Proteome of the Luminal Surface of the Blood–Brain Barrier
Abstract
:1. Introduction
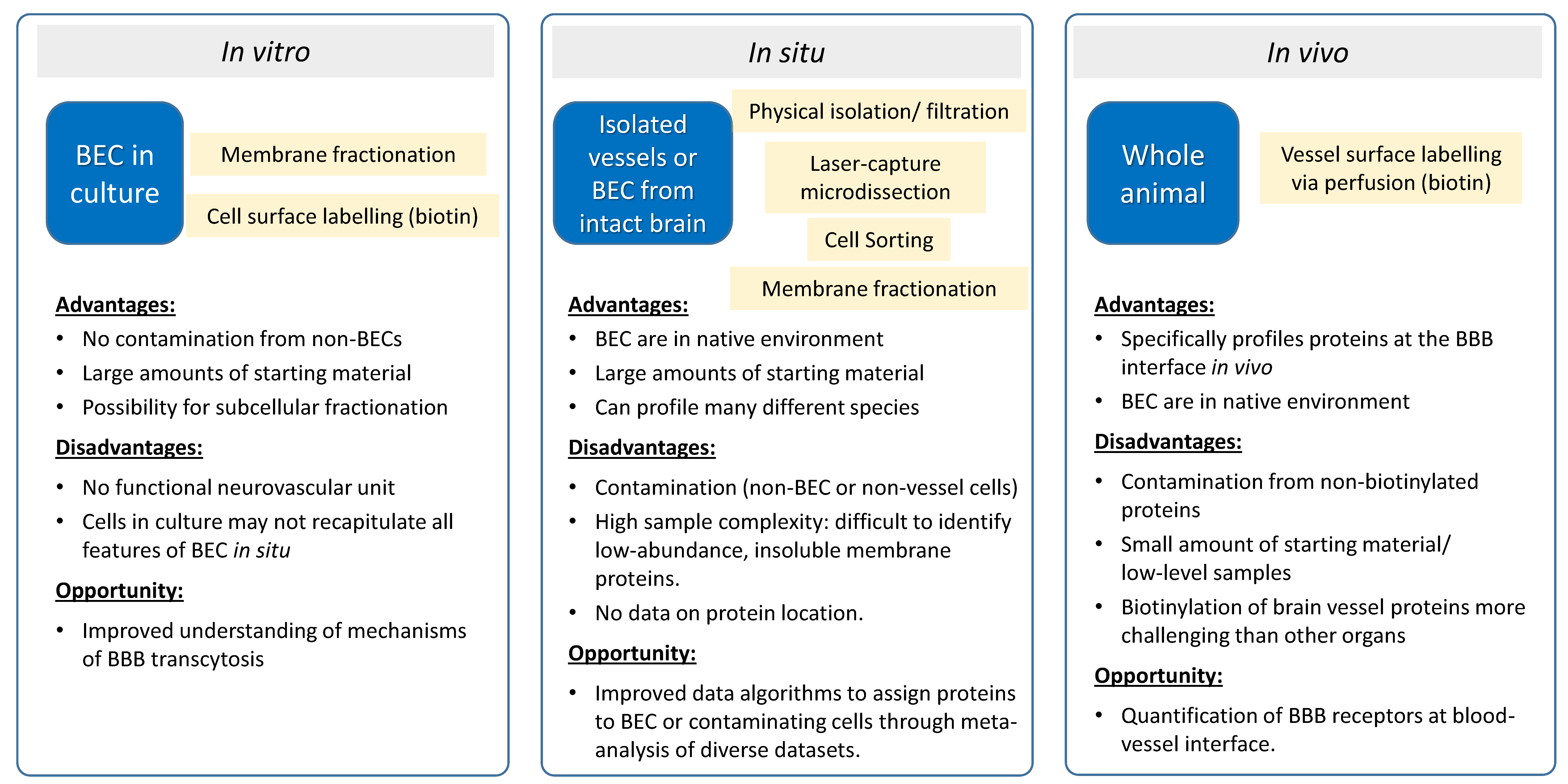
2. Proteomics of In Vitro BBB Models
3. Vessel Proteomics In Situ
3.1. Brain Vessel Isolation
3.2. Laser-Capture Microdissection of Brain Vessels
3.3. Isolated Brain Endothelial Cells
4. Luminal Proteomics In Vivo
4.1. Vessel Membrane Fractionation
4.2. Perfusion-Based Chemical Labelling Methods
5. Beyond Proteomic Profiling for In Vivo Luminal Protein Profiling in Vessels: Alternative Methods to Identify Potential Luminal RMT Targets In Vivo
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Stanimirovic, D.B.; Friedman, A. Pathophysiology of the Neurovascular Unit: Disease Cause or Consequence? J. Cereb. Blood Flow Metab. 2012, 32, 1207–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enerson, B.E.; Drewes, L.R. The Rat Blood–Brain Barrier Transcriptome. J. Cereb. Blood Flow Metab. 2005, 26, 959–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntley, M.A.; Ebien-Ly, N.; Edaneman, R.; Watts, R.J. Dissecting gene expression at the blood-brain barrier. Front. Neurosci. 2014, 8, 355. [Google Scholar] [CrossRef] [Green Version]
- Haseloff, R.F.; Krause, E.; Bigl, M.; Mikoteit, K.; Stanimirovic, D.; Blasig, I.E. Differential protein expression in brain capillary endothelial cells induced by hypoxia and posthypoxic reoxygenation. Proteomics 2006, 6, 1803–1809. [Google Scholar] [CrossRef] [PubMed]
- Haqqani, A.S.; Nesic, M.; Preston, E.; Baumann, E.; Kelly, J.; Stanimirovic, D. Characterization of vascular protein expression patterns in cerebral ischemia/reperfusion using laser capture microdissection and ICAT-nanoLC-MS/MS. FASEB J. 2005, 19, 1809–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.M.; Houghton, S.; Magdeldin, T.; Durán, J.G.B.; Minotti, A.P.; Snead, A.; Sproul, A.; Nguyen, D.-H.T.; Xiang, J.; Fine, H.A.; et al. Pluripotent stem cell-derived epithelium misidentified as brain microvascular endothelium requires ETS factors to acquire vascular fate. Proc. Natl. Acad. Sci. USA 2021, 118, e2016950118. [Google Scholar] [CrossRef]
- Lippmann, E.S.; Al-Ahmad, A.; Palecek, S.P.; Shusta, E.V. Modeling the blood–brain barrier using stem cell sources. Fluids Barriers CNS 2013, 10, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribecco-Lutkiewicz, M.; Sodja, C.; Haukenfrers, J.; Haqqani, A.S.; Ly, D.; Zachar, P.; Baumann, E.; Ball, M.; Huang, J.; Rukhlova, M.; et al. A novel human induced pluripotent stem cell blood-brain barrier model: Applicability to study antibody-triggered receptor-mediated transcytosis. Sci. Rep. 2018, 8, 1873. [Google Scholar] [CrossRef]
