Comparison of Sample Preparation Methods for Shotgun Proteomic Studies in Aquaculture Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Protein Extraction
2.3. Sample Preparation for LC-MS Analysis
2.3.1. Filter-Aided Sample Preparation (FASP)
2.3.2. Single-Pot, Solid-Phase-Enhanced Sample Preparation (SP3)
2.3.3. S-Trap Sample Preparation
2.4. LC–MS Analysis
2.5. Protein Identification and Quantification
2.6. Functional Classification of Proteins and Pathway Analysis
2.7. Protein Quantification
3. Results
3.1. Turbot, Scophthalmus Maximus
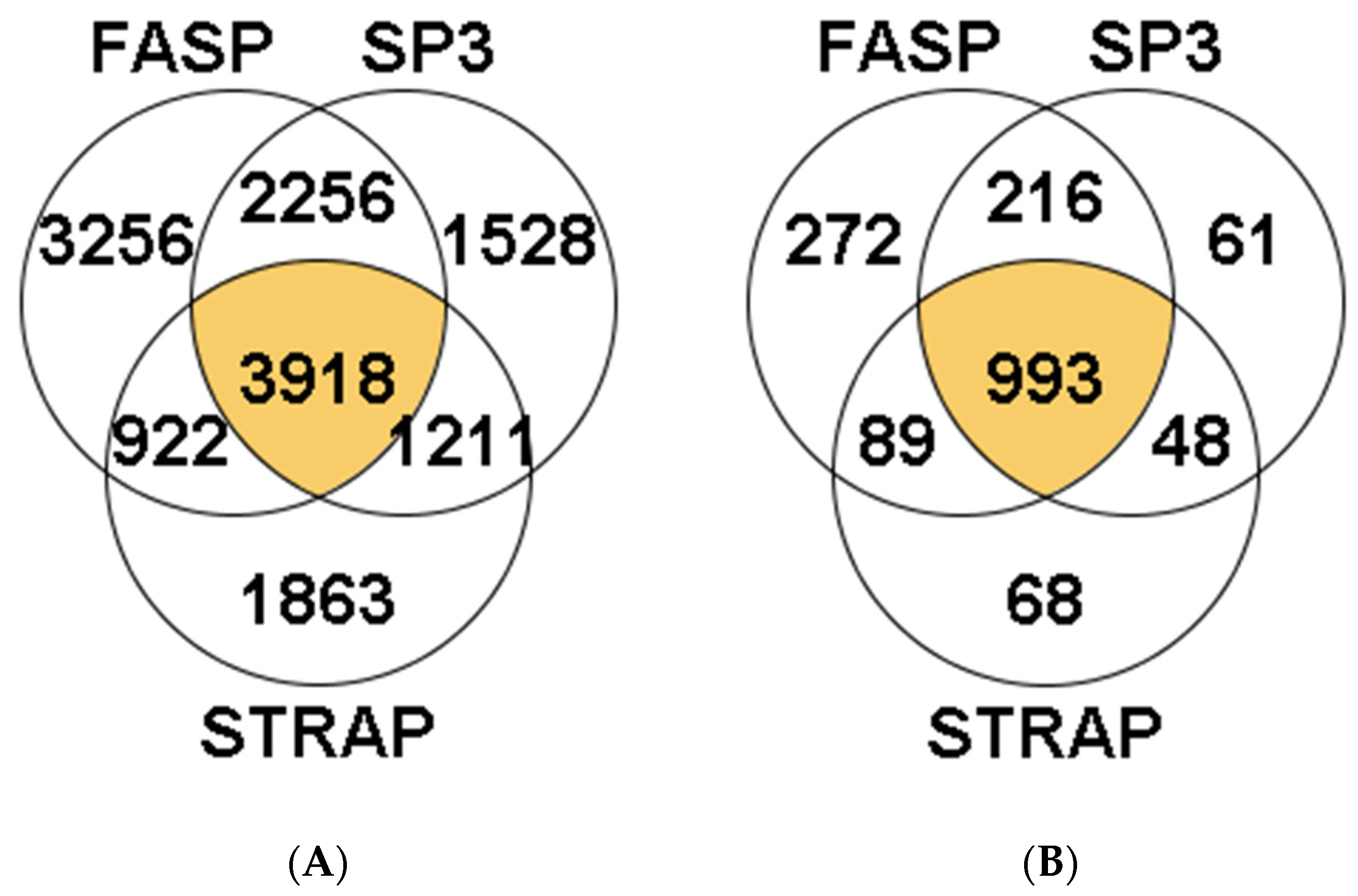
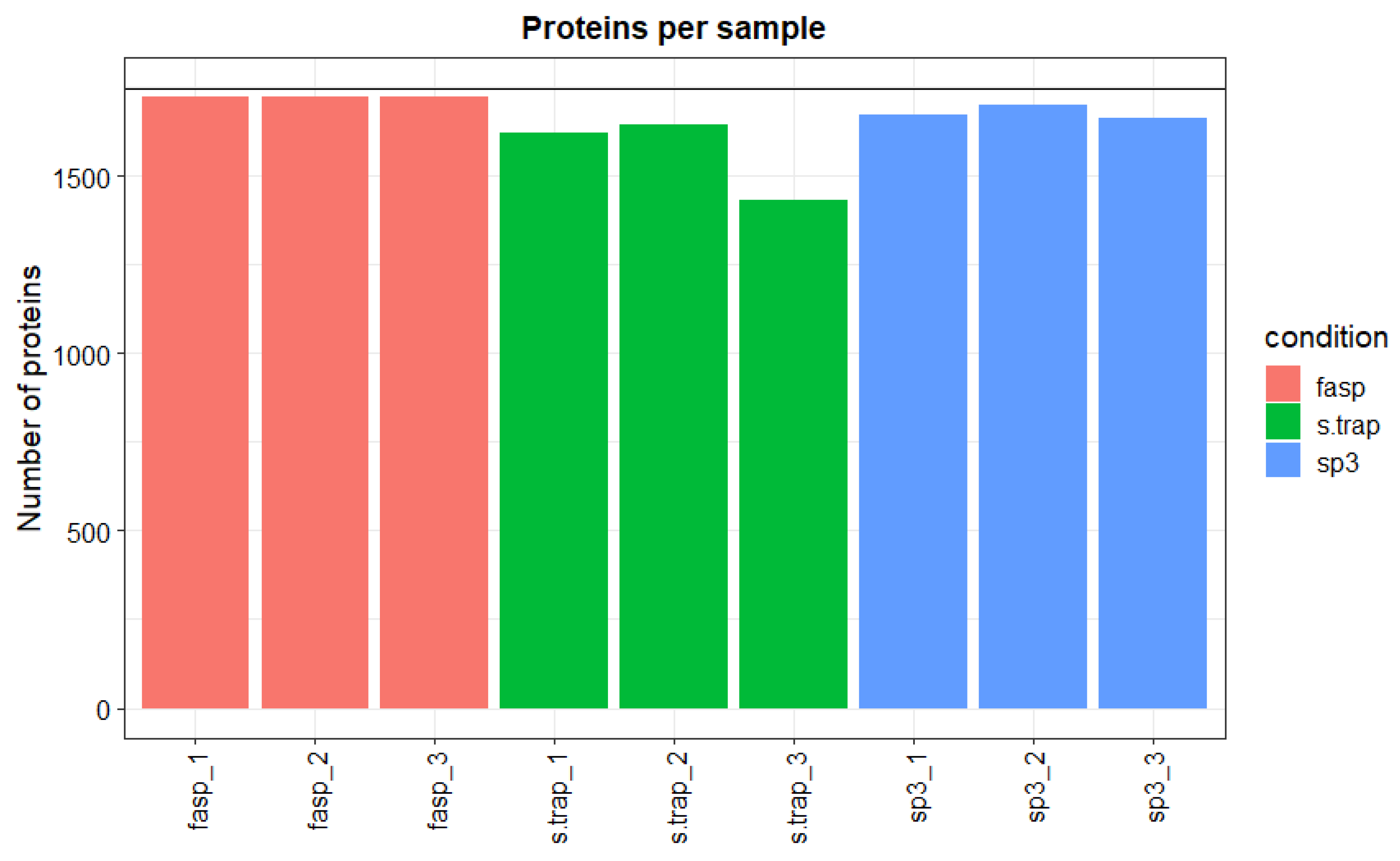
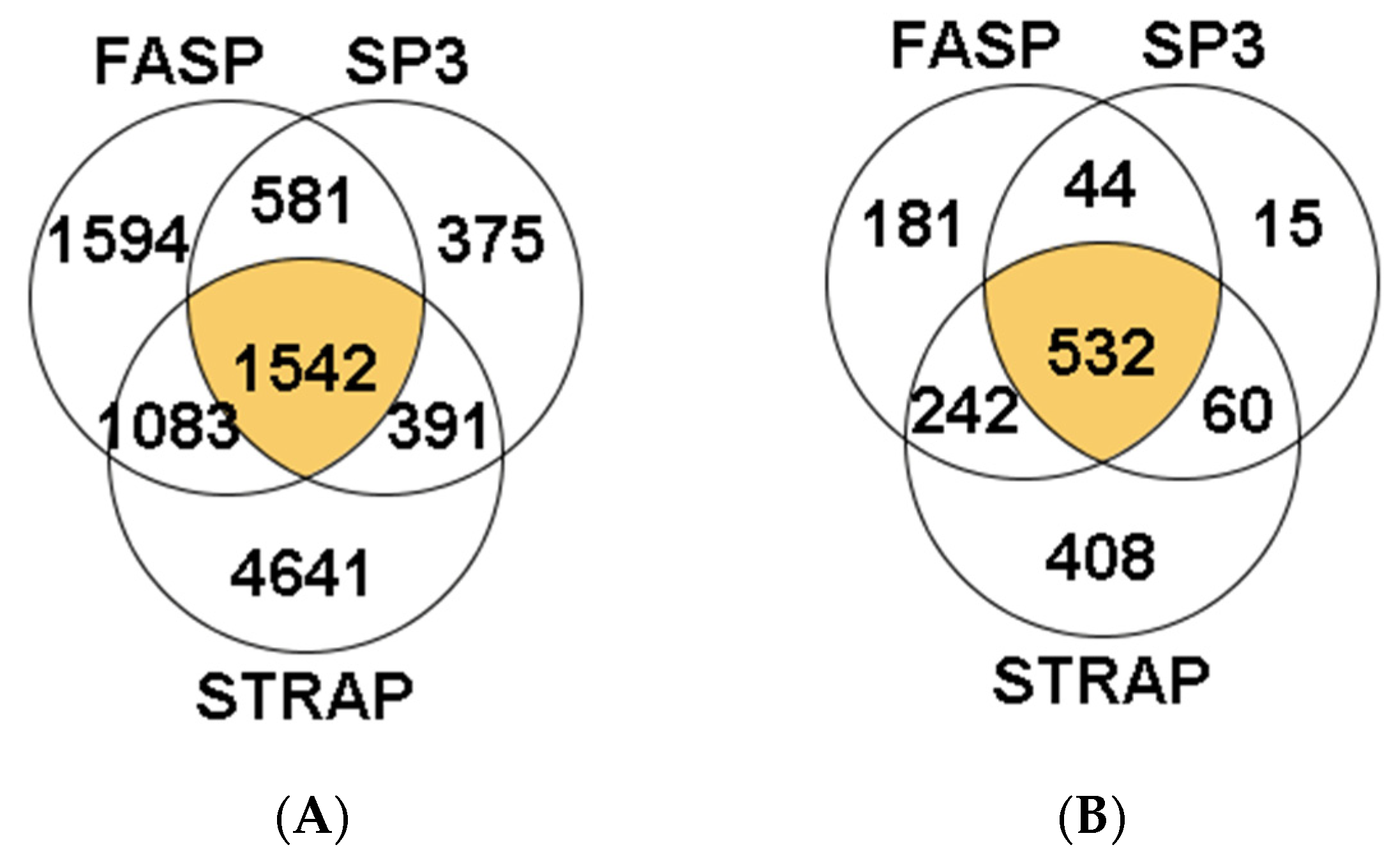
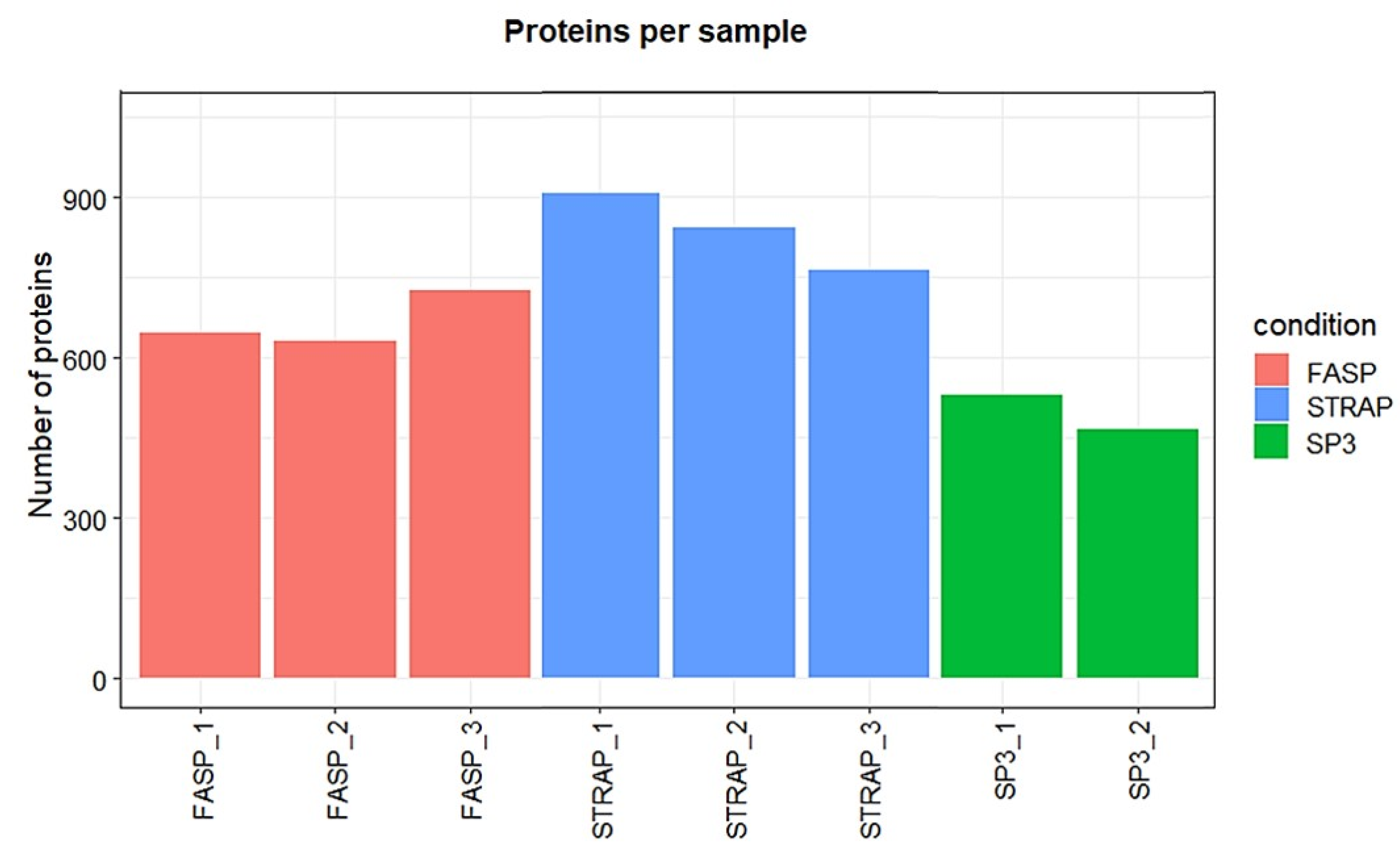
3.1.1. Peptide and Protein Identification
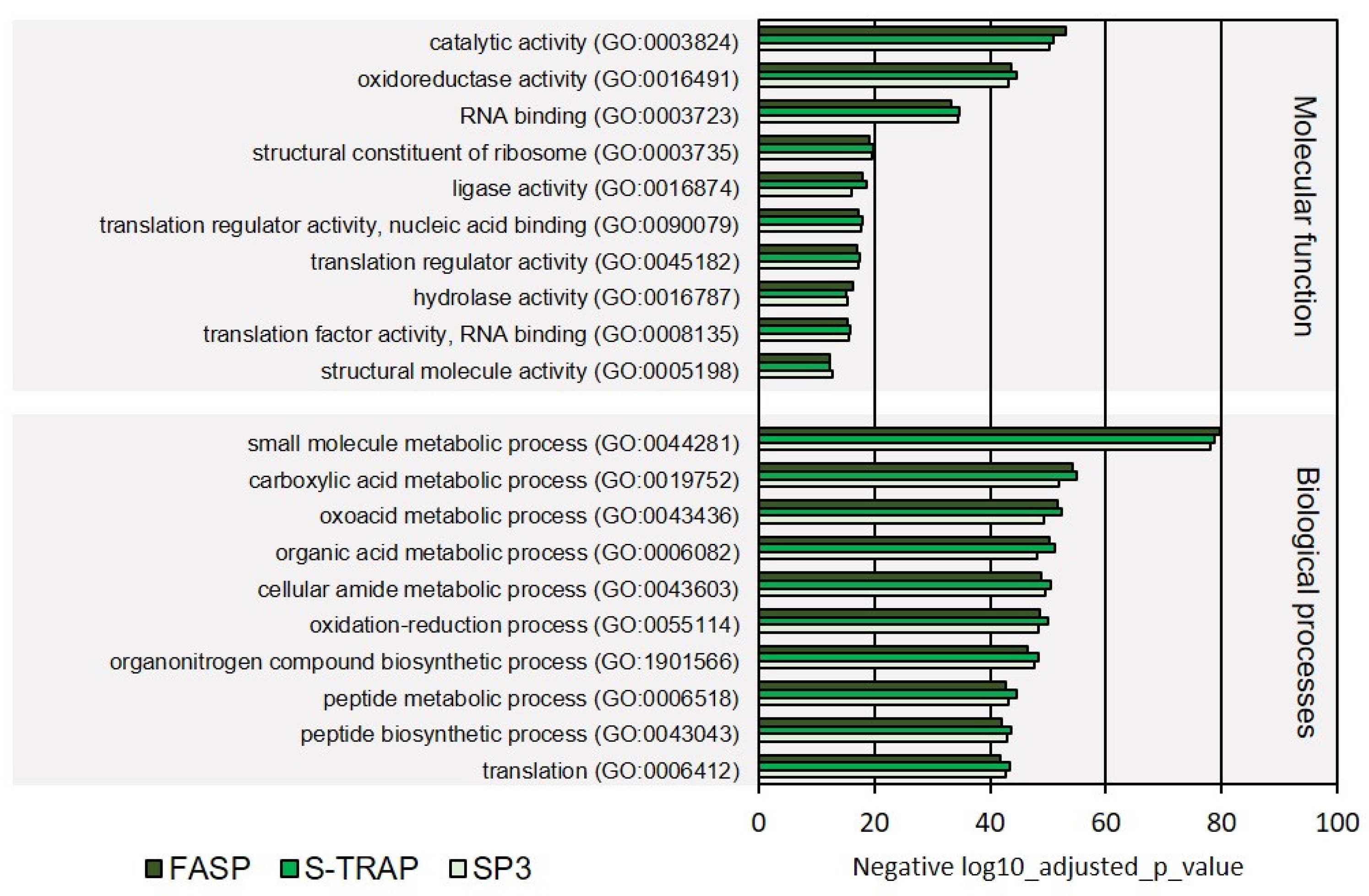
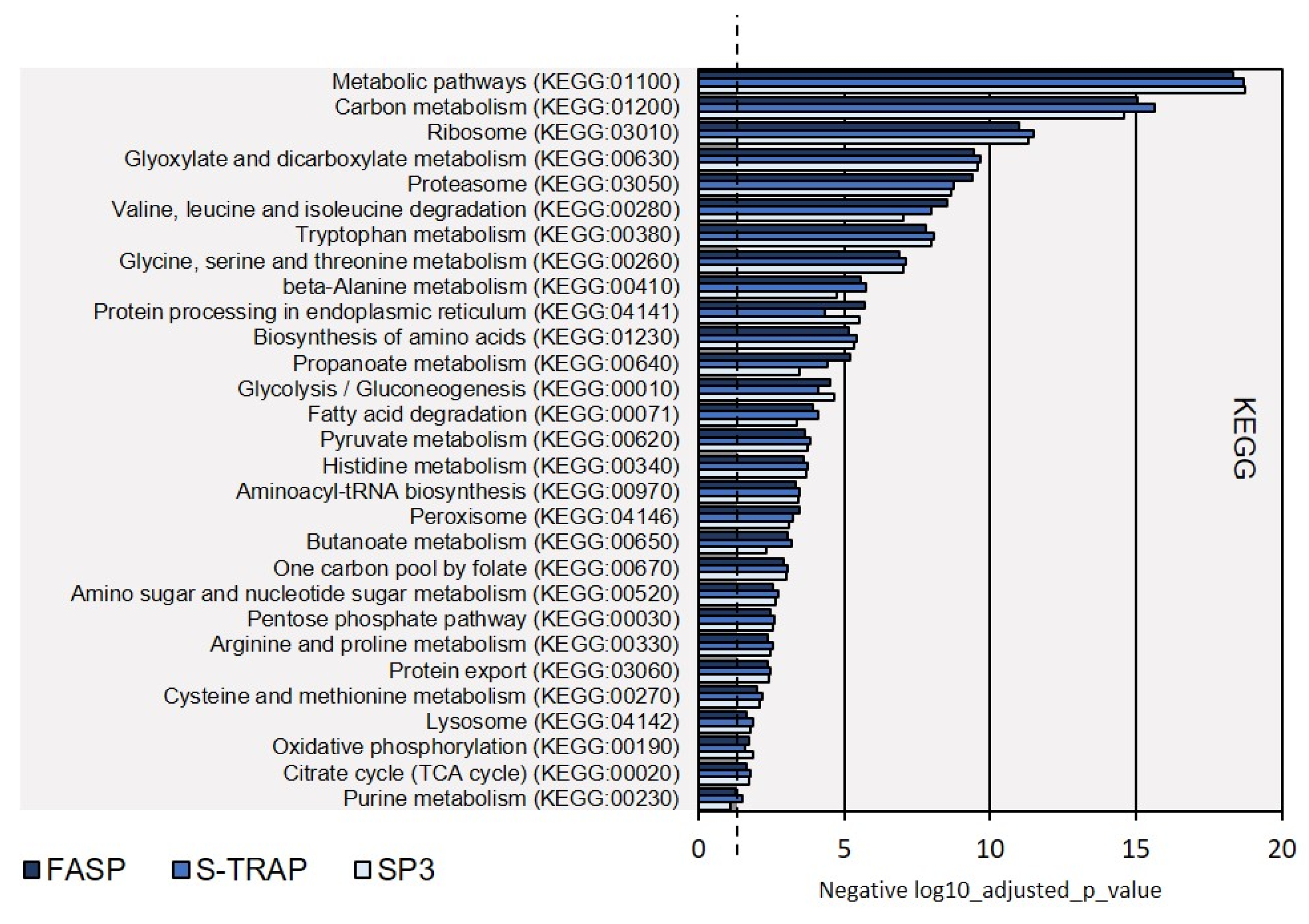
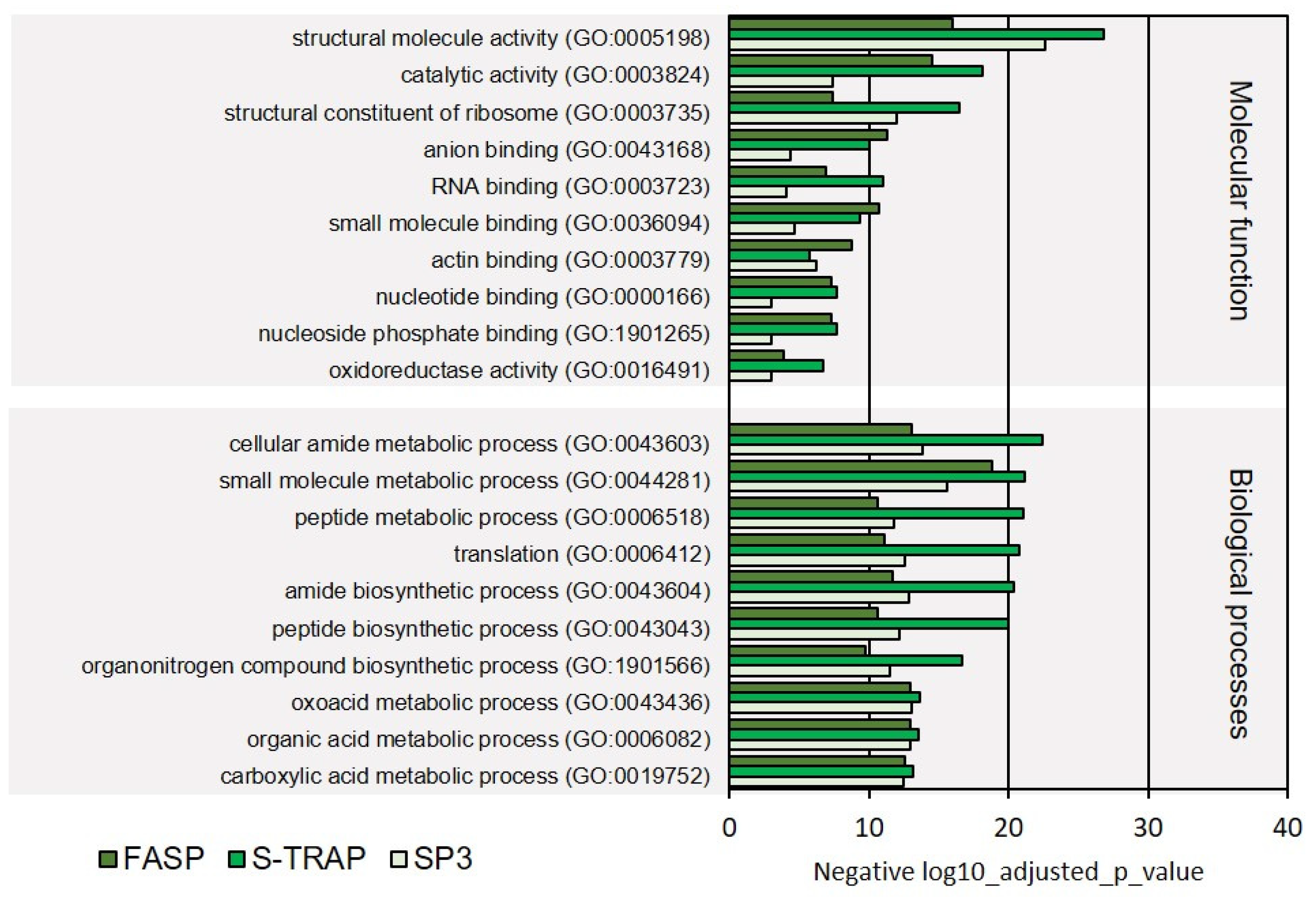
3.1.2. Functional Analysis
3.2. Mediterranean Mussel, Mytilus Galloprovincialis
3.2.1. Peptide and Protein Identification
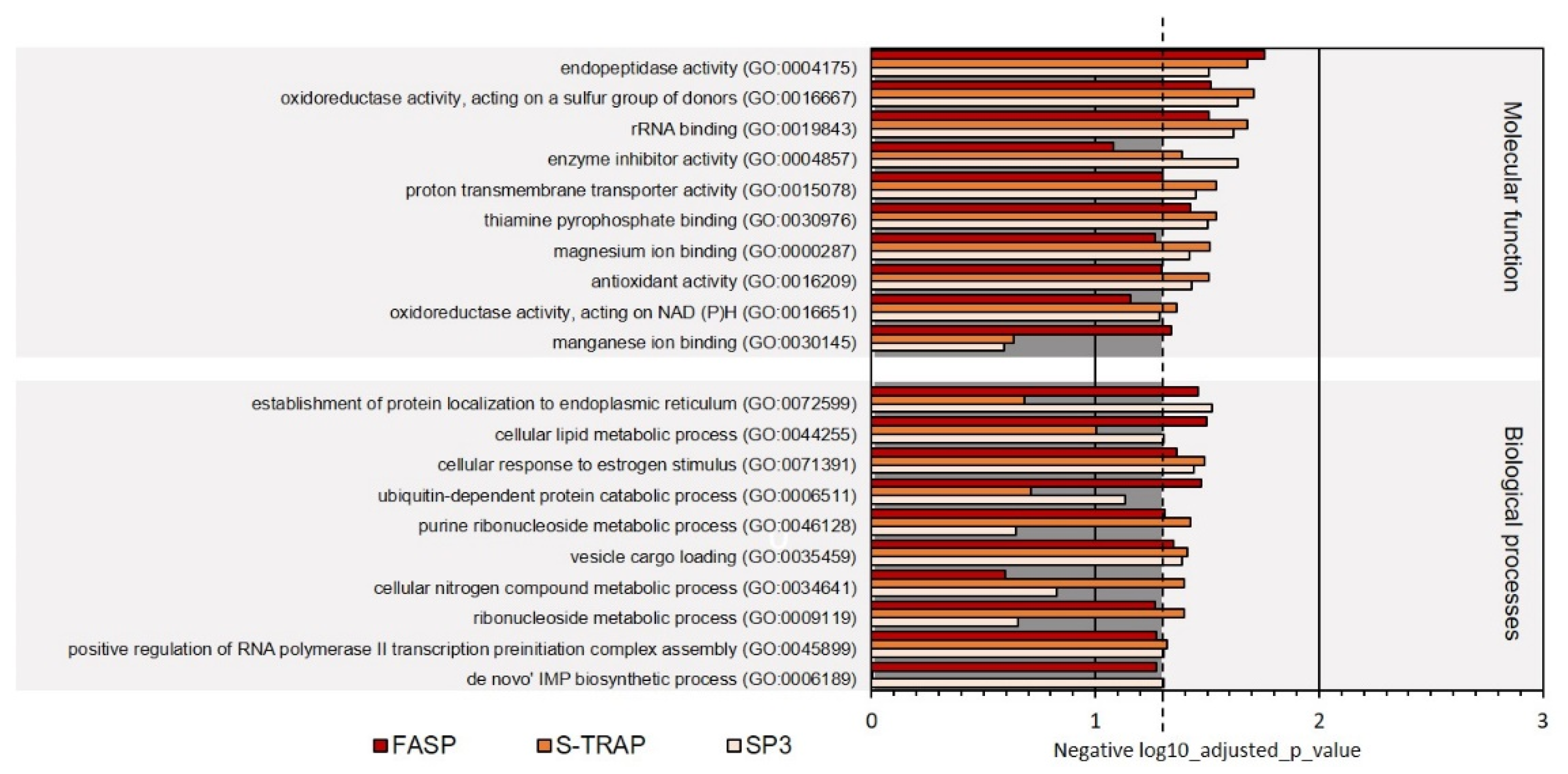
3.2.2. Functional Analysis
3.3. Protein Markers of Oxidative Stress and Response to Toxic Substances
