1. Introduction
In recent years, artificial intelligence technologies have been used to solve various problems [
1], which dictates the importance of storing data and information. With continued advances in science, a plethora of enormous datasets, including a large number of features, are being stored in different fields, such as business, text mining, biology, and medicine. Since medical datasets are often gathered for different purposes and from different sources, they may have challenges and complexities, such as structural and type heterogeneity, high dimensional, outliers, missing values, skewness, integration, and irrelevant and redundant features [
2,
3,
4]. The existence of irrelevant and redundant features may degrade the accuracy of the classifier and bring additional computational costs [
5]. To tackle this issue, many effective methods have been proposed to select effective features by reducing such disadvantageous features [
6,
7,
8,
9,
10]. Feature selection (FS) is employed in a wide range of real-world applications, including disease diagnosis [
11,
12,
13,
14,
15], text clustering [
16,
17], intrusion detection systems [
18,
19,
20,
21], e-mail spam detection [
22,
23,
24,
25], and genomic analysis [
25,
26,
27,
28,
29].
The FS algorithms are broadly classified into filter-based, wrapper-based, and embedded-based methods [
30,
31,
32]. The filter-based methods assess and rank features of datasets based on principle criteria such as distance, information, similarity, consistency, and statistical measures [
33,
34]. Although filter-based methods demand lower computational costs than other methods, they cannot provide satisfactory performance. The wrapper-based methods search for an optimal feature subset using a predetermined learning algorithm for evaluating the feature subsets. The advantages of both filter-based and wrapper-based methods are combined in embedded-based methods. These methods incorporate the search for an optimal feature subset as part of the classifier training process [
32]. The wrapper-based methods can generally provide greater classification accuracy than other methods by using a machine-learning algorithm to assess possible solutions [
6,
35]. Since assessing 2
N subsets of problem space with
N features is an NP-hard issue, near-optimal subsets are discovered using approximate algorithms that heuristically search for an optimal subset [
36,
37,
38].
Metaheuristic algorithms are a subset of approximate algorithms that have been used for solving many NP-hard problems in different fields of science, such as engineering design [
39,
40,
41,
42,
43,
44,
45,
46,
47,
48,
49,
50], task scheduling [
51,
52,
53], engineering prediction [
54,
55,
56,
57,
58], and optimal power flow [
59,
60,
61,
62,
63,
64] problems. When tackling the FS problem, metaheuristic algorithms have shown outstanding results in prior studies [
65,
66,
67,
68]. For instance, Emary et al. [
69] introduced two versions of binary grey wolf optimizer (bGWO) to solve the FS problem as a wrapper-based method. The first approach was developed by performing stochastic crossover among the three best solutions, while in the second approach, the authors applied the S-shaped transfer function to convert continuous solutions of GWO to binary ones. Mafarja et al. [
70] proposed a binary grasshopper optimization algorithm (BGOA) to tackle the feature selection problem within a wrapper-based framework by applying S-shaped and V-shaped transfer functions as the first mechanism. The second mechanism employs a new method that combines the finest solutions found so far. Furthermore, a mutation operator is used in the BGOA algorithm to improve the exploration phase.
Sindhu et al. [
71] proposed an improved sine cosine algorithm (ISCA) that includes a feature selection elitism technique and a new best solution update method to select the best features and increase the classification accuracy. Dhiman et al. [
72] developed eight binary versions of the emperor penguin optimizer to solve the FS problem by employing S-shaped and V-shaped transfer functions. In this study, 25 standard benchmark functions have been used to validate the results of the developed algorithms. The results revealed that the V
4 transfer function provides better solutions than other transfer functions. A binary farmland fertility algorithm (BFFA) has been proposed by Naseri et al. [
18] to tackle feature selection problems in intrusion detection systems using a V-shaped transfer function. Although many metaheuristic algorithms have been developed in the FS domain, most of them are not scalable enough to overcome small and large datasets.
Quantum-based avian navigation optimizer algorithm (QANA) [
73] is a recently introduced evolutionary algorithm inspired by the navigation mechanism of migratory birds during long-distance aerial paths for solving continuous optimization problems. The QANA provides competitive results by employing several operators, including population partitioning, self-adaptive quantum orientation, a qubit-crossover, and two mutation strategies. Moreover, the gained information is shared among search agents using a V-echelon communication topology. The experimental evaluations reveal that QANA is scalable for solving high-dimensional problems. It is worth mentioning that when working with high-dimensional datasets, tackling optimization problems becomes particularly difficult due to the curse of dimensionality problems [
74,
75].
This paper aims to extend our earlier study [
73] by using our proposed binary QANA (BQANA) to overcome the curse of dimensionality difficulties in the FS domain and generate high-quality solutions using two approaches. In the first approach, the canonical QANA is converted to binary using 20 different transfer functions from five categories of S-shaped [
76], V-shaped [
77], U-shaped [
78], Z-shaped [
79], and quadratic [
80,
81] to solve FS problem in medical datasets. The transfer functions are discussed in this paper, and then they are paired with the QANA to develop several binary QANA models. In the second approach, a threshold is assigned for each dimension to map the continuous solutions of QANA to binary and develop BQANA without any further computational cost. The effectiveness of the proposed approaches is investigated on 10 medical datasets with various scales. To validate the proposed algorithms, the results of the winner version of binary QANA named BQANA were compared with the results of nine well-known metaheuristic algorithms, including binary particle swarm optimization (BPSO) [
82], ant colony optimization (ACO) [
83], binary differential evolution (BDE) [
84], binary bat algorithm (BBA) [
85], feature selection based on whale optimization algorithm (FSWOA) [
11], binary ant lion optimizer (BALO) [
86], binary dragonfly algorithm (BDA) [
87], quadratic binary Harris hawk optimization (QBHHO) [
80], and binary atom search optimization (BASO) [
88]. The convergence behavior, the average number of selected features, and the accuracy of the proposed BQANA and comparative algorithms were visualized and investigated for all datasets. Moreover, the BQANA is statistically assessed by the Friedman test to demonstrate the algorithm’s superiority. The main contributions of this study are summarized as follows.