- Pen, A.; Moreno, M.J.; Martin, J.; Stanimirovic, D.B. Molecular markers of extracellular matrix remodeling in glioblastoma vessels: Microarray study of laser-captured glioblastoma vessels. Glia 2007, 55, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Demarest, T.G.; Murugesan, N.; Shrestha, B.; Pachter, J.S. Rapid expression profiling of brain microvascular endothelial cells by immuno-laser capture microdissection coupled to TaqMan® Low Density Array. J. Neurosci. Methods 2012, 206, 200–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.W.; Foreman, K.L.; Gastfriend, B.D.; Kuo, J.S.; Palecek, S.P.; Shusta, E.V. Transcriptomic comparison of human and mouse brain microvessels. Sci. Rep. 2020, 10, 12358. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Q.Y.; Haqqani, A.S.; Leclerc, S.; Liu, Z.; Fauteux, F.; Baumann, E.; Delaney, C.E.; Ly, D.; Star, A.T.; et al. Differential expression of receptors mediating receptor-mediated transcytosis (RMT) in brain microvessels, brain parenchyma and peripheral tissues of the mouse and the human. Fluids Barriers CNS 2020, 17, 47. [Google Scholar] [CrossRef]
- Chen, M.B.; Yang, A.C.; Yousef, H.; Lee, D.; Chen, W.; Schaum, N.; Lehallier, B.; Quake, S.R.; Wyss-Coray, T. Brain Endothelial Cells Are Exquisite Sensors of Age-Related Circulatory Cues. Cell Rep. 2020, 30, 4418–4432.e4. [Google Scholar] [CrossRef] [PubMed]
- Mäe, M.A.; He, L.; Nordling, S.; Vazquez-Liebanas, E.; Nahar, K.; Jung, B.; Li, X.; Tan, B.C.; Foo, J.C.; Cazenave-Gassiot, A.; et al. Single-Cell Analysis of Blood-Brain Barrier Response to Pericyte Loss. Circ. Res. 2021, 128, 46. [Google Scholar] [CrossRef] [PubMed]
- Kalucka, J.; de Rooij, L.P.; Goveia, J.; Rohlenova, K.; Dumas, S.; Meta, E.; Conchinha, N.; Taverna, F.; Teuwen, L.-A.; Veys, K.; et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 2020, 180, 764–779.e20. [Google Scholar] [CrossRef] [PubMed]
- Dusart, P.; Hallström, B.M.; Renné, T.; Odeberg, J.; Uhlén, M.; Butler, L.M. A Systems-Based Map of Human Brain Cell-Type Enriched Genes and Malignancy-Associated Endothelial Changes. Cell Rep. 2019, 29, 1690–1706.e4. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Pan, S.; Zhang, Z.; Jia, L.; Chen, W.-H.; Zhao, X.-M. STAB: A spatio-temporal cell atlas of the human brain. Nucleic Acids Res. 2021, 49, D1029–D1037. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Rozanova, S.; Barkovits, K.; Nikolov, M.; Schmidt, C.; Urlaub, H.; Marcus, K. Quantitative Mass Spectrometry-Based Proteomics: An Overview. Methods Mol. Biol. 2021, 2228, 85–116. [Google Scholar] [CrossRef]
- Sinha, A.; Mann, M. A beginner’s guide to mass spectrometry-based proteomics. Biochemist 2020, 42, 64–69. [Google Scholar] [CrossRef]
- Zhang, F.; Ge, W.; Ruan, G.; Cai, X.; Guo, T. Data-Independent Acquisition Mass Spectrometry-Based Proteomics and Software Tools: A Glimpse in 2020. Proteomics 2020, 20, e1900276. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, Y.; Uchida, Y.; Tachikawa, M.; Inoue, T.; Ohtsuki, S.; Terasaki, T. Quantitative Atlas of Blood–Brain Barrier Transporters, Receptors, and Tight Junction Proteins in Rats and Common Marmoset. J. Pharm. Sci. 2013, 102, 3343–3355. [Google Scholar] [CrossRef] [PubMed]