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Nilo-Poyanco, R.; Moraga, C.; Benedetto, G.; Orellana, A.; Almeida, A.M. Shotgun Proteomics of Peach Fruit Reveals Major Metabolic Pathways Associated to Ripening. BMC Genom. 2021, 22, 17. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, J.M.; Washburn, M.P. Advances in shotgun proteomics and the analysis of membrane proteomes. J. Proteom. 2010, 73, 2078–2091. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Danielsson, G.; Farinha, A.P.; Kuruvilla, J.; Warholm, P.; Cristobal, S. Shotgun proteomics to unravel marine mussel (Mytilus edulis) response to long-term exposure to low salinity and propranolol in a Baltic Sea microcosm. J. Proteom. 2016, 137, 97–106. [Google Scholar] [CrossRef] [PubMed]
- De Felice, B.; Parolini, M. Can Proteomics Be Considered as a Valuable Tool to Assess the Toxicity of Nanoparticles in Marine Bivalves? J. Mar. Sci. Eng. 2020, 8, 1033. [Google Scholar] [CrossRef]
- Farinha, A.P.; Schrama, D.; Silva, T.; Conceição, L.E.; Colen, R.; Engrola, S.; Rodrigues, P.; Cerqueira, M. Evaluating the impact of methionine-enriched diets in the liver of European seabass through label-free shotgun proteomics. J. Proteom. 2020, 232, 104047. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Campos, A.; Kuruvilla, J.; Schrama, D.; Cristobal, S. Chapter 17—Proteomics in Aquaculture: Quality and Safety. In Proteomics in Food Science; Colgrave, M.L., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 279–295. [Google Scholar]