Introducing binary approaches of quantum-based avian navigation optimizer algorithm (QANA) to select effective features from high-dimensional medical datasets.
The binary QANA variants have been developed by adapting the main components of the standard QANA.
Comparing the behavior of QANA with different transfer functions from five different categories, S-shaped, V-shaped, U-shaped, Z-shaped, and quadratic, to develop 20 versions of binary QANA based on the first approach.
Applying the second approach as a low-cost and effective method to develop the superior version of binary QANA named BQANA by assigning a threshold for each variable.
Dimensionality reduction, generating solutions with high accuracy and a minimum number of features are obtained by the second approach.
The experiments prove that the BQANA developed by the second approach provides superior results compared to the first approach and nine other comparative algorithms in terms of increasing classification accuracy and minimizing the number of features for 10 medical datasets with various scales.
2. Related Works
The Fs is an NP-hard problem with discrete search space, in which the number of potential solutions grows exponentially as the number of features grows. Hence, metaheuristic algorithms are known as powerful optimizers in the literature. Ant colony optimization (ACO) [
89] is a discrete metaheuristic algorithm inspired by the behavior of some ants in nature that has been applied for solving FS problems in different fields such as text categorization [
90], image feature selection [
91], intrusion detection system [
92], and email spam classification [
93]. As most of the metaheuristic algorithms are proposed for continuous search spaces, many researchers applied metaheuristic algorithms to discover an optimal feature subset by converting the continuous solutions into a binary form using logical operators or different transfer functions [
94]. Logical operators have been applied for producing binary solutions due to their low computational costs [
95]. Boolean particle swarm optimization was first proposed by Marandi et al. [
96] to solve antenna design by converting continuous particle swarm optimization (PSO) into a binary form using three Boolean operators. In [
97], the authors proposed a binary form of the Jaya algorithm named Jayax using the xor logical operator and incorporating a local search module to boost the algorithm’s performance.
Many researchers apply different transfer functions to convert continuous values of optimizer algorithms into binary ones. The most well-known transfer functions used in the literature are S-shaped [
76], V-shaped [
77,
98], U-shaped [
78], Z-shaped [
78], and quadratic [
81] transfer functions. In 1997, Kennedy and Eberhart [
76] introduced a binary version of the particle swarm optimization (BPSO) algorithm by applying the sigmoid transfer function to solve discrete optimization problems [
82,
99]. The sigmoid function is known as the S-shaped transfer function and has been applied to many other metaheuristic algorithms. Gong et al. [
84] proposed binary differential evolution (BDE) algorithm to apply the differential evolution algorithm in discrete search space. To construct binary-adapted DE operators, DE operator templates are explicitly specified through the forma analysis. In [
85] authors proposed a binary bat algorithm (BBA) for solving FS by applying the S-shaped transfer function to restrict the new search agent’s position to only binary values. A new binary algorithm named feature selection based on whale optimization algorithm (FSWOA) was proposed by Zamani et al. [
11] to handle the dimensionality of medical data using the whale optimization algorithm (WOA). To map continuous solutions of WOA to binary ones, the authors applied the S-shaped transfer function. The binary dragonfly algorithm (BDA) is the binary version of the dragonfly algorithm proposed by Mirjalili [
87] which mimics the static and dynamic swarming behaviors of dragonflies in nature. The exploration and exploitation of the algorithm are modeled by the social interaction of dragonflies in searching for foods, avoiding enemies, and navigating when swarming dynamically or statistically.
The V-shaped transfer function introduced by Rashdi et al. [
77] is a symmetric transfer function initially used in binary gravity search algorithm (BGSA) to map continuous values of GSA into binary ones. Emary et al. [
86] proposed a binary antlion optimizer (BALO) for finding optimal feature subsets by applying S-shaped and V-shaped transfer functions. The findings indicate that the developed binary algorithm based on V-shaped transfer functions outperforms the S-shaped transfer functions. In a comparative study, Mirjalili et al. [
98] evaluated six variants of S-shaped and V-shaped transfer functions on the traditional BPSO algorithm. The results indicate that the newly presented V-shaped family of transfer functions significantly outperforms the original BPSO. Too et al. [
88] proposed eight versions of the binary atom search optimization (BASO) algorithm to effectively select an optimal feature subset by applying S-shaped and V-shaped transfer functions. In comparison to other BASO versions, the results showed that BASO with S-shaped transfer function (S
1) is highly capable of finding effective features.
Mirjalili et al. [
68] proposed a new U-shaped transfer function for the PSO algorithm to convert continuous values of velocity to binary solutions. The obtained results reveal that the U-shaped transfer functions greatly enhance the performance of BPSO. The DEOSA proposed by Guha et al. [
100] is a discrete combination of equilibrium optimizer and simulated annealing for selecting optimal features. This algorithm uses a U-shaped transfer function to convert continuous values into binary. Nadimi-Shahraki et al. [
101] proposed an enhanced version of the whale optimization algorithm named E-WOA to solve continuous optimization problems using a pooling mechanism and three robust search strategies. To address the FS problem, the solutions of E-WOA are converted to binary form using a U-shaped (U
2) transfer function.