- Uchida, Y.; Yagi, Y.; Takao, M.; Tano, M.; Umetsu, M.; Hirano, S.; Usui, T.; Tachikawa, M.; Terasaki, T. Comparison of Absolute Protein Abundances of Transporters and Receptors among Blood–Brain Barriers at Different Cerebral Regions and the Blood–Spinal Cord Barrier in Humans and Rats. Mol. Pharm. 2020, 17, 2006–2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Uchida, Y.; Hirano, S.; Ando, D.; Kubo, Y.; Auriola, S.; Akanuma, S.-I.; Hosoya, K.-I.; Urtti, A.; Terasaki, T.; et al. Inner Blood–Retinal Barrier Dominantly Expresses Breast Cancer Resistance Protein: Comparative Quantitative Targeted Absolute Proteomics Study of CNS Barriers in Pig. Mol. Pharm. 2017, 14, 3729–3738. [Google Scholar] [CrossRef]
- Braun, C.; Sakamoto, A.; Fuchs, H.; Ishiguro, N.; Suzuki, S.; Cui, Y.; Klinder, K.; Watanabe, M.; Terasaki, T.; Sauer, A. Quantification of Transporter and Receptor Proteins in Dog Brain Capillaries and Choroid Plexus: Relevance for the Distribution in Brain and CSF of Selected BCRP and P-gp Substrates. Mol. Pharm. 2017, 14, 3436–3447. [Google Scholar] [CrossRef]
- Karamanos, Y.; Gosselet, F.; Dehouck, M.-P.; Cecchelli, R. Blood–Brain Barrier Proteomics: Towards the Understanding of Neurodegenerative Diseases. Arch. Med. Res. 2014, 45, 730–737. [Google Scholar] [CrossRef]
- Uchida, Y. Quantitative Proteomics-Based Blood–Brain Barrier Study. Biol. Pharm. Bull. 2021, 44, 465–473. [Google Scholar] [CrossRef]
- Badhwar, A.; Brown, R.; Stanimirovic, D.B.; Haqqani, A.S.; Hamel, E. Proteomic differences in brain vessels of Alzheimer’s disease mice: Normalization by PPARγ agonist pioglitazone. J. Cereb. Blood Flow Metab. 2016, 37, 1120–1136. [Google Scholar] [CrossRef] [Green Version]
- Stanimirovic, D.B.; Bani-Yaghoub, M.; Perkins, M.; Haqqani, A.S. Blood–brain barrier models:in vitrotoin vivotranslation in preclinical development of CNS-targeting biotherapeutics. Expert Opin. Drug Discov. 2014, 10, 141–155. [Google Scholar] [CrossRef]
- Ito, S.; Oishi, M.; Ogata, S.; Uemura, T.; Couraud, P.-O.; Masuda, T.; Ohtsuki, S. Identification of Cell-Surface Proteins Endocytosed by Human Brain Microvascular Endothelial Cells in vitro. Pharmaceutics 2020, 12, 579. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Hoshiyama, T.; Uemura, T.; Hirayama-Kurogi, M.; Ogata, S.; Furukawa, A.; Couraud, P.-O.; Furihata, T.; Ito, S.; Ohtsuki, S. Large-Scale Quantitative Comparison of Plasma Transmembrane Proteins between Two Human Blood–Brain Barrier Model Cell Lines, hCMEC/D3 and HBMEC/ciβ. Mol. Pharm. 2019, 16, 2162–2171. [Google Scholar] [CrossRef] [PubMed]