- Rodrigues, P.M.; Schrama, D.; Campos, A.; Osório, H.; Freitas, M. Applications of Proteomics in Aquaculture. In Agricultural Proteomics Volume 1: Crops, Horticulture, Farm Animals, Food, Insect and Microorganisms; Salekdeh, G.H., Ed.; Springer International Publishing: Cham, Denmark, 2016; pp. 175–209. [Google Scholar]
- Martins, J.C.; Domínguez-Pérez, D.; Azevedo, C.; Braga, A.C.; Costa, P.R.; Osório, H.; Vasconcelos, V.; Campos, A. Molecular Responses of Mussel Mytilus galloprovincialis Associated to Accumulation and Depuration of Marine Biotoxins Okadaic Acid and Dinophysistoxin-1 Revealed by Shotgun Proteomics. Front. Mar. Sci. 2020, 7, 1101. [Google Scholar] [CrossRef]
- Xie, D.; Wu, J.; Wu, Q.; Zhang, X.; Zhou, D.; Dai, W.; Zhu, M.; Wang, D. Integrating Proteomic, Lipidomic and Metabolomic Data to Construct a Global Metabolic Network of Lethal Ventricular Tachyarrhythmias (Lvta) Induced by Aconitine. J. Proteom. 2021, 232, 104043. