.; Brown, K.; Mulvihill, J.D.; Boucher, K.M.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. The Clinical Utility of CA 19-9 in Pancreatic Adenocarcinoma: Diagnostic and Prognostic Updates. Curr. Mol. Med. 2013, 13, 340–351. [Google Scholar] [PubMed]
- Steinberg, W. The Clinical Utility of the CA 19-9 Tumor-Associated Antigen. Am. J. Gastroenterol. 1990, 85, 350–355. [Google Scholar] [PubMed]
- Baryeh, K.; Takalkar, S.; Lund, M.; Liu, G. Development of quantitative immunochromatographic assay for rapid and sensitive detection of carbohydrate antigen 19-9 (CA 19-9) in human plasma. J. Pharm. Biomed. Anal. 2017, 146, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Passerini, R.; Cassatella, M.C.; Boveri, S.; Salvatici, M.; Radice, D.; Zorzino, L.; Galli, C.; Sandri, M.T. The pitfalls of CA19-9: Routine testing and comparison of two automated immunoassays in a reference oncology center. Am. J. Clin. Pathol. 2012, 138, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ozsoz, M.; Liu, G. Gold nanocage-based lateral flow immunoassay for immunoglobulin G. Mikrochim. Acta 2017, 184, 2023–2029. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhang, Y.; Wei, D.; Wang, X.; Yan, T.; Du, B.; Wei, Q. Label-free photoelectrochemical immunosensor for carcinoembryonic antigen detection based on g-C3N4 nanosheets hybridized with Zn0.1Cd0.9S nanocrystals. Sens. Actuators B Chem. 2018, 256, 812–819. [Google Scholar] [CrossRef]
- Chung, J.W.; Bernhardt, R.; Pyun, J.C. Additive assay of cancer marker CA 19-9 by SPR biosensor. Sens. Actuators B Chem. 2006, 118, 28–32. [Google Scholar] [CrossRef]
- Kirmani, S.A.M.; Gudagunti, F.D.; Velmanickam, L.; Nawarathna, D.; Lima, I.T., Jr. Dielectrophoretic spectroscopy using a microscopic electrode array. In Proceedings of the SPIE—BiOS Photonics West, San Francisco, CA, USA, 28 January–2 February 2017; SPIE: San Francisco, CA, USA, 2017; Volume 10068, p. 100680Z. [Google Scholar]
- Kirmani, S.A.M.; Gudagunti, F.D.; Velmanickam, L.; Nawarathna, D.; Lima, I.T., Jr. Negative dielectrophoresis spectroscopy for rare analyte quantification in biological samples. J. Biomed. Opt. 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Abd Rahman, N.; Ibrahim, F.; Yafouz, B. Dielectrophoresis for Biomedical Sciences Applications: A Review. Sensors 2017, 17, 449. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.B. Basic theory of dielectrophoresis and electrorotation. IEEE Eng. Med. Biol. Mag. 2003, 22, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Huang, H.; Chen, L.; Li, X.; Ge, Z.; Chen, T.; Yang, Z.; Sun, L. Dielectrophoresis for Bioparticle Manipulation. Int. J. Mol. Sci. 2014, 15, 18281–18309. [Google Scholar] [CrossRef] [PubMed]
- Pethig, R. Dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics 2010, 4, 1–35. [Google Scholar] [CrossRef]
- Viefhues, M.; Eichhorn, R. DNA dielectrophoresis: Theory and applications a review. Electrophoresis 2017, 38, 1483–1506. [Google Scholar] [CrossRef] [PubMed]
- Lima, I.T., Jr.; Nawarathna, D.; Gudagunti, F.D.; Velmanickam, L. Method for Detecting and Quantifying Biological Molecules Using Dielectrophoresis. Provisional. U.S. Patent Ser. No 62/622,508, 26 January 2018. [Google Scholar]
- Khoshmanesh, K.; Nahavandi, S.; Baratchi, S.; Mitchell, A.; Kalantar-zadeh, K. Dielectrophoretic platforms for bio-microfluidic systems. Biosens. Bioelectron. 2011, 26, 1800–1814. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, T.; Washizu, M. Dielectrophoretic detection of molecular bindings. IEEE Trans. Ind. Appl. 2001, 37, 1625–1633. [Google Scholar] [CrossRef]
- Ramón-Azcón, J.; Yasukawa, T.; Mizutani, F. Sensitive and spatially multiplexed detection system based on dielectrophoretic manipulation of DNA-encoded particles used as immunoreactions platform. Anal. Chem. 2011, 83, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Viefhues, M.; Regtmeier, J.; Anselmetti, D. Fast and continuous-flow separation of DNA-complexes and topological DNA variants in microfluidic chip format. Analyst 2013, 138, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Velmanickam, L.; Laudenbach, D.; Nawarathna, D. Dielectrophoretic label-free immunoassay for rare-analyte quantification in biological samples. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2016, 94, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Pethig, R.; Smith, S. Introductory Bioelectronics for Engineers and Physical Scientists; Willey: West Sussex, UK, 2013; ISBN 9781119970873. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dackson Gudagunti, F.; Velmanickam, L.; Nawarathna, D.; Lima, I.T., Jr. Label-Free Biosensing Method for the Detection of a Pancreatic Cancer Biomarker Based on Dielectrophoresis Spectroscopy. Chemosensors 2018, 6, 33. https://doi.org/10.3390/chemosensors6030033
Dackson Gudagunti F, Velmanickam L, Nawarathna D, Lima IT Jr. Label-Free Biosensing Method for the Detection of a Pancreatic Cancer Biomarker Based on Dielectrophoresis Spectroscopy. Chemosensors. 2018; 6(3):33. https://doi.org/10.3390/chemosensors6030033
Chicago/Turabian StyleDackson Gudagunti, Fleming, Logeeshan Velmanickam, Dharmakeerthi Nawarathna, and Ivan T. Lima, Jr. 2018. "Label-Free Biosensing Method for the Detection of a Pancreatic Cancer Biomarker Based on Dielectrophoresis Spectroscopy" Chemosensors 6, no. 3: 33. https://doi.org/10.3390/chemosensors6030033
APA StyleDackson Gudagunti, F., Velmanickam, L., Nawarathna, D., & Lima, I. T., Jr. (2018). Label-Free Biosensing Method for the Detection of a Pancreatic Cancer Biomarker Based on Dielectrophoresis Spectroscopy. Chemosensors, 6(3), 33. https://doi.org/10.3390/chemosensors6030033