- Simonian, M.; Loo, R.R.O.; Rannulu, N.; Loo, J.A.; Molloy, M.; Stoodley, M.A. Identification of protein targets in cerebral endothelial cells for brain arteriovenous malformation (AVMs) molecular therapies. Clin. Proteom. 2017, 14, 17. [Google Scholar] [CrossRef] [PubMed]
- Del Pino, M.S.; Hawkins, R.A.; Peterson, D.R. Biochemical Discrimination between Luminal and Abluminal Enzyme and Transport Activities of the Blood-Brain Barrier. J. Biol. Chem. 1995, 270, 14907–14912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betz, A.L.; Firth, J.A.; Goldstein, G.W. Polarity of the blood-brain barrier: Distribution of enzymes between the luminal and antiluminal membranes of brain capillary endothelial cells. Brain Res. 1980, 192, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Bamji-Mirza, M.; Chang, N.; Haqqani, A.S.; Stanimirovic, D.B. The blood-brain barrier in health and disease. Volume 1, Morphology, biology and immune function. In The Blood-Brain Barrier in Health and Disease; Dorovini-Zis, K., Ed.; Morphology, Biology and Immune Function; CRC Press: Boca Raton, FL, USA, 2015; Volume 1, pp. 172–214. ISBN 9781498727051. [Google Scholar]
- Haqqani, A.S.; Hill, J.J.; Mullen, J.; Stanimirovic, D.B. Methods to Study Glycoproteins at the Blood-Brain Barrier Using Mass Spectrometry. Methods Mol. Biol. 2010, 686, 337–353. [Google Scholar] [CrossRef]
- Haqqani, A.S.; Delaney, C.E.; Brunette, E.; Baumann, E.; Farrington, G.K.; Sisk, W.; Eldredge, J.; Ding, W.; Tremblay, T.-L.; Stanimirovic, D.B. Endosomal trafficking regulates receptor-mediated transcytosis of antibodies across the blood brain barrier. J. Cereb. Blood Flow Metab. 2018, 38, 727–740. [Google Scholar] [CrossRef]
- Haqqani, A.S.; Thom, G.; Burrell, M.; Delaney, C.E.; Brunette, E.; Baumann, E.; Sodja, C.; Jezierski, A.; Webster, C.; Stanimirovic, D.B. Intracellular sorting and transcytosis of the rat transferrin receptor antibody OX26 across the blood-brain barrierin vitrois dependent on its binding affinity. J. Neurochem. 2018, 146, 735–752. [Google Scholar] [CrossRef] [Green Version]
- Haqqani, A.S.; E Delaney, C.; Tremblay, T.-L.; Sodja, C.; Sandhu, J.K.; Stanimirovic, D.B. Method for isolation and molecular characterization of extracellular microvesicles released from brain endothelial cells. Fluids Barriers CNS 2013, 10, 4. [Google Scholar] [CrossRef]
- Kutuzov, N.; Flyvbjerg, H.; Lauritzen, M. Contributions of the glycocalyx, endothelium, and extravascular compartment to the blood-brain barrier. Proc. Natl. Acad. Sci. USA 2018, 115, E9429–E9438. [Google Scholar] [CrossRef] [Green Version]
- Durr, E.; Yu, J.; Krasinska, K.M.; A Carver, L.; Yates, J.R.; E Testa, J.; Oh, P.; E Schnitzer, J. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat. Biotechnol. 2004, 22, 985–992. [Google Scholar] [CrossRef]
- Pardridge, W.M. The Isolated Brain Microvessel: A Versatile Experimental Model of the Blood-Brain Barrier. Front. Physiol. 2020, 11, 398. [Google Scholar] [CrossRef]
