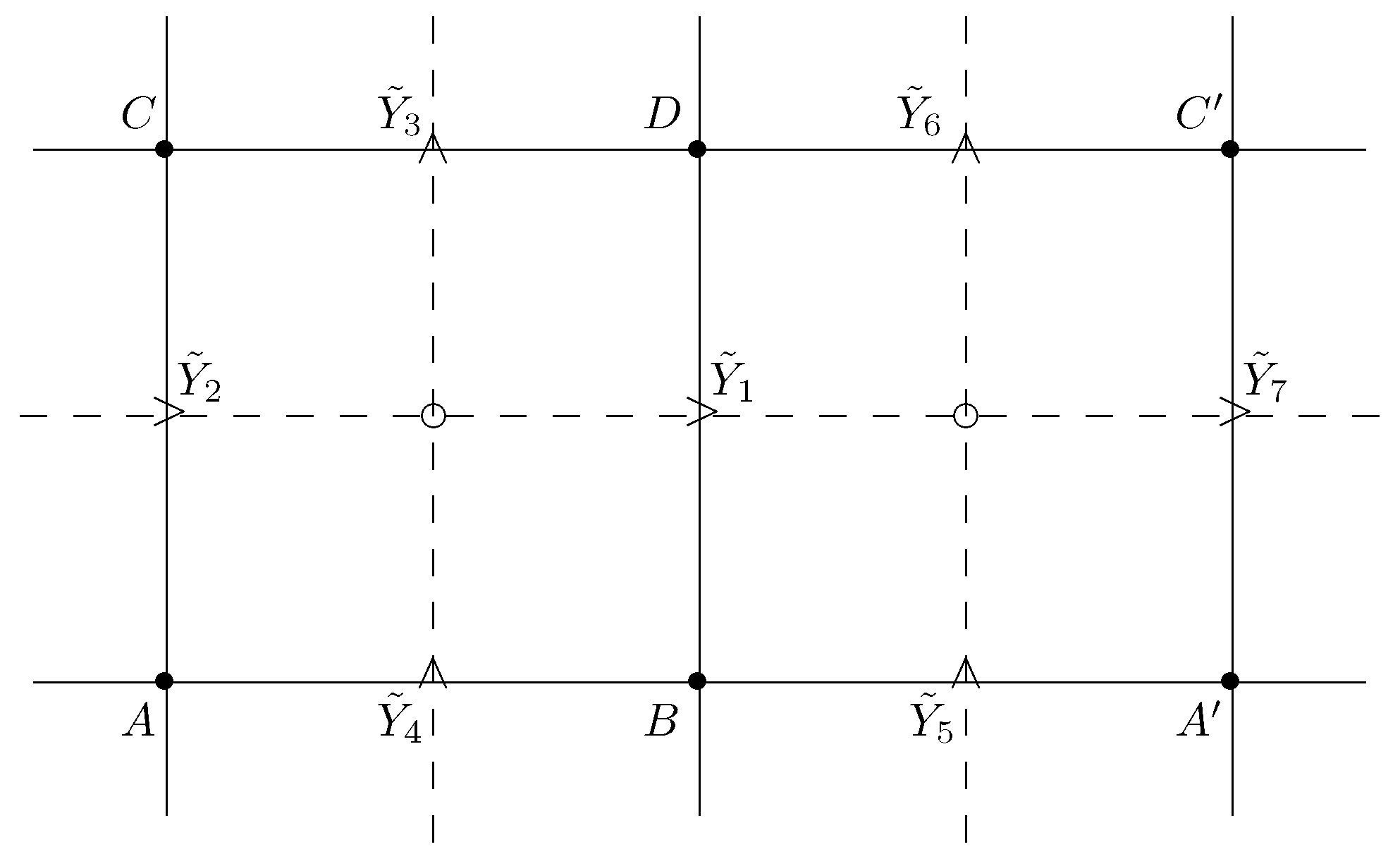1. Introduction
A
Bayesian network is a probabilistic graphical model that represents a set of variables and their conditional dependencies via a directed acyclic graph [Wikipedia]. Bayesian networks are a fundamental concept in learning and artificial intelligence, see [
1,
2,
3]. Formally, a Bayesian network is a random field
indexed by sites
of a directed acyclic graph (DAG)
whose probability distribution writes as a product of conditional probabilities of
given its parent variables
, where
is the set of parents of
v.
A Bayesian network is a special case of Markov random field on a (generally undirected) graph
. Markov random fields include nearest-neighbor Gibbs random fields and play an important role in many applied sciences including statistical physics and image analysis [
4,
5,
6,
7].