The Z-shaped [
79] and quadratic [
81] transfer functions are two recently proposed transfer functions for mapping continuous solutions to binary ones. The quadratic binary Harris hawk optimization (QBHHO) [
80] algorithm is a binary version of the Harris hawk optimization algorithm developed by applying quadratic transfer functions for converting continuous solutions to binary. Considering a threshold for each variable is another efficient method to map continuous solutions to binary ones. Hafez et al. [
102] utilized the sine cosine algorithm (SCA) to address the FS problem by assigning a variable threshold (0.5) to convert solutions to binary form. In [
103], the authors proposed a PSO-based FS algorithm with a variable-length representation called VLPSO. The results showed that the variable-length representation enhances the scalability of PSO. The algorithm uses a predefined threshold (0.6) to map solutions into binary form.
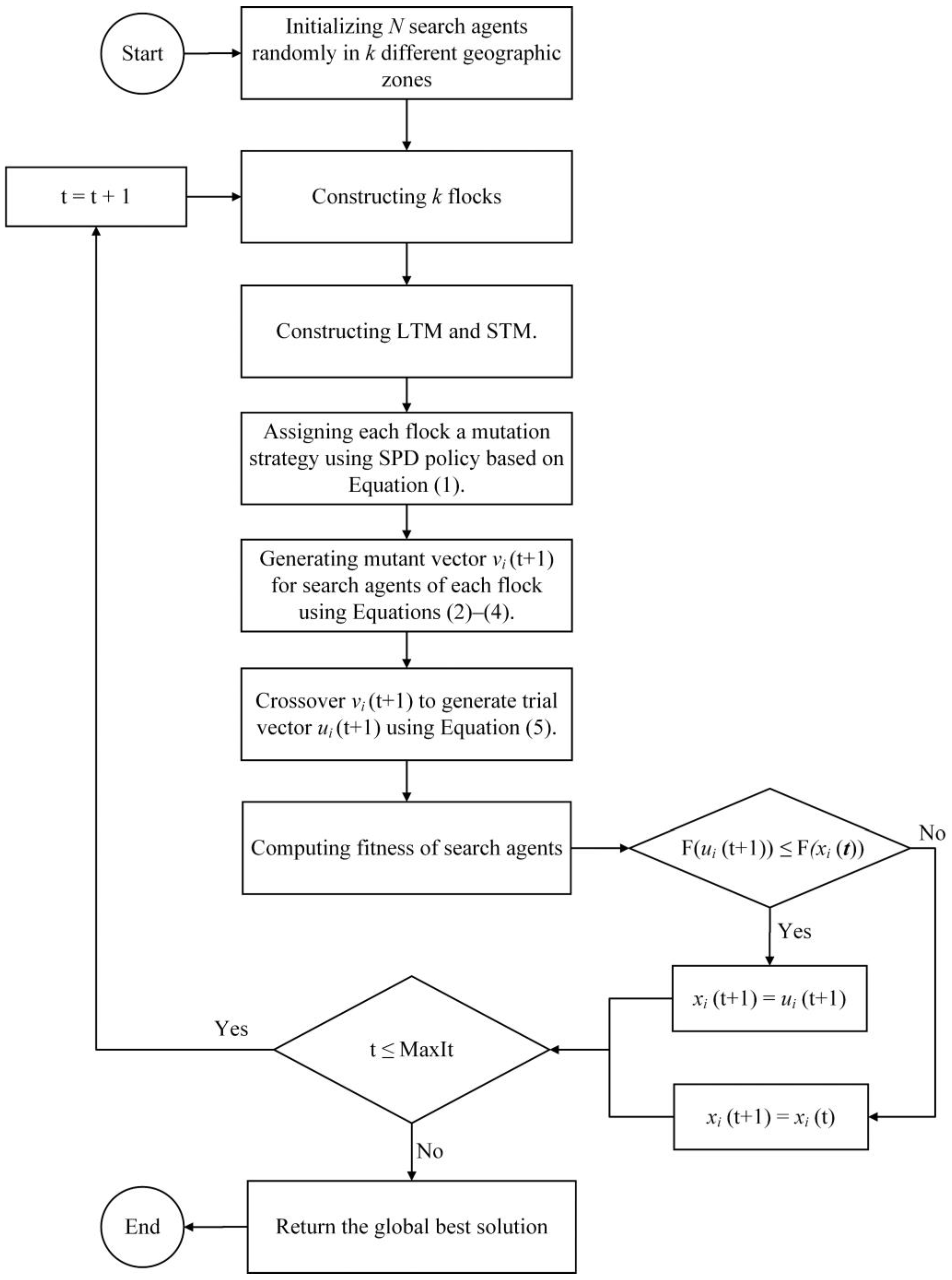
4. The Proposed Binary QANA
According to the previous study [
73], QANA outperforms other well-known optimizers in various continuous search space benchmark tests. In comparison to its competitors, QANA outperforms them in terms of exploration and exploitation abilities. Hence, the main components of the standard QANA are utilized to develop binary QANA for solving the FS problem. To develop binary QANA, initially, solutions are randomly generated in the range [0, 1]. The iterative procedure is continued after initialization until the stopping condition (maximum number of iterations) is met. In each iteration, the positions of search agents are mapped to binary using the transfer function (first approach) or by assigning a threshold for each variable (second approach). In the first approach, 20 different transfer functions from five categories, S-shaped, V-shaped, U-shaped, Z-shaped, and quadratic, are applied to map the continuous solutions of the canonical QANA to binary ones. While in the second approach, the QANA is converted to binary by simply assigning a threshold for each dimension. Both approaches are described and investigated in more detail in the following subsections.
4.1. Binary QANA Development Using Different Transfer Functions
In accordance with the literature, the transfer function has a crucial role in mapping continuous solutions to discrete space. The output of a transfer function is in the range of [0, 1]. The value of the search agent’s position determines the probability of changing the solution of the previous iteration, where the transfer function has to provide a large enough probability of changing the previous solution for a higher value of the search agent’s position. On the other hand, the computed probability of changing the solution should also be low if the value is low. Based on the above discussion, choosing a suitable transfer function will enhance the algorithm’s performance in solving the FS problem. Hence, in this study, the four versions of each transfer function S-shaped, V-shaped, U-shaped, Z-shaped, and quadratic are discussed and applied to develop different variants of binary QANA.