- Torbett, B.E.; Baird, A.; Eliceiri, B.P. Understanding the rules of the road: Proteomic approaches to interrogate the blood brain barrier. Front. Neurosci. 2015, 9, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, H.B.; Scott, M.; Niessen, S.; Hoover, H.; Baird, A.; Yates, J.; E Torbett, B.; Eliceiri, B.P. The Proteome of Mouse Brain Microvessel Membranes and Basal Lamina. J. Cereb. Blood Flow Metab. 2011, 31, 2267–2281. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Goldmann, U.; Müller, S.; van Oostrum, M.; Müller, M.; Schubert, O.T.; Wollscheid, B. The in silico human surfaceome. Proc. Natl. Acad. Sci. USA 2018, 115, E10988–E10997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Feteisi, H.; Al-Majdoub, Z.M.; Achour, B.; Couto, N.; Rostami-Hodjegan, A.; Barber, J. Identification and quantification of blood-brain barrier transporters in isolated rat brain microvessels. J. Neurochem. 2018, 146, 670–685. [Google Scholar] [CrossRef] [PubMed]
- Badhwar, A.; Stanimirovic, D.B.; Hamel, E.; Haqqani, A.S. The Proteome of Mouse Cerebral Arteries. J. Cereb. Blood Flow Metab. 2014, 34, 1033–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campeau, A.; Mills, R.H.; Blanchette, M.; Bajc, K.; Malfavon, M.; Munji, R.N.; Deng, L.; Hancock, B.; Patras, K.A.; Olson, J.; et al. Multidimensional Proteome Profiling of Blood-Brain Barrier Perturbation by Group B Streptococcus. mSystems 2020, 5, e00368-20. [Google Scholar] [CrossRef]
- Murugesan, N.; Macdonald, J.A.; Lu, Q.; Wu, S.; Hancock, W.S.; Pachter, J.S. Analysis of Mouse Brain Microvascular Endothe-lium Using Laser Capture Microdissection Coupled with Proteomics. In The Blood-Brain and Other Neural Barriers; Methods in Molecular Biology (Methods and Protocols); Humana Press: New York, NY, USA, 2011; Volume 686. [Google Scholar]
- Zajec, M.; Kros, J.M.; Dekker-Nijholt, D.A.T.; Dekker, L.J.; Stingl, C.; Van Der Weiden, M.; Bosch, T.P.P.V.D.; Mustafa, D.A.M.; Luider, T.M. Identification of Blood–Brain Barrier-Associated Proteins in the Human Brain. J. Proteome Res. 2021, 20, 531–537. [Google Scholar] [CrossRef]
- Tasic, B.; Yao, Z.; Graybuck, L.T.; Smith, K.A.; Nguyen, T.N.; Bertagnolli, D.; Goldy, J.; Garren, E.; Economo, M.N.; Viswanathan, S.; et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 2018, 563, 72–78. [Google Scholar] [CrossRef]
- Orsenigo, F.; Conze, L.L.; Jauhiainen, S.; Corada, M.; Lazzaroni, F.; Malinverno, M.; Sundell, V.; Cunha, S.I.; Brännström, J.; Globisch, M.A.; et al. Mapping endothelial-cell diversity in cerebral cavernous malformations at single-cell resolution. Elife 2020, 9, e61413. [Google Scholar] [CrossRef]
- Vanlandewijck, M.; Lebouvier, T.; Mäe, M.A.; Nahar, K.; Betsholtz, C. Primary isolation of vascular cells from murine brain for single cell sequencing. Protoc. Exch. 2018, 1–11. [Google Scholar] [CrossRef]