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Ostasiewicz, P.; Mann, M. High Recovery FASP Applied to the Proteomic Analysis of Microdissected Formalin Fixed Paraffin Embedded Cancer Tissues Retrieves Known Colon Cancer Markers. J. Proteome Res. 2011, 10, 3040–3049. [Google Scholar] [CrossRef]
- Campos, A.; Tedesco, S.; Vasconcelos, V.; Cristobal, S. Proteomic Research in Bivalves: Towards the Identification of Molecular Markers of Aquatic Pollution. J. Proteom. 2012, 75, 4346–4359. [Google Scholar] [CrossRef]
- Causey, D.R.; Pohl, M.A.N.; Stead, D.; Martin, S.A.M.; Secombes, C.J.; MacQueen, D.J. High-throughput proteomic profiling of the fish liver following bacterial infection. BMC Genom. 2018, 19, 719. [Google Scholar] [CrossRef]
- Boersema, P.J.; Kahraman, A.; Picotti, P. Proteomics beyond large-scale protein expression analysis. Curr. Opin. Biotechnol. 2015, 34, 162–170. [Google Scholar] [CrossRef]
- Mastroleo, F.; Leroy, B.; Van Houdt, R.; Heeren, C.S.; Mergeay, M.; Hendrickx, L.; Wattiez, R. Shotgun Proteome Analysis of Rhodospirillum rubrum S1H: Integrating Data from Gel-Free and Gel-Based Peptides Fractionation Methods. J. Proteome Res. 2009, 8, 2530–2541. [Google Scholar] [CrossRef] [PubMed]