4.1.1. Binary QANA Using S-Shaped Transfer Function (S-BQANA)
The S-shaped transfer function first used in BPSO [
76] employs the sigmoid function (S
2) to map continuous position into binary form based on Equation (7),
where
is the position of the
i-th search agent in the
d-th dimension at the current iteration. The new position of the search agent is then updated using Equation (8), where
r is a random number in [0, 1].
S
2 and three other variants of S-shaped transfer functions are presented visually in
Figure 3 and mathematically in
Table 1. As shown in
Figure 3, S
1 sharply grows and hits saturation as the value of the position increases substantially higher than S
2, while S
3 and S
4 saturations begin later than S
2. Hence, among these four versions of the S-shaped transfer function, S
1 generates the highest probability for the same value, while S
4 returns the lowest value.
The S-shaped transfer function has certain flaws. In SI algorithms, if the value is 0, the next solution remains the same as the present position. To put it another way, the 0 value indicates that the new location should not be modified. However, with a chance of 0.5, the new position in the S-shaped transfer function may be modified to 0 or 1. Also, in the SI algorithms, there is no difference between large positive or negative values, as a large absolute position value implies that the present search agent’s location is insufficient and that a significant movement is necessary to attain the ideal position. A tiny absolute value also indicates that the present search agent’s location is near the ideal solution and that only a small distance is required to reach it. However, in S-shaped transfer functions, a positive value results in a higher likelihood (probability of 1), whereas a negative value results in a probability of 0 for the following particles’ location, which is in contrast with the natural movements of SI algorithms.
4.1.2. Binary QANA Using V-Shaped Transfer Function (V-BQANA)
The V-shaped transfer function (hyperbolic) is a symmetric function introduced by Rashdi et al. [
77] to develop a binary version of the gravity search algorithm named BGSA. According to Equation (9), this function calculates the probability of changing the value of each dimension based on the position of each search agent in continuous search space,
where
indicates the position value of the
i-th search agent in the
d-th dimension at the current iteration. As illustrated in Equation (10), the position updating rules of V-shaped transfer functions are quite different from S-shaped transfer functions,
where
and
represent the binary position and continuous position value of the
i-th search agent in the
d-th dimension at the current iteration,
is the complement of
, and
r denotes a random value in [0, 1]. Based on this rule, if the value obtained from the transfer function is equal to or greater than
r, the value of the
d-th dimension will change to the complement of the current binary position as the continuous value is high enough to change the current position. In contrast, the binary position of the
d-th dimension remains constant if the value obtained from the transfer function is less than
r. As can be seen in
Table 1, three new transfer functions have been introduced by implementing different mathematical equations. According to
Figure 3, transfer functions V
1, V
2, V
3, and V
4 provide the highest probability of switching search agents’ positions, respectively.
Unlike the S-shaped transfer functions, V-shaped transfer functions do not require search agents to take 0 or 1 values, as they let search agents with low values remain at their current positions or switch to their complements if their value is high enough. Also, the V-shaped transfer functions solve the shortcomings of the S-shaped transfer functions by assigning 0 probability of changing the position of a search agent with zero value and considering the absolute value of the continuous position in the equations to avoid assigning a probability of 0 for search agents with negative values. Moreover, in another study, Mirjalili et al. [
98] evaluated and compared different versions of sigmoid and hyperbolic functions, which showed the relative superiority of hyperbolic family functions in solving the FS problem.
4.1.3. Binary QANA Using U-Shaped Transfer Function (U-BQANA)
In a recent study, Mirjalili et al. [
78] proposed a new U-shaped transfer function for the PSO algorithm to map continuous solutions to binary ones. This transfer function comes with two control parameters to modify the range of exploration and exploitation. As can be seen in
Figure 3, similar to the V-shaped transfer function, U-shaped is a symmetric function, which means that it assigns 0 probability of changing the position of a search agent with 0 value. Also, as the absolute value of continuous position is considered in this transfer function, there are no differences between the positive and negative values. The mathematical formulation of the U-shaped transfer function is presented in Equations (11) and (12),
where
α and
β are two control parameters for determining the slope and saturation point of the U-shaped transfer function.
and
represent the binary and continuous positions of the
i-th search agent in the
d-th dimension, respectively.
r is a uniform random value in [0, 1].
Table 1 and
Figure 3 illustrate different versions of the U-shaped transfer function labeled U
1, U
2, U
3, and U
4 that were established using different values of control parameters. The
α control parameter determines the U-shaped curve’s saturation point. In contrast,
β modifies the exploration range of the transfer function by changing the width of the U-shaped transfer function’s basin. Hence, it is noticeable that U
4 provides a higher exploration range than other variations. It is also noticeable that all U-shaped variants offer higher exploration than V-shaped ones.
4.1.4. Binary QANA Using Z-Shaped Transfer Function (Z-BQANA)
The Z-shaped transfer function proposed by Guo et al. [
79] is a symmetric transfer function applied to denote the probability that an element of the position vector will change from 0 to 1 in the BPSO algorithm. Based on this transfer function, when the continuous position value is 0, the probability of change should be low because when the particle reaches the best value, the continuous position value should be lowered to 0, and the probability of the particle’s position change should be 0. The Z-shaped transfer function is defined mathematically based on Equations (13) and (14),
where
and
represent the binary and continuous positions of the
i-th search agent in the
d-th dimension, respectively, and
a denotes a positive integer. A collection of Z-shaped function families is generated by modifying the value of
a, the formulas and figures of which are presented in
Table 2 and
Figure 4, respectively. The Z-shaped transfer function is an asymmetric mapping function, as seen in
Figure 4. The asymmetric mapping function essentially fulfills the absolute value to calculate the mapping probability of the particle position vector variation, resulting in a quick convergence. The function’s slope varies when the parameter
Dparticle = DFunction × 15 is changed. The lesser the slope of the function, the greater
Dparticle = DFunction × 15. Hence, when the value remains constant, the probability of obtaining small changes in the location of the
i-th particles is greater.