As noted in [
8,
9,
10] and elsewhere, manipulating the marginals (e.g., computing the mean or the covariance function) of a Gibbs random field is generally very hard when
V is large. A class of Markov random fields on
which avoids this difficulty was proposed by Pickard [
10,
11] (the main idea of their construction belongs to Verhagen [
12], see [
11] (p. 718)). Pickard and related unilateral random fields have found useful applications in image analysis, coding, information theory, crystallography and other scientific areas, not least because they allow for efficient simulation procedures [
8,
9,
13,
14,
15,
16].
A Pickard model
(rigorously defined in
Section 2) is a family of Bayesian networks on rectangular graphs
enjoying the important (Kolmogorov) consistency property: for any rectangles
we have that
where
is the distribution of
and
is the restriction of marginal of
on
. Property (
1) implies that the Pickard model extends to a stationary random field on
whose marginals coincide with
and form Markov chains on each horizontal or vertical line.
Arak et al. [
17,
18] introduced a class of polygonal Markov graphs and random fields
indexed by continuous argument
satisfying a similar consistency property: for any bounded convex domains
, the restriction of
to
coincides in distribution with
. The above models allow a Gibbs representation but are constructed via equilibrium evolution of a one-dimensional particle system with random piece-wise constant Markov velocities, with birth, death and branching. Particles move independently and interact only at collisions. Similarity between polygonal and Pickard models was noted in [
19]. Several papers [
20,
21,
22] used polygonal graphs in landscape modeling and random tesselation problems.
The present paper discusses an extension of polygonal graphs and the Pickard model, satisfying a consistency property similar to (
1) but not directly related to lattices or rectangles. The original idea of the construction (communicated to the author by Taivo Arak in 1992) referred to a particle evolution on the three-dimensional lattice
. In this paper, the above Arak model is extended to any dimension and discussed in
Section 4 in detail. It is a special case of a
Markov edge process with probability distribution
introduced in
Section 3 and defined on edges (arcs)
of a directed acyclic graph (DAG)
as the product over
of the conditional clique probabilities
, somewhat similarly to a Bayesian network, albeit the conditional probabilities refer to collection
of ‘outgoing’ clique variables and not to a single ‘child’
as in a Bayesian network. Markov random fields indexed by edges discussed in the literature [
5,
6,
7] are usually defined on undirected graphs through Gibbs representation and experience a similar difficulty of computing their marginal distributions as Gibbsian site models. On the other hand, ‘directed’ Markov edge processes might be useful in causal analysis where edges of DAG
represent ‘decisions’ and
can be interpreted as the (random) ‘cost’ of ‘decision’
.
The main result of this work is Theorem 2, providing sufficient conditions for consistency
of a family
of Markov edge processes defined on DAGs,
, with the partial order
given by
The sufficient conditions for (
2) are expressed in terms of clique distributions
and essentially reduce to the
marginal independence of incoming and outgoing clique variables (i.e., collections
and
), see (
33) and (
34).
The class
of DAGs
satisfying (
2) is of special interest. In Theorem 2,
is the class of all sub-DAGs obtained from a given DAG
by a
mesh dismantling algorithm (MDA). The above algorithm starts by erasing any edge that leads to a sink
or comes from a source
, and proceeds in the same way and in an arbitrary order, see Definition 4. The class
contains several interesting classes of ‘one-dimensional’ and ‘multi-dimensional’ sub-DAGs
on which the restriction in (
2) is identified as a Markov chain or a sequence of independent r.v.s (Corollary 2).
Section 4 discusses consistent Markov edge processes on ‘rectangular’ subgraphs of
endowed with nearest-neighbor edges directed in the lexicographic order. We discuss properties of such edge processes and present several examples of ‘generic’ (common) clique distributions, with particular attention paid to the binary case (referred to as the Arak model). In dimension
we provide a detailed description of the Arak model in terms of a particle system moving along the edges of
. Finally,
Section 5 establishes a relation between the Pickard and Arak models, the latter model identified in Theorem 3 as a contour model of the former model under an additional symmetry assumption.
We expect that the present work can be extended in several directions. Similarly to [
8,
9] and some other related studies, the discussion is limited to discrete probability distributions, albeit continuous distributions (e.g., Gaussian edge processes) are of interest. A major challenge is the application of Markov edge processes to causal analysis and Bayesian inference, bearing in mind the extensive research in the case of Bayesian networks [
1,
2]. Consistent Markov edge processes on regular lattices may be of interest to pattern recognition and information theory. Some open problems are mentioned in Remark 7.