- Hodge, R.D.; Bakken, T.E.; Miller, J.A.; Smith, K.A.; Barkan, E.R.; Graybuck, L.T.; Close, J.L.; Long, B.; Johansen, N.; Penn, O.; et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 2019, 573, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Zuchero, Y.J.Y.; Chen, X.; Bien-Ly, N.; Bumbaca, D.; Tong, R.K.; Gao, X.; Zhang, S.; Hoyte, K.; Luk, W.; Huntley, M.A.; et al. Discovery of Novel Blood-Brain Barrier Targets to Enhance Brain Uptake of Therapeutic Antibodies. Neuron 2016, 89, 70–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, L.; Black, D.; Raman, C.; Woodford, K.; Zhou, M.; Haggerty, J.; Yan, A.; Cwirla, S.; Grindstaff, K. Subcellular localization of transporters along the rat blood–brain barrier and blood–cerebral-spinal fluid barrier by in vivo biotinylation. Neuroscience 2008, 155, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Kubo, Y.; Ohtsuki, S.; Uchida, Y.; Terasaki, T. Quantitative Determination of Luminal and Abluminal Membrane Distributions of Transporters in Porcine Brain Capillaries by Plasma Membrane Fractionation and Quantitative Targeted Proteomics. J. Pharm. Sci. 2015, 104, 3060–3068. [Google Scholar] [CrossRef] [Green Version]
- Gage, G.J.; Kipke, D.R.; Shain, W. Whole Animal Perfusion Fixation for Rodents. J. Vis. Exp. 2012, 30, e3564. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, B.S.; E Schnitzer, J.; McCaffery, M.; E Palade, G. Isolation and partial characterization of the luminal plasmalemma of microvascular endothelium from rat lungs. Eur. J. Cell Biol. 1992, 58, 296–306. [Google Scholar]
- Li, Y.; Yu, J.; Wang, Y.; Griffin, N.M.; Long, F.; Shore, S.; Oh, P.; Schnitzer, J.E. Enhancing Identifications of Lipid-embedded Proteins by Mass Spectrometry for Improved Mapping of Endothelial Plasma Membranes in vivo. Mol. Cell. Proteom. 2009, 8, 1219–1235. [Google Scholar] [CrossRef] [Green Version]
- Oh, P.; Li, Y.; Yu, J.; Durr, E.; Krasinska, K.M.; Carver, L.A.; Testa, J.E.; Schnitzer, J.E. Subtractive proteomic mapping of the endothelial surface in lung and solid tumours for tissue-specific therapy. Nature 2004, 429, 629–635. [Google Scholar] [CrossRef]
- Rybak, J.-N.; Ettorre, A.; Kaissling, B.; Giavazzi, R.; Neri, D.; Elia, G. In vivo protein biotinylation for identification of organ-specific antigens accessible from the vasculature. Nat. Methods 2005, 2, 291–298. [Google Scholar] [CrossRef]
- McCann, L.A.; Haywood, M.C.; Ren, B.H.; Simpson, A.M.; Guilhaus, M.; Wasinger, V.; Raftery, M.J.; Davey, R.A. Identification of Vascular Surface Proteins by in vivo Biotinylation: A Method Sufficiently Sensitive To Detect Changes in Rat Liver 2 Weeks after Partial Hepatectomy. J. Proteome Res. 2007, 6, 3108–3113. [Google Scholar] [CrossRef] [PubMed]