4.1.5. Binary QANA Using Quadratic Transfer Function (Q-BQANA)
The quadratic transfer function proposed by Rezaee Jordehi [
81] is a recent transfer function used for converting continuous solutions of the PSO to binary ones based on Equations (15) and (16),
where
TFQ denotes the quadratic transfer function and
and
represent the binary and continuous positions of the
i-th search agent in the
d-th dimension, respectively. The variable
r is a random number in [0, 1]. The three other variants of the quadratic transfer function [
80] are presented mathematically in
Table 2 and visualized in
Figure 4.
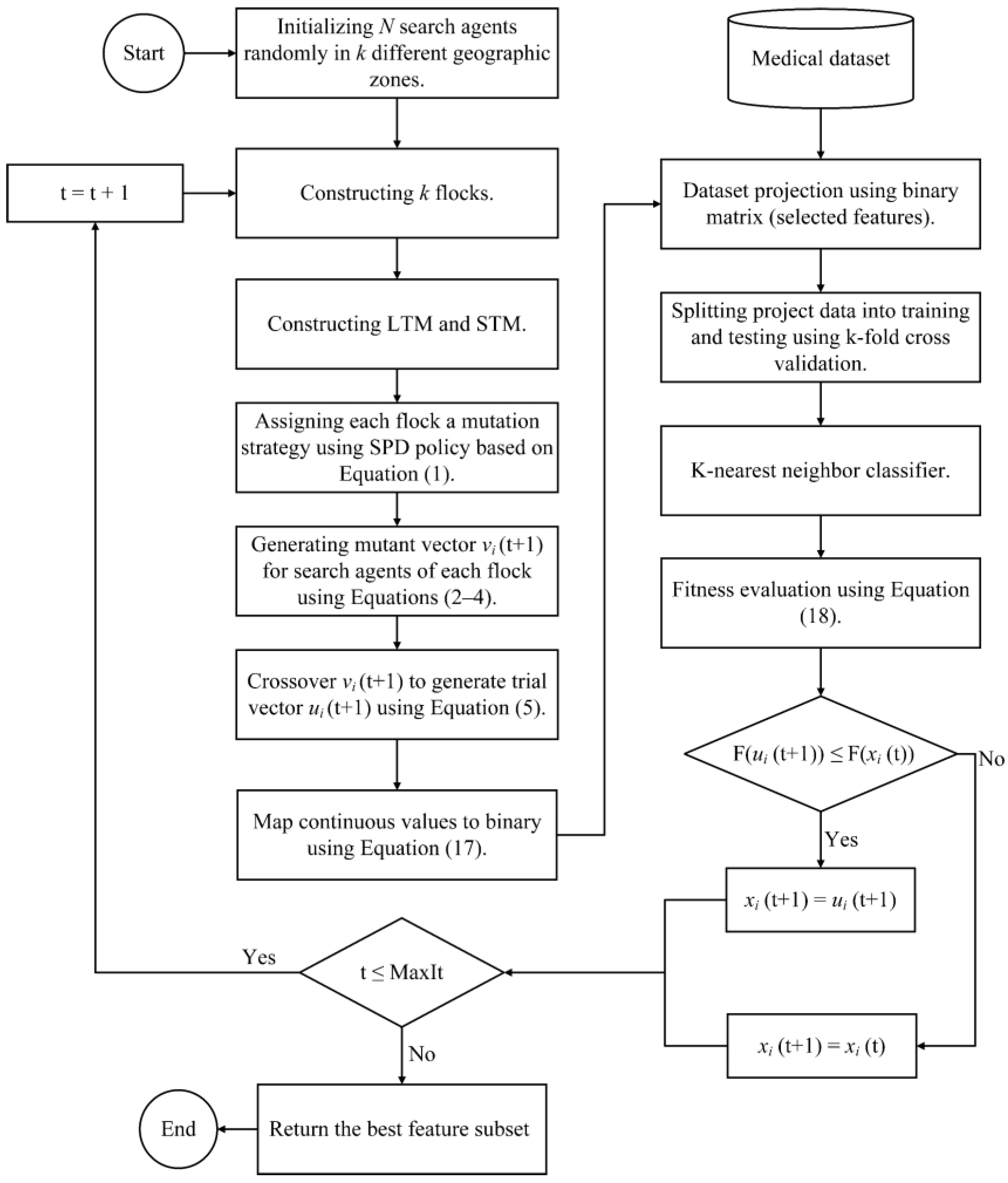
4.2. Binary QANA Development Using Variable Threshold (BQANA)
The previous subsections introduced different variants of binary QANA based on five different categories of transfer functions. Although transfer functions are widely used in the literature of FS, they impose an additional computational cost. Furthermore, transfer functions cannot provide superior results for every metaheuristic algorithm, especially for high-dimensional datasets. On the other hand, the QANA proved to be an effective problem solver in solving high-dimensional problems as it provides adequate search space coverage [
73]. It is expected that the BQANA developed based on the second approach can generate suitable candidates for solving the FS problem. Hence, this section proposes the superior version of binary QANA, named BQANA, by simply using a threshold for assigning continuous solutions of the QANA into binary. In this approach, the generated continuous solutions are converted to binary form based on Equation (17),
where
is the binary solution of the
i-th search agent in the
d-th dimension,
denotes the continuous position of the
i-th search agent in the
d-th dimension at iteration
t. The general procedure of selecting effective features with BQANA is illustrated in
Figure 5, where the algorithm receives the dataset with all features as input and returns an optimal feature subset as output.