2. Bayesian Networks and Pickard Random Fields
Let
be a given DAG. A directed edge from
to
, is denoted as
. With any vertex
we can associate three sets of edges
representing the
incoming edges,
outgoing edges and all edges incident to
v. A vertex
is called a
source or a
sink if
or
, respectively. The partial order (reachability relation)
on
G means that there is a directed path on this DAG from
to
. We write
if
The above partial order carries over to edges of
G, namely,
Following [
2], the set of vertices
is called the
parents of v, whereas
is called the
family of v.
Definition 1. Let be a given DAG. We call family distribution at
any discrete probability distribution , that is, a sequence of positive numbers summing up to 1:The set of all family distributions at is denoted by . For
, the conditional probability of
given parent configuration
writes as
with
if
and
if
.
Definition 2. Let be a set of family distributions on DAG . We call a Bayesian network on
G corresponding to
a random process indexed by vertices (sites) of G and such that for any configuration We remark that the terminology ‘family distribution’ is not commonplace, whereas the definition of Bayesian network varies in the literature [
1,
2,
23], being equivalent to (
5) under the positivity condition
In the simplest case of chain graph
,
(Example 2 below), a Bayesian network
is a Markov chain with transition probabilities
; particularly, any non-Markovian site process
on the above
G is not a Bayesian network. In the general case
, the conditional (family) distributions
in (
5) are most intuitive and often used instead of
. A Bayesian network satisfies several Markov conditions [
2], particularly, the
ordered Markov condition:
The Markov properties of site models on directed (not necessary acyclic) graphs are discussed in Lauritzen et al. [
24] and other works.
In the rest of this subsection,
is a directed subgraph of the infinite DAG
with lexicographic partial order
and edge set
The class of ‘rectangular’ subDAGs
with
will be denoted
. Given two ‘rectangular’ DAGs
, we write
if
. For the generic family in
Figure 1, right, the family distribution is is indexed by the elements of the table
and written as the joint distribution
of four r.v.s
with
,
. The marginal distributions of these r.v.s are designated by the corresponding subsets of the table (
8), e.g.,
is the distribution of r.v.
A. The conditional distributions of these r.v.s are denoted as in (
4); viz.,
is the conditional distribution of
B given a value of
A.
Definition 3. We call a Pickard model a Bayesian network on DAG with family distribution independent of and satisfying the following conditions:(stationarity) and(conditional independence). In a Pickard model, the family
of
consists of four points, see
Figure 1, except when
v belongs to the left lower boundary
of rectangle
V in (
7) and
; for
it consists of the single point
. Accordingly, the family distributions
are marginals of the generic distribution
in Definition 3. We note that Pickard model (also called Pickard random field) is defined in [
8,
9,
10,
16] in somewhat different ways, which are equivalent to Definition 3. Let
be the class of all rectangles
V in (
7).
Theorem 1 ([
10]).
The family of Pickard models on DAGs and corresponding to the same generic family distribution ϕ in (9) and (10) is consistent; in other words, for any we have that Remark 1. The consistency property in (11) implies that for in (7), and any , the restriction of a Pickard model on the horizontal interval is a Markov chainwith initial distribution and transition probabilities . Indeed, is the set of vertices of , and the corresponding ‘one-dimensional’ Pickard model on is defined by (12). In a similar way, the restriction of a Pickard model on a vertical interval is a Markov chainwith initial distribution and transition probabilities . Ref. [10] (Thm. 3) extend the Markov chain identifications in (12) and (13) to any non-increasing undirected path in V. This fact and the discussion in Section 3 and Section 5 suggest that the Pickard model and the consistency property in (11) can be extended to non-rectangular index sets . Remark 2. In the binary case or , the distribution ϕ is completely determined by probabilitiesIn terms of (14), conditions (9) and (10) translate tosee [10] (p. 666). The special cases , and correspond to a degenerated Pickard model, the latter two implying and , respectively, and leading to a random field which takes constant values on each horizontal (or vertical) line in . Hence, a binary Pickard model can be parametrized by seven parameters,as in the non-degenerated case can be found from :The parameters in (16) satisfy natural constraints resulting from their definition as probabilities, particularly, . 3. Markov Edge Process: General Properties and Consistency
It is convenient to allow
G to have isolated vertices, which of course can be removed w.l.g., our interest primarily being focused on edges. The
directed line graph of DAG
is defined as the graph whose set of vertices is
is the same as that of an undirected line graph, and the set of (directed) edges is
Note that
is acyclic, and hence a DAG.
Given a DAG , we denote as the class of all non-empty sub-DAGs (subgraphs) with .
Definition 4. A transformation is said to be a top mesh dismantling algorithm (top MDA) if it erases an edge leading to a sink ; in other words, if withSimilarly, a transformation is said to be a bottom mesh dismantling algorithm (bottom MDA) if it erases an edge starting from a source , in other words, if with We denote as
and
the classes of DAGs containing
G and the subDAGs
which can be obtained from
G by succesively applying a top MDA, bottom MDA, or both types of MDA in an arbitrary order:
Note that the above transformations may lead to a subDAG
containing isolated vertices which can be removed from
w.l.g.