- Roesli, C.; Neri, D.; Rybak, J.-N. In vivo protein biotinylation and sample preparation for the proteomic identification of organ- and disease-specific antigens accessible from the vasculature. Nat. Protoc. 2006, 1, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Borgia, B.; Roesli, C.; Fugmann, T.; Schliemann, C.; Cesca, M.; Neri, D.; Giavazzi, R. A Proteomic Approach for the Identification of Vascular Markers of Liver Metastasis. Cancer Res. 2010, 70, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.-J.; Wang, S.-Q.; Zhang, J.; Zhang, W.; Bi, F.; Guo, Z.-G.; Ding, B.-S.; Kumar, P.; Liu, J.-N.; Tan, X.-Y. A novel method to isolate and map endothelial membrane proteins from pulmonary vasculature. Am. J. Physiol. Cell Physiol. 2005, 288, C950–C956. [Google Scholar] [CrossRef] [Green Version]
- McRobb, L.S.; Lee, V.S.; Simonian, M.; Zhao, Z.; Thomas, S.G.; Wiedmann, M.; Raj, J.V.A.; Grace, M.; Moutrie, V.; McKay, M.; et al. Radiosurgery Alters the Endothelial Surface Proteome: Externalized Intracellular Molecules as Potential Vascular Targets in Irradiated Brain Arteriovenous Malformations. Radiat. Res. 2017, 187, 66–78. [Google Scholar] [CrossRef]
- Soulet, F.; Kilarski, W.W.; Roux-Dalvai, F.; Herbert, J.M.J.; Sacewicz-Hofman, I.; Mouton-Barbosa, E.; Bicknell, R.; Lalor, P.; Monsarrat, B.; Bikfalvi, A. Mapping the Extracellular and Membrane Proteome Associated with the Vasculature and the Stroma in the Embryo. Mol. Cell. Proteom. 2013, 12, 2293–2312. [Google Scholar] [CrossRef] [Green Version]
- Toledo, A.G.; Golden, G.; Campos, A.R.; Cuello, H.; Sorrentino, J.; Lewis, N.; Varki, N.; Nizet, V.; Smith, J.W.; Esko, J.D. Proteomic atlas of organ vasculopathies triggered by Staphylococcus aureus sepsis. Nat. Commun. 2019, 10, 4656. [Google Scholar] [CrossRef] [Green Version]
- Roesli, C.; Fugmann, T.; Borgia, B.; Schliemann, C.; Neri, D.; Jucker, M. The accessible cerebral vascular proteome in a mouse model of cerebral β-amyloidosis. J. Proteom. 2011, 74, 539–546. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Lee, S.J.; Kwon, S.; Gatti, J.R.; Korcari, E.; Gresser, T.E.; Felix, P.C.; Keep, S.; Pasquale, K.C.; Bai, T.; Blanchett-Anderson, S.A.; et al. Large-scale identification of human cerebrovascular proteins: Inter-tissue and intracerebral vascular protein diversity. PLoS ONE 2017, 12, e0188540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, A.C.; Stevens, M.Y.; Chen, M.B.; Lee, D.P.; Stähli, D.; Gate, D.; Contrepois, K.; Chen, W.; Iram, T.; Zhang, L.; et al. Physiological blood–brain transport is impaired with age by a shift in transcytosis. Nature 2020, 583, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Pardridge, W.M. The Blood-Brain Barrier and Neurotherapeutics. NeuroRX 2005, 2, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.V.; Prescher, J.A.; Sletten, E.M.; Baskin, J.M.; Miller, I.A.; Agard, N.J.; Lo, A.; Bertozzi, C.R. Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. USA 2010, 107, 1821–1826. [Google Scholar] [CrossRef] [Green Version]