- Dowell, J.A.; Frost, D.; Zhang, J.; Li, L. Comparison of Two-Dimensional Fractionation Techniques for Shotgun Proteomics. Anal. Chem. 2008, 80, 6715–6723. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, P.Y.; Saraygord-Afshari, N.; Low, T.Y. The evolution of two-dimensional gel electrophoresis—From proteomics to emerging alternative applications. J. Chromatogr. A 2019, 1615, 460763. [Google Scholar] [CrossRef]
- Domínguez-Pérez, D.; Lippolis, J.; Dennis, M.; Miller, B.; Tiley, K.; Vasconcelos, V.; de Almeida, A.M.; Campos, A. The Queen Conch (Lobatus gigas) Proteome: A Valuable Tool for Biological Studies in Marine Gastropods. Protein J. 2019, 38, 628–639. [Google Scholar] [CrossRef]
- Matallana-Surget, S.; Leroy, B.; Wattiez, R. Shotgun Proteomics: Concept, Key Points and Data Mining. Expert Rev. Proteom. 2010, 7, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- HaileMariam, M.; Eguez, R.V.; Singh, H.; Bekele, S.; Ameni, G.; Pieper, R.; Yu, Y. S-Trap, an Ultrafast Sample-Preparation Approach for Shotgun Proteomics. J. Proteome Res. 2018, 17, 2917–2924. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.S.; Moggridge, S.; Müller, T.; Sorensen, P.H.; Morin, G.B.; Krijgsveld, J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019, 14, 68–85. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zielinska, D.F.; Mann, M. Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal. Biochem. 2011, 410, 307–309. [Google Scholar] [CrossRef]