Figure 2 illustrates that
in general.
Definition 5. A subDAG is said to be the following:
- (i)
for some ;
- (ii)
A chain DAG if , for some
- (iii)
A source-to-sink DAG if any edge connects a source of to a sink of .
The corresponding classes of subDAGs in Definition 5 (i)–(iii) will be denoted by and .
Proposition 1. For any DAG we have that Proof. The proof of each inclusion in (
20) proceeds by induction on the number
of edges of
G. Clearly, the proposition holds for
. Assume it holds for
; we will show that it holds for
The induction step
for the three relation in (
20) is proved as follows.
- (i)
Fix
and
to be as in (
19). Let
be the set of sinks of
G. If
is a sink and
(or
), then
belongs to
by definition of the last class. If
is a sink and
, we can dismantle an edge
, and the remaining graph
in (
18) has
edges and contains
, meaning the inductive assumption applies, proving
.
Next, let
. Then there is a path in
G from
to at least one of these sinks. If
, we apply a top MDA to
and see that
in (
18) has the number of edges
and contains the interval graph
in (
19), so that the inductive assumption applies to
and consequently to
G as well, as before, proving the induction step in case (i).
- (ii)
Let
be a chain between
and
, since
belongs to
by definition. Let
be the set of sinks of
G. If
is a sink and
then
G contains an edge
which does not belong to
. Then, by removing
e from
G by top MDA we see that
in (
18) contains the chain
viz.,
, and therefore
by the inductive assumption. If
is a sink and
, we remove from
G any edge leading to
and arrive at the same conclusion. If
is not a sink, we remove any edge leading to a sink and apply the inductive assumption to the remaining graph
having
edges. This proves the induction step in case (ii).
- (iii)
Let
,
where
and
are the sets of sources and sinks of
, respectively. Let
and
be the sets of sources and sinks of
. If
, i.e.,
G is a source-to-sink DAG, we can remove from it an edge
and the remaining graph
in (
18) contains
and satisfies the inductive assumption. If
G is not a source-to-sink DAG, it contains a sink
or a source
In the first case, there is
, which can be removed from
G, and the remaining graph
contains
and satisfies the inductive assumption. The second case follows from the first one by DAG reversion. This proves the induction step
in case (iii), hence the proposition.
□
An edge process on DAG is a family of discrete r.v.s indexed by edges of G. It is identified with a (discrete) probability distribution
Definition 6. Let be a given DAG. A clique distribution at is any discrete probability distribution , that is, a family of positive numbers summing up to 1:The set of all clique distributions at is denoted by . Given
, the conditional probabilities of out-configuration
given in-configuration
write as
with
and
for
for
.
Definition 7. A Markov edge process on a DAG corresponding to a given family of clique distributions is a random process indexed by edges of G and such that for any configuration An edge process on a DAG can be viewed as a site process on the line graph of , with
Corollary 1. A Markov edge process in (21) is a Bayesian network on the line DAG if Condition (
22) can be rephrased as the statement that ’outgoing’ variables
are conditionally independent given ‘ingoing’ variables
for each node
. Analogously,
in (
21) is a Bayesian network on the reversed line graph
under a symmetric condition that ‘ingoing’ variables
are conditionally independent given ‘outgoing’ variables
, for each node
.
Definition 8. Let be a DAG and be a family of subDAGs of G. A family of edge processes is said consistent if Given edge process
in (
21), we define its restriction on a subDAG
as
In general,
is not a Markov edge process on
, as shown in the following example.
Example 1. Let Then andThe subDAG is composed of two edges going from different sources 1 and 2 to the same sink 3. By definition, a Markov edge process on corresponds to independent , viz.,It is easy to see that the two probabilities in (25) and (26) are generally different (however, they are equal if is a product distribution and , in which case and In Example 1, the restriction to is a Markov edge process with . The following proposition shows that a similar fact holds in a general case for subgraphs obtained from G by applying a top MDA.
Proposition 2. Let be a Markov edge process on DAG . Then for any , the restriction is a Markov edge process on with clique distribution given bywhich is the restriction of clique distributions to (sub-clique) Proof. It suffices to prove the proposition for a one-step top MDA, or
in (
18). Let
,
,
. From the definitions in (
21) and (
27),
□
Example 2 (Markov chain).