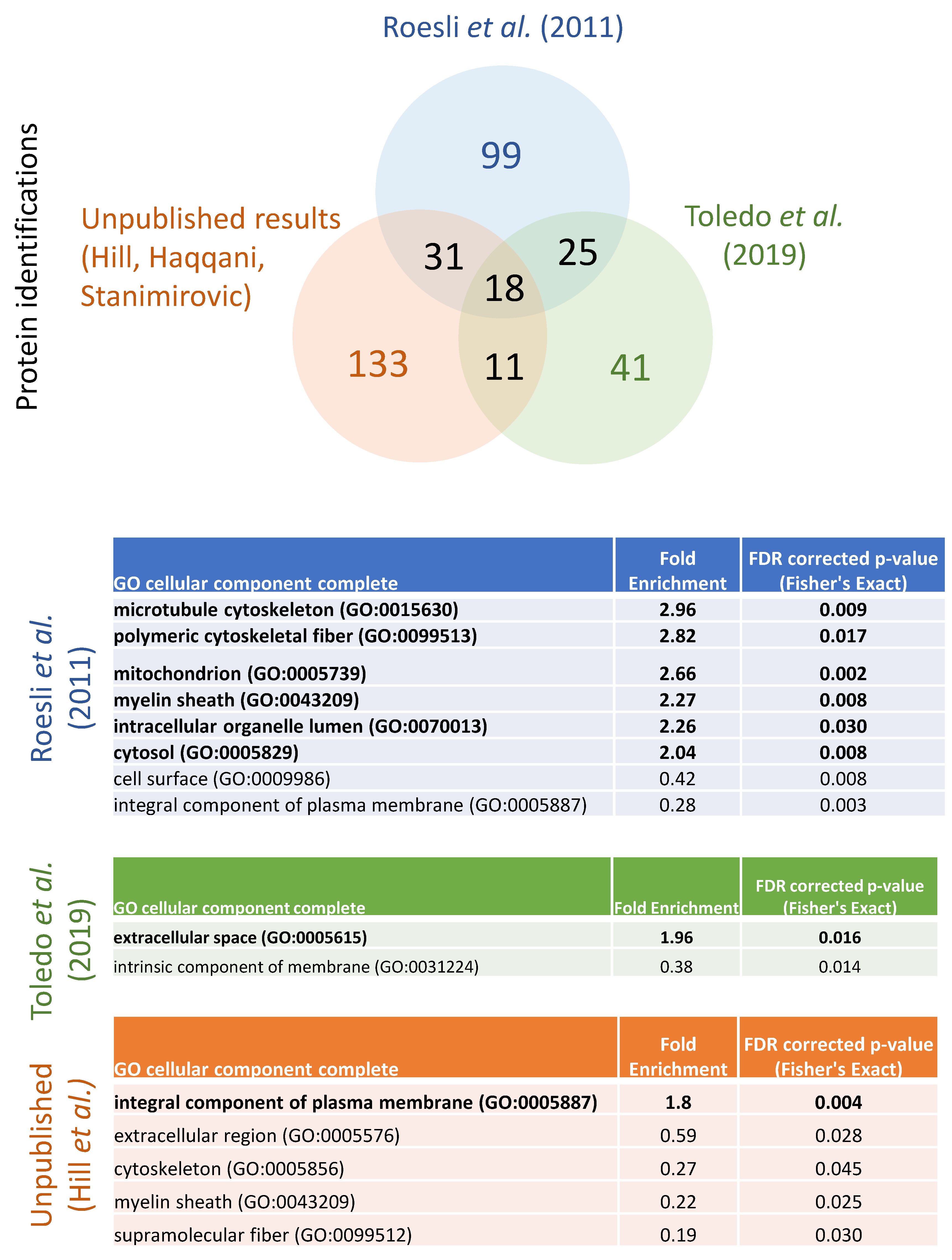
| Study | Sample | # Unique Proteins Identified | % Cell Surface Proteins a | RMT Receptors (HGNC Symbol) b | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| INSR | IGF1R | TFRC | SLC2A1 | LRP1 | TMEM30A | SLC3A2 | FCGRT | BSG | LEPR | LRP8 | LDLR | ||||
| Proteomics of isolated vessels * (membrane focus or post-2015) | |||||||||||||||
| Chun et al., (2011) | enriched mouse brain vessels followed by membrane isolation | 1143 | 14% | ? | Y | Y | Y | Y | ? | Y | Y | Y | N | N | N |
| Badhwar et al., (2014, 2017) | surgical enrichment of mouse brain arteries | 6630 | 11% | ? | N | N | Y | ? | ? | Y | ? | N | N | N | N |
| Al Feteisi et al., (2018) | enriched rat brain vessels | 1897 c | 10% | N | N | Y | Y | Y | Y | Y | Y | Y | N | N | N |
| Campeau et al., (2020) | enriched mouse brain vessels | 3511 c,d | 10% | Y | Y | Y | Y | Y | N | Y | Y | Y | N | N | ? |
| Zajec et al., (2021) | LCM of human brain vessels | 1882 | 8% | N | N | N | Y | Y | N | Y | N | Y | N | N | N |
| Proteomics of perfusion labelled blood-accessible vessel proteins | |||||||||||||||
| Roesli et al., (2011) | perfusion of sulfo-NHS-LC-biotin; enrichment of labelled proteins from mouse brain | 177 | 25% | N | Y | N | Y | N | N | Y | N | Y | N | N | N |
| Toledo et al., (2019) | perfusion of sulfo-NHS-biotin; enrichment of labelled proteins from mouse brain | 96 | 13% | N | N | N | N | N | N | N | N | N | N | N | N |
| Unpublished results (JJH, ASH, DBS-2021) | perfusion-based labelling approach, vessel enrichment, and enrichment of labelled proteins from mouse and rat brain | 193 | 75% | Y | Y | Y | Y | Y | N | Y | N | Y | N | N | N |
| Alternative ‘omics approaches | |||||||||||||||
| Yang et al., (2020) | Top 1% genes showing positive correlation between scRNAseq expression and amount of plasma protein uptake in brain ECs | 199 | 20% | Y | Y | Y | Y | N | N | Y | N | Y | N | Y | N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hill, J.J.; Haqqani, A.S.; Stanimirovic, D.B. Proteome of the Luminal Surface of the Blood–Brain Barrier. Proteomes 2021, 9, 45. https://doi.org/10.3390/proteomes9040045
Hill JJ, Haqqani AS, Stanimirovic DB. Proteome of the Luminal Surface of the Blood–Brain Barrier. Proteomes. 2021; 9(4):45. https://doi.org/10.3390/proteomes9040045
Chicago/Turabian StyleHill, Jennifer J., Arsalan S. Haqqani, and Danica B. Stanimirovic. 2021. "Proteome of the Luminal Surface of the Blood–Brain Barrier" Proteomes 9, no. 4: 45. https://doi.org/10.3390/proteomes9040045
APA StyleHill, J. J., Haqqani, A. S., & Stanimirovic, D. B. (2021). Proteome of the Luminal Surface of the Blood–Brain Barrier. Proteomes, 9(4), 45. https://doi.org/10.3390/proteomes9040045