- Ni, M.W.; Wang, L.; Chen, W.; Mou, H.Z.; Zhou, J.; Zheng, Z.G. Modified Filter-Aided Sample Preparation (Fasp) Method Increases Peptide and Protein Identifications for Shotgun Proteomics. Rapid Commun. Mass Spectrom. 2017, 31, 171–178. [Google Scholar] [CrossRef]
- Nel, A.J.M.; Garnett, S.; Blackburn, J.M.; Soares, N.C. Comparative Reevaluation of FASP and Enhanced FASP Methods by LC–MS/MS. J. Proteome Res. 2015, 14, 1637–1642. [Google Scholar] [CrossRef]
- Olkowicz, M.; Jablonska, P.; Rogowski, J.; Smolenski, R.T. Simultaneous Accurate Quantification of Ho-1, Cd39, and Cd73 in Human Calcified Aortic Valves Using Multiple Enzyme Digestion—Filter Aided Sample Pretreatment (Med-Fasp) Method and Targeted Proteomics. Talanta 2018, 182, 492–499. [Google Scholar] [CrossRef]
- Wang, W.; Wu, X.; Liu, Z.; Zheng, H.; Cheng, Y. Insights into Hepatopancreatic Functions for Nutrition Metabolism and Ovarian Development in the Crab Portunus Trituberculatus: Gene Discovery in the Comparative Transcriptome of Different Hepatopancreas Stages. PLoS ONE 2014, 9, e84921. [Google Scholar]