Let be a chain from 0 to n. The classes and consist respectively of all chains from 0 to , from to n and from i to j ( A Markov edge process on the above graph is a Markov chain with the probability distributionwhere is a (discrete) univariate and are bivariate probability distributions; are conditional or transitional probabilities. It is clear that the restriction on is a Markov chain with ; in other words, it satisfies Proposition 2 and (27). However, the restriction to or is a Markov chain with the initial distributionwhich is generally different from . We conclude that Proposition 2 ((27) in particular) fails for subDAGs of G obtained by a bottom MDA. On the other hand, hold for the above provided the values satisfy the additional compatibility condition: Proposition 3. Let be a Markov edge process on DAG in (21). Then for any edge andwith satisfying . Proof. Relation (
28) follows from
and the definition in (
21), since the products
cancel in the numerator and the denominator of (
30).
Consider (
29). We use Propositions 1 and 2, according to which the interval DAGs
belong to
. The ‘intermediate’ DAG
, constructed from
by adding the single edge
, viz.,
, also belongs to
since it can be attained from
by dismantling all edges with the exception of
. Note that
. Therefore, by Proposition 2,
where
is a Markov edge process on
with clique distributions
The expression in (
29) follows from (
31) and (
28) with
E replaced by
by noting that
is a sink in
; hence,
, whereas
. □
Remark 3. Note that in the conditional probability on the r.h.s. of (28) are nearest neighbors of e in the line graph (DAG) Theorem 2. Let be a given DAG and a Markov edge process in (21) with clique distributions satisfying the compatibility conditionand two marginal independence conditions:andThen the family of Markov edge process with clique distributions given in (27) is consistent, viz., Remark 4. - (i)
Conditions (33) and (34) do not imply the mutual independence of the in- and out-clique variables and under . - (ii)
Conditions (33) and (34) are automatically satisfied if (Markov chain). - (iii)
Conditions (33) and (34) are symmetric with regard to DAG reversion (all directions reversed). - (iv)
In the binary case (), the value can be interpreted as the presence of a ‘particle’ and as its absence on edge . ‘Particles’ ‘move’ on a DAG in the direction of arrows. ‘Particles’ ‘collide’, ‘annihilate’ or ‘branch’ at nodes , with probabilities determined by clique distribution . See Section 4 for a detailed description of particle evolution for the Arak model on .
Proof of Theorem 2. It suffices to prove (
35) for a one-step MDA:
which remove a single edge
coming from a source
and a single edge
going to a sink
, respectively. Moreover, it suffices to consider
only. The proof for
follows from Proposition 2 and does not require (
32)–(
34). (It also follows from
by DAG reversion.) Then, by the marginal independence of
and
,
From the definition of
in (
27), we have that
, and therefore
leading to
and proving (
35) for
defined above, or the statement of the theorem for a one-step MDA
. □
Corollary 2. Let be a Markov edge process on DAG satisfying the conditions of Theorem 2:
- (i)
The restriction of on a chain DAG , is a Markov chain, viz., - (ii)
The restriction of on a source-to-sink DAG is a sequence of independent r.v.s, viz., where is the set of sources of and is the set of edges coming from a source and ending into a sink of ,
Proof. - (i)
By Proposition 1,
belongs to
so that Theorem 2 applies with
in (
27) given by (
31), resulting in (
36).
- (ii)
By Proposition 1,
belongs to
so that Theorem 2 applies with
in (
27) given by
(the second equality holds by (
33)), resulting in (
37).
□
Remark 5. A natural generalization of chain and source-to-sink DAGs is a source–chain–sink
DAG with the property that any sink is reachable from a source by a single chain (directed path). We conjecture that for a source–chain–sink DAG , the restriction of in Theorem 2 is a product of independent Markov chains on disjointed directed paths of , in agreement with representations (36) and (37) of Corollary 2. Gibbsian representation of Markov edge processes. Gibbsian representation is fundamental in the study of Markov random fields [
4,
6]. Gibbsian representation of Pickard random fields was discussed in [
8,
9,
11,
12]. The following Corollary 3 provides Gibbsian representation of the consistent Markov edge process in Theorem 2. Accordingly, the set of vertices of DAG
is written as
, where the boundary
consists of sinks and sources of
G and the interior
of the remaining sites.
Corollary 3. Let be a Markov edge process on DAG satisfying the conditions of Theorem 2 and the positivity condition . Thenwhere the inner and boundary potentials are given by Formula (
38) follows by writing (
21) as
and rearranging terms in the exponent using (
32)–(
34). Note (
38) is invariant with regard to graph reversal (direction of all edges reversed). Formally, the inner potentials
in (
39) do not depend on the orientation of
G, raising the question of the necessity of conditions (
33) and (
34) in Theorem 2. An interesting perspective seems to be the study of the Markov evolution
of Markov edge processes on DAG
with invariant Gibbs distribution in (
38).