5. Experimental Assessment
In this section, the performance of the proposed binary QANA approaches for solving the FS problem is experimentally assessed on 10 medical datasets of various sizes, which are described in
Table 3. Also, the parameters of the algorithms used in this experiment are shown in
Table 4. In the first approach, the canonical QANA is converted to binary using 20 different transfer functions from five categories of S-shaped [
76], V-shaped [
77], U-shaped [
78], Z-shaped [
79], and quadratic [
80,
81] to solve FS problem. The comparison results related to different variants of the first approach are tabulated in
Table A1,
Table A2,
Table A3,
Table A4 and
Table A5. In the second approach, the QANA is converted to binary by assigning a threshold for each dimension to map the continuous solutions into binary without further computational cost. To select the best algorithms from the first approach, one algorithm is considered representative of each transfer function category. Then, the five selected algorithms are compared against the BQANA developed based on the second approach in
Table 5. Ultimately,
Table 6 presents a comparison between the proposed BQANA and nine well-known metaheuristic algorithms introduced in the literature, including binary particle swarm optimization (BPSO) [
82], ant colony optimization (ACO) [
83], binary deferential evolution (BDE) [
84], binary bat algorithm (BBA) [
85], feature selection based on whale optimization algorithm (FSWOA) [
11], binary ant lion optimizer (BALO) [
86], binary dragonfly algorithm (BDA) [
87], quadratic binary Harris hawk optimization (QBHHO) [
80], and binary atom search optimization (BASO) [
88]. In
Table 5,
Table 6,
Table A1,
Table A2,
Table A3,
Table A4 and
Table A5, the bold values indicate the winning algorithms, and at the end of each table, the overall comparisons are shown based on the numbers of the wins (W), ties (T), and losses (L).