- Dupree, E.J.; Crimmins, B.S.; Holsen, T.M.; Darie, C.C. Proteomic Analysis of the Lake Trout (Salvelinus namaycush) Liver Identifies Proteins from Evolutionarily Close and -Distant Fish Relatives. Proteomics 2019, 19, e1800429. [Google Scholar] [CrossRef] [PubMed]
- Dupree, E.J.; Crimmins, B.S.; Holsen, T.M.; Darie, C.C. Developing Well-Annotated Species-Specific Protein Databases Using Comparative Proteogenomics. Adv. Exp. Med. Biol. 2019, 1140, 389–400. [Google Scholar]
- Nuez-Ortín, W.G.; Carter, C.G.; Nichols, P.D.; Cooke, I.R.; Wilson, R. Liver proteome response of pre-harvest Atlantic salmon following exposure to elevated temperature. BMC Genom. 2018, 19, 133. [Google Scholar] [CrossRef] [PubMed]
- Nhu, T.Q.; Hang, B.T.B.; Cornet, V.; Oger, M.; Bach, L.T.; Dao, N.L.A.; Huong, D.T.T.; Quetin-Leclercq, J.; Scippo, M.L.; Phuong, N.T.; et al. Single or Combined Dietary Supply of Psidium guajava and Phyllanthus amarus Extracts Differentially Modulate Immune Responses and Liver Proteome in Striped Catfish (Pangasianodon hyphophthalmus). Front. Immunol. 2020, 11, 797. [Google Scholar] [CrossRef] [PubMed]
- Ayobahan, S.U.; Eilebrecht, S.; Baumann, L.; Teigeler, M.; Hollert, H.; Kalkhof, S.; Eilebrecht, E.; Schäfers, C. Detection of biomarkers to differentiate endocrine disruption from hepatotoxicity in zebrafish (Danio rerio) using proteomics. Chemosphere 2019, 240, 124970. [Google Scholar] [CrossRef]
- Campos, A.; Puerto, M.; Prieto, A.; Cameán, A.; Almeida, A.M.; Coelho, A.V.; Vasconcelos, V. Protein Extraction and Two-Dimensional Gel Electrophoresis of Proteins in the Marine Mussel Mytilus galloprovincialis: An Important Tool for Protein Expression Studies, Food Quality and Safety Assessment. J. Sci. Food Agric. 2013, 93, 1779–1787. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Apraiz, I.; da Fonseca, R.R.; Cristobal, S. Shotgun Analysis of the Marine Mussel Mytilus Edulis Hemolymph Proteome and Mapping the Innate Immunity Elements. Proteomics 2015, 15, 4021–4029. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway Enrichment Analysis and Visualization of Omics Data Using G:Profiler, Gsea, Cytoscape and Enrichmentmap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Dessau, R.B.; Pipper, C.B. “R”—Project for Statistical Computing. Ugeskr Laeger 2008, 170, 328–330. [Google Scholar] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.gbif.org/pt/tool/81287/r-a-language-andenvironment-for-statistical-computing (accessed on 30 December 2020).
- Murgarella, M.