4. Consistent Markov Edge Process on and Arak Model
Let
be an infinite DAG whose vertices are points of regular lattice
and whose edges are pairs
directed in the lexicographic order
if and only if
. We write
if
. A
-dimensional hyperplane
can be identified with
for any fixed
.
Let
denote the class of all finite ‘rectangular’ subgraphs of
, viz.,
if
for some
and
We denote
the class of all subgraphs
of
formed by intersection
with a
-dimensional hyperplane
that can be identified with an element of
. Particularly, for
and any
,
and the subgraph
can be identified with a chain
of length
. Similarly, for any
,
and the subgraph
of
is a planar ‘rectangular’ graph belonging to
. Note that any subgraph
is an interval subDAG of
in the sense of Definition 5 (i). From Proposition 1 we obtain the following corollary.
Corollary 4. Let . Any subgraph can be obtained by applying an MDA to G.
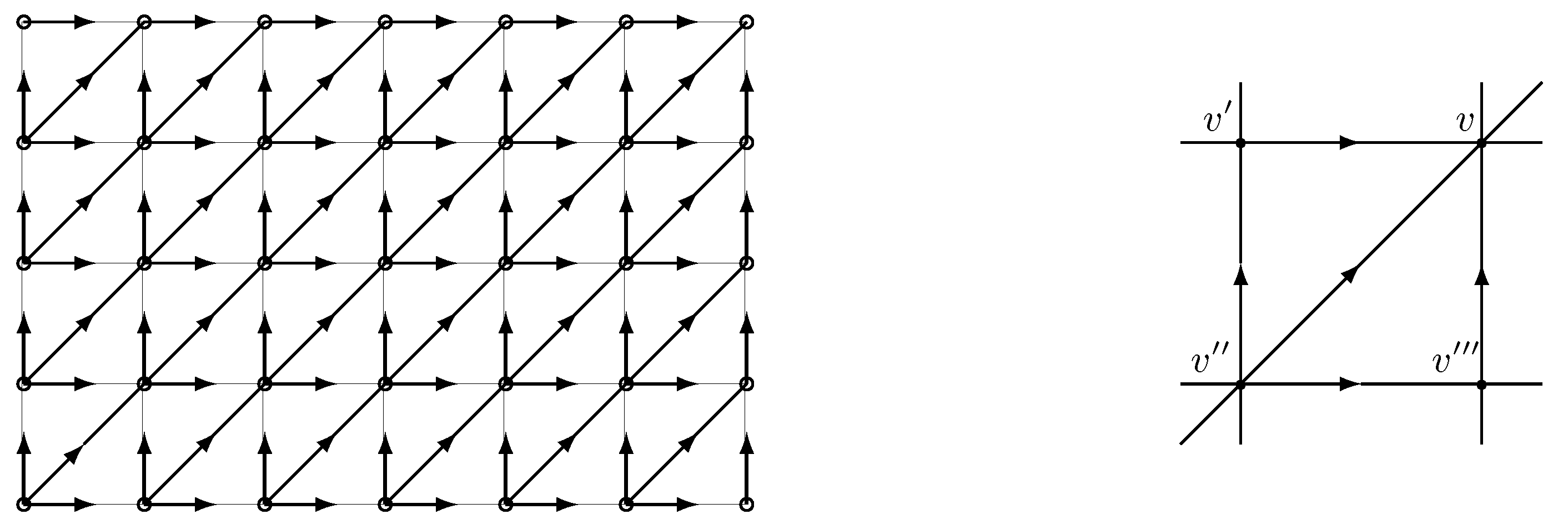
We designate
‘outgoing’ and
‘incoming’ clique variables (see
Figure 3). Let
be a generic clique distribution of random vector
on incident edges of
. We use the notation
.
The compatibility and consistency relations in Theorem 2 write as follows for any
and
Let
be the class of all discrete probability distributions
on
satisfying (
45) and (
46).
Corollary 5. Let be a (consistent) Markov edge process on DAG with clique distribution . The restrictionof on any -dimensional hyperplane in (40) is a consistent Markov edge process on DAG with generic clique distribution . Particularly, the restrictionof to a one-dimensional hyperplane in (40) is a reversible Markov chain with the probability distributionwhere and are the conditional probabilities. Example 3. Broken line process. Let and take integer values and have a joint distribution, where is a parameter. Note that for and the last equality is valid for , too. Therefore, π in (50) is a probability distribution on with (marginals) and written as the product of the geometric distribution with parameter . We see that (50) belongs to and satisfies (45) and (46) of Corollary 5. The corresponding edge process called the discrete broken line process was studied by Rollo et al. [25] and Sidoravicius et al. [26] in connection with planar Bernoulli first passage percolation. The discrete broken line process can be described as an evolution of particles moving (horizontally or vertically) with constant velocity until collision with another particle and dying upon collision, with independent immigration of pairs of particles. An interesting and challenging open problem is the extension of the broken line process to higher dimensions, particularly to or . Definition 9. We call an Arak model a binary Markov edge process on a DAG with clique distribution .