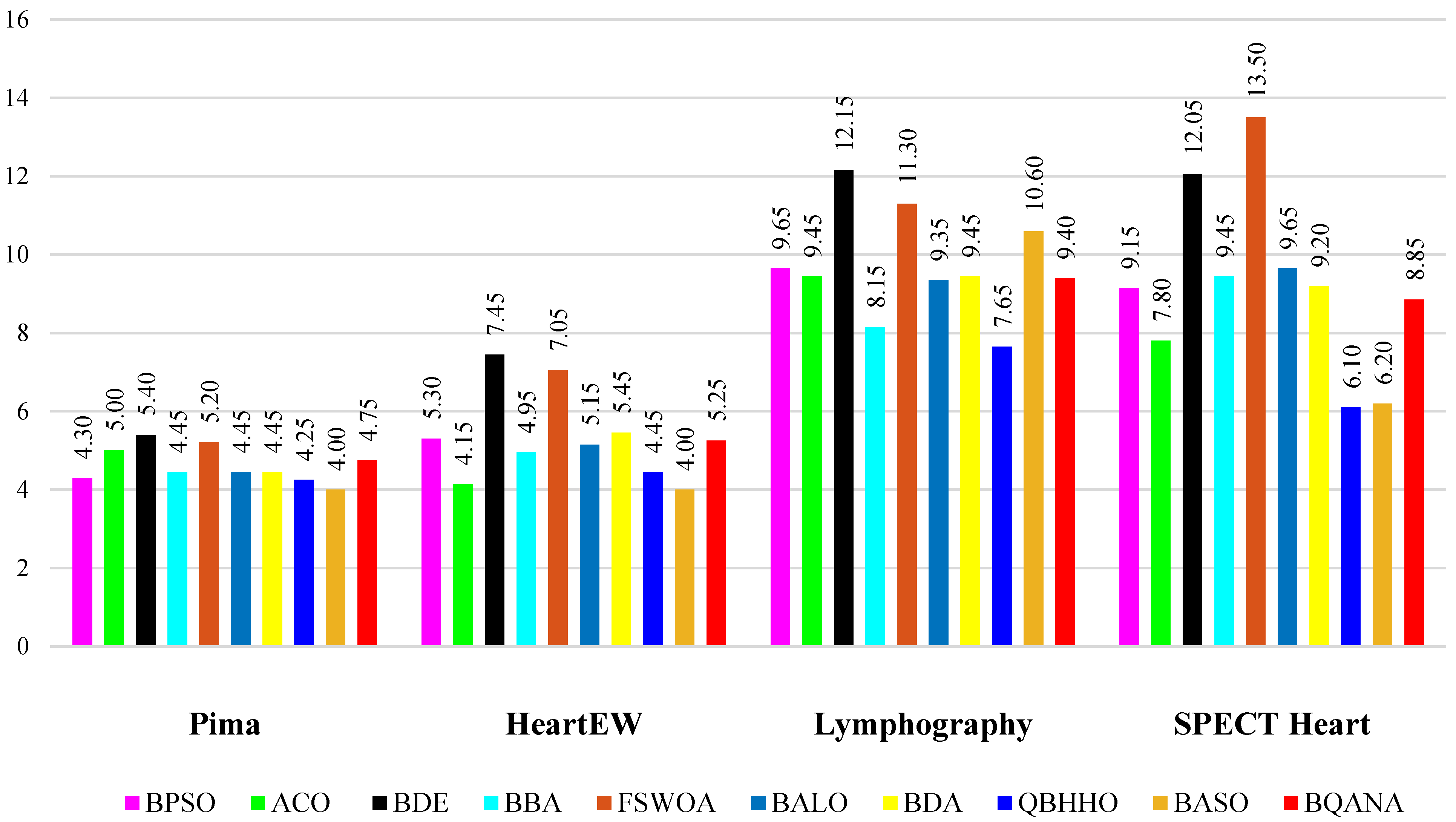
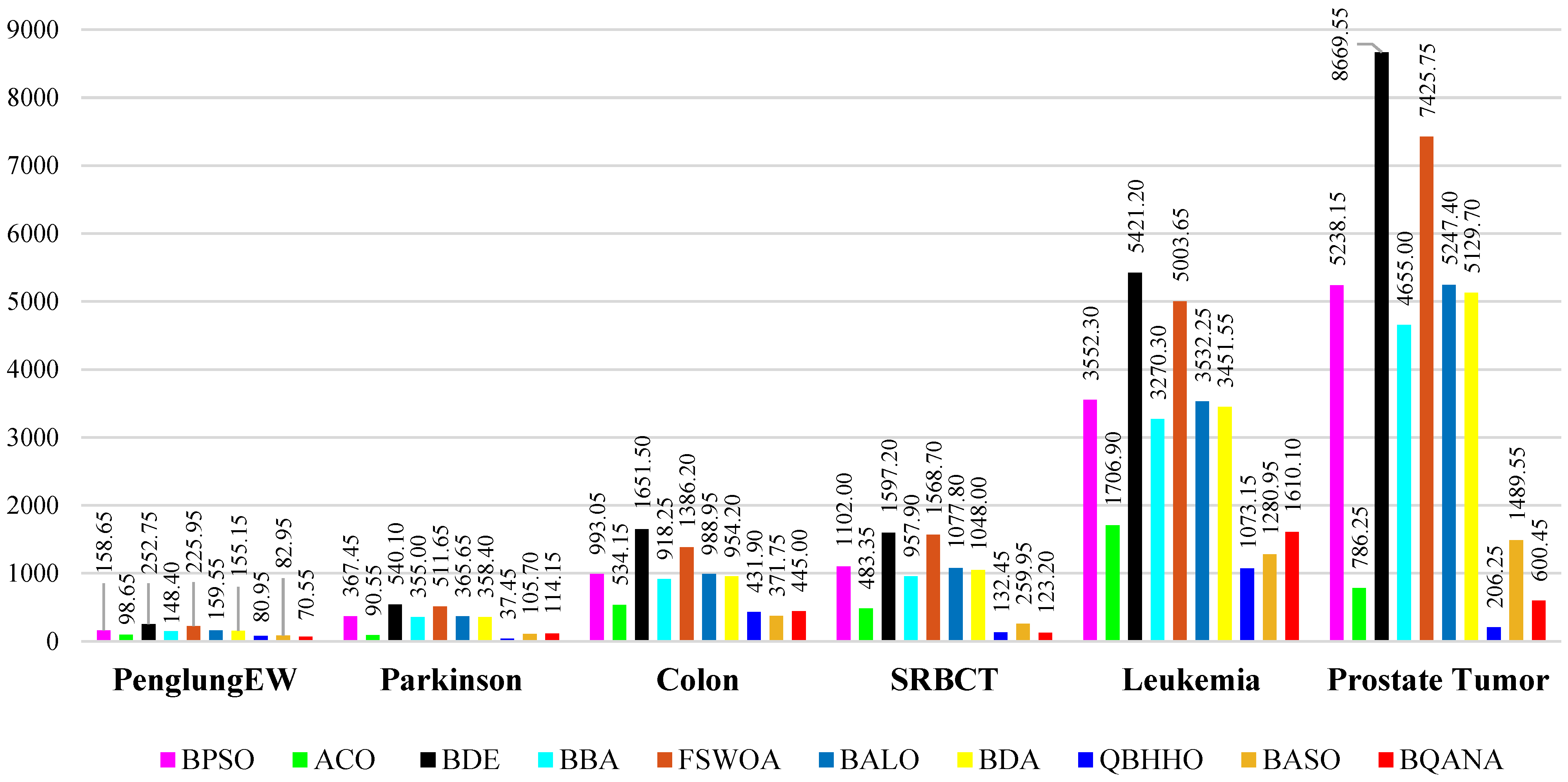
The comparison tables show the average fitness, minimum fitness, average classification accuracy, maximum classification accuracy, average number of selected features, and minimum number of selected features obtained by each algorithm. The average number of selected features by each algorithm from different datasets with various sizes is visualized in
Figure 6 and
Figure 7. Also, as classification accuracy is the most important criterion in medical datasets, the boxplot results of 10 different algorithms are exhibited in
Figure 8. Furthermore, the convergence curves of fitness values obtained during the optimization process are visualized in
Figure 9. Ultimately, the nonparametric Friedman test [
105] was used to rank the significance of the algorithms based on their performance in minimizing the fitness, as is shown in
Table 7 and
Figure 10.
5.1. Medical Datasets Description
In this study, 10 medical benchmark datasets, mostly from the UCI machine learning repository, are utilized to evaluate the performance of the proposed BQANA and comparative algorithms in solving the FS problem. The benchmark datasets utilized in the experimental evaluation of this study are on an ordinal scale, as common in the literature. Datasets with non-ordinal features can be encoded in the pre-processing stage [
106].
Table 3 provides the specifics of the utilized datasets in terms of the number of samples, total number of features, number of classes, and size that is considered small if
Nf < 300, medium if 300 ≤
Nf < 1000, and considered large if
Nf ≥ 1000, where
Nf is the number of features. To avoid overfitting problems, the K-fold cross-validation method divides datasets into
k folds where
kfold = 10. In this method, the classifier uses one fold as the testing set and
k − 1 folds as the training sets.