We also use the same terminology for the restriction of an Arak model on a subDAG
, and speak about an
Arak model on since
extends to infinite DAG
; the extension
is a stationary binary random field whose restriction coincides with
. Note that when
, the clique distribution
is determined by
probabilities
Particularly, for
the compatibility and consistency relations in (
45) and (
46) read as
The resulting consistent Markov edge process on
depends on
parameters. In the lattice isotropic case it depends on nine parameters
satisfying two consistency equations,
and
, resulting in a seven-parameter isotropic binary edge process communicated to the author by T. Arak.
Denote
unit vectors in
,
—edges parallel to vectors
The following corollary is a consequence of Corollary 5 and the formula for the transition probabilities of a binary Markov chain in Feller [
27] (Ch.16.2).
Corollary 6. The covariance function of an Arak model. Let be an Arak model on DAG . Then for any , ,where are as in (51). Remark 6. The conditional probabilities in Corollary 1 (22) for an Arak model can be expressed through in (51). Particularly, an Arak model in dimension is a Bayesian network on the line graph of if In the two-dimensional case
, the Arak model on DAG
is determined by the clique distribution
of
in
Figure 3, middle, with probabilities
satisfying four conditions:
The resulting edge process depends on 11 = (15 − 4) parameters. In the lattice isotropic case, we have five parameters
satisfying a single condition
and leading to a four-parameter consistent isotropic binary Markov edge process on
. Some special cases of parameters in (
54) are discussed in Examples 4–6 below.
The evolution of the particle system. Below, we describe the binary edge process
in Remark 2 on DAG
in terms of particle system evolution. The description becomes somewhat simpler by embedding
G into
, where
where
and
are boundary sites (sources or sinks belonging of the extended graph
), as shown in
Figure 4. The edge process
is obtained from
as
The presence of a particle on edge
(in the sense explained in
Section 2) is identified with
. A particle moving on a horizontal or vertical edge is termed horizontal or vertical, respectively. The evolution of particles is described by the following rules:
- (p0)
Particles enter the boundary edges (vertically) and (horizontally) independently of each other with respective probabilities and .
- (p1)
Particles move along directed edges of independently of each other until collision with another moving particle. A vertical particle entering an empty site will undergoes one of the following transformations.
- (i)
Leaves v as a vertical particle with probability ;
- (ii)
Changes the direction at v to horizontal with probability
- (iii)
Branches at v into two particles moving into different directions, with probability ;
- (iv)
Dies at v, with probability .
Similarly, a horizontal particle entering an empty site exhibits transformations as in (i)–(iv) with respective probabilities , and .
- (p2)
Two (horizontal and vertical) particles entering , either
- (i)
both die with probability or
- (ii)
the horizontal one survives and the vertical one dies with probability . or
- (iii)
the vertical one survives and the horizontal one dies with probability . or
- (iv)
both particles survive with probability .
- (p3)
At an empty site (no particle enters v), one of the following events occurs:
- (i)
A single horizontal particle is born, with probability
- (ii)
A single vertical particle is born, with probability
- (iii)
No particles are born, with probability ;
- (iv)
Two (horizontal and vertical particles) are born, with probability .
The above list provides a complete description of the particle transformations and transition probabilities of the edge process in terms of parameters
Example 4. are independent r.v.s, . Accordingly, take independent values on edges of a ’rectangular’ graph , with generally different probabilities and for horizontal and vertical edges. In terms of the particle evolution, this means the ‘outgoing’ particles at each site being independent and independent of the ‘incoming’ ones.
Example 5. , implying and for any horizontal shift of . The corresponding edge process in this case takes constant values on each horizontal line of the rectangle V. Similarly, case leads to taking constant values on each vertical line of V.
Example 6. , meaning that none of the four binary edge variables can be different from the remaining three. This implies orand orThe resulting four-parameter model is determined by . In the isotropic case we have two parameters since This case does not allow for the death, birth or branching of a single particle; particles move independently until collision with another moving particle, upon which both colliding particles die or cross each other with probabilities defined in (p1)–(p3) above. 5. Contour Edge Process Induced by Pickard Model
Consider a binary site model
on rectangular graph
Let
be the shifted lattice,
and
be the rectangular graph with
For any horizontal or vertical edge
we designate
the edge of
, which ‘perpendicularly crosses
e at the middle’ and is defined formally by
A Boolean function
takes two values, 0 or 1. The class of such Boolean functions has
elements.