The Pima Indian Diabetes dataset aims to diagnose diabetes based on medical examination of females at least 21 years old and being tested for diabetes [
107]. The HeartEW dataset [
108] predicts the absence or presence of heart disease based on data gathered from 270 samples, 120 samples with a heart problem, and the remaining are healthy. The Lymphography dataset [
108] has 18 predictor features and 148 cases, with four distinct values for the class label. The aim of diagnosing cardiac single proton emission computed tomography (SPECT) heart dataset is to discriminate between the normal and abnormal function of patients’ hearts using 267 image sets. The PenglungEW is a medium dataset consisting of 73 samples and 325 features with seven different classes. The Parkinson’s dataset describes diagnosing healthy persons from those with Parkinson’s disease. This dataset is built up of various biological voice measurements with 22 features. The Colon dataset aims to classify tissues as cancerous or normal based on data gathered from 62 colon tissue samples with 2000 genes [
109]. There are 83 samples in the small round blue-cell tumor (SRBCT) dataset, each containing 2308 genes. The four classes of this dataset are the Ewing family of tumors (EWS), Burkitt lymphoma (BL) tumors, rhabdomyosarcoma (RMS) tumors, and neuroblastoma (NB) tumors [
110]. The data for the Leukemia dataset came from publicly accessible microarray gene data [
111]. The bone marrow expressions of 72 samples with 7128 genes are included. The dataset contains two different kinds of Leukemia classifications. The prostate tumor [
112] is the largest dataset used in our experiments that contains 10,509 genes from 52 prostate cancers and 50 non-tumor prostate tissues.
5.2. Parameter Settings
In this study, the error rate is calculated using the k-nearest neighbor (KNN) algorithm with Euclidean distance and
k = 5 to evaluate the effectiveness of selected feature subsets. The objectives of this study are evaluated by a fitness function presented in Equation (18), where
CE denotes the classification error,
α is the significance of classification quality,
Nsf and
Ntf are the numbers of selected features, and the total features of the dataset, respectively. As classification accuracy is the most important factor for medical datasets, we considered
α = 0.99 for this study.
To verify that the comparisons are accurate and fair, all experiments are conducted 20 times independently on a laptop with an Intel Core i7-10750H CPU and 24.0 GB of RAM using MATLAB R2022a. The maximum number of iterations (
MaxIt) and the population size (
N) were set to 300 and 20, respectively. Furthermore, the competing algorithms’ parameters were adjusted to the same values as the stated settings in their works, shown in
Table 4.
6. Discussion
Table A1,
Table A2,
Table A3,
Table A4 and
Table A5 show the comparison results of applying different transfer functions to develop different binary versions of the QANA based on the first approach. The results indicate that S4, V2, U4, Z1, and Q3 transfer functions provide superior results compared to other family members.
Table 5 compares the results of five selected algorithms developed by the first approach with the BQANA developed based on the second approach. Clearly, the BQANA developed using the second approach overcomes the binary algorithms belonging to the first approach.
Table 6 further investigates the proposed BQANA’s effectiveness by comparing it with nine well-known optimization algorithms of the feature selection domain. The results show that the BQANA achieves superior results in terms of average fitness for most of the datasets. Regarding the BQANA’s results shown in
Figure 6, it has an average performance in minimizing the number of features, while turning to
Figure 7, it is clear that the BQANA and the QBHHO provide the minimum number of features for most of the datasets among the competitors. The boxplots in
Figure 8 illustrate the spread of the classification accuracy distribution obtained by each algorithm, in which the BQANA is predominantly the superior algorithm in terms of obtaining the highest accuracy and normal distribution.
Convergence curves plotted in
Figure 9 generally suggest that the BQANA has the fastest convergence toward optimum solutions compared to comparative algorithms for most cases. Furthermore, it is noticeable that the BQANA consistently improves the solutions until the final iterations. Overall, the BQANA is fairly scalable as it can find better feature subsets for different scales of medical datasets by maintaining a balance between exploration and exploitation. Based on the results of the Friedman test reported in
Table 7, the BQANA is superior to comparative algorithms in feature selection from different scales of medical data. For further statistical evaluation,
Figure 10 provides the exploratory data analysis in a radar chart format. It is noticeable in the radar chart that the BQANA surrounds the center of the radar chart for most of the datasets, which shows the superiority of the BQANA over the comparative algorithms.
7. Conclusions
The advancement of information storage technologies in medical science has resulted in the generation of massive amounts of raw datasets with many irrelevant or redundant features. Selecting desirable features will reduce the computational costs and improve the algorithms’ accuracy in the data-driven decision-maker software. Although many metaheuristic algorithms have been developed to select effective features, a few can maintain their performance when the number of features increases. This paper introduces an efficient binary version of the quantum-based avian navigation optimizer algorithm (QANA), called BQANA, to select effective features from various scales of medical datasets. The study consists of two approaches for mapping continuous solutions of QANA into binary. In the first approach, 20 different transfer functions from five distinct categories, S-shaped, V-shaped, U-shaped, Z-shaped, and quadratic, were applied to develop different variants of the binary QANA. According to the results, transfer functions cannot generate optimal binary solutions for every metaheuristic algorithm in the FS domain. Moreover, using transfer functions imposes additional computational costs on the optimization algorithms.
In the second approach, a simple threshold with minimum computational costs is used to assign continuous QANA solutions into binary ones to develop the BQANA. All variants were experimentally evaluated on 10 medical datasets to identify the winner variant. The experimental results reveal that the BQANA developed by the second approach generates better solutions than the other variants. Then, the results of the BQANA were compared with results obtained from nine well-known metaheuristic algorithms: BPSO, ACO, BDE, BBA, FSWOA, BALO, BDA, QBHHO, and BASO. Furthermore, the Friedman test was applied to rank the algorithms based on their performance. The experimental results and statistical analysis revealed that the BQANA developed by the second approach outperforms comparative algorithms in selecting effective feature subsets from different scales of medical datasets. In the future, the BQANA can be enhanced by improving its search strategy and using novel and more effective transfer functions. Moreover, the BQANA can be applied to solve real-world applications and other discreet problems such as nurse scheduling.