Definition 10. Let and be two Boolean functions. A contour edge process induced by a binary site model on in (57) and corresponding to is a binary edge process on defined by Probably the most natural contour process occurs in the case of Boolean functions
visualized by ‘drawing an edge’
across neighboring ‘occupied’ and ‘empty’ sites
in the site process. The introduced ‘edges’ form ‘contours’ between connected components of the random set
. Contour edge processes are usually defined for site models on undirected lattices and are well-known in statistical physics [
28] (e.g., the Ising model), where they represent boundaries between
‘spins’ and are very helpful in rigorous study of phase transitions. In our paper, a contour process is viewed as a bridge between binary site and edge processes on DAG
in (
57), particularly, between Pickard and Arak models. Even in this special case, our results are limited to the Boolean functions in (
60), raising many open questions for future work. Some of these questions are mentioned at the end of the paper.
Theorem 3. The contour edge process in (59) and (60) induced by a non-degenerate Pickard model coincides with the Arak model on if and only if is symmetric with regard to . The last condition is equivalent toMoreover, coincides with the Arak model in Example 6 with Proof. Necessity. Let
in (
59) and (
60) agree with the Arak model. Accordingly, the generic clique distribution
is determined by a generic family distribution
as
see
Figure 3. From (
15), we find that
The two first equations in (
54) are satisfied by (
64). Let us show that (
64) implies the last two equations in (
54), viz.,
Using (
10), (
64) and the same notation as in (
17), we see that
is equivalent to the equation
which factorizes as
Hence,
, since
and
are excluded by the non-degeneracy of
.
Let us show the necessity of the two other conditions in (
61). Let
be a
rectangle as in
Figure 4 and
be the corresponding edge process, where
are as in (
63) and
The last fact implies that
and
are conditionally independent given
. Particularly,
Figure 4.
Contour edge variables in Theorem 3.
Figure 4.
Contour edge variables in Theorem 3.
and
From
and (
15) we find that
and
Hence, (
65) for
leads to
, or
Next, consider (
66). We have
Hence, (
66) for
writes as
yielding
and
Equations (
67) and (
68) prove (
61).
Finally, let us show that (
61) implies symmetry of family distribution
and Pickard model
. Indeed,
is equivalent to the equality of moment functions:
The relation
implies the coincidence of moment functions up to order 2:
so that (
69) reduces to
The relation
follows from (
15), whereas the remaining three relations in (
70) use (
15) and (
61).
It remains to show the symmetry of Pickard model
. Write
for configuration of the transformed Pickard model
. Then by the definition of Bayesian network in (
5),
where
provided
have the symmetry property. The latter property is valid for our family distribution
, implying
and ending the proof of the necessity part of Theorem 3.
Sufficiency. As shown above, the symmetry
implies (
61) and (
54) for the distribution
of the quadruple
in (
63). We need to show that
in (
59) and (
60) is a Markov edge process with clique distribution
following Definition 7.
Let
be the left bottom point of
V in (
57) and
be the set of all configurations of the contour model. It is clear that any
uniquely determines
in (
72) up to the symmetry transformation: there exist two and only two configurations,
, satisfying (
72). Then, by the definition of the Pickard model and the symmetry of
,
where
for
and
having a four-point family as in
Figure 1, right. From the definitions in (
63), we see that
This and (
74) yield
where
are out-edges and
in-edges of
. An analogous relation to (
75) holds for
, whereas for
we have
, cancelling with factor 2 on the r.h.s. of (
73). The above argument leads to the desired expression
of the contour model as the Arak model with clique distribution
given by the distribution of
in (
63).
Let us show that the latter model coincides with Example 6 and its parameters satisfy (
62), (
55) and (
56). Indeed, (
63) implies
and hence (
55) since
follow from the second equation in (
61). Finally, (
62) is a consequence of (
17). Theorem 3 is proved. □
Remark 7. - (i)
The Boolean functions in (60) are invariant under symmetry . This fact seems to be related to the symmetry of the Pickard model in Theorem 3. It is of interest to extend Theorem 3 to non-symmetric Boolean functions, in an attempt to completely clarify the relation between the Pickard and Arak models in dimension 2. - (ii)
A contour model in dimension is usually formed by drawing a -dimensional plaquette perpendicularly to the edge between neighboring sites of a site model X in [28,29]. It is possible that a natural extension of the Pickard model in higher dimensions is a plaquette model (i.e., a random field indexed by plaquettes rather than sites in ), which satisfies a similar consistency property as in (11) and is related to the Arak model in . Plaquette models may be a useful and realistic alternative to site models in crystallography [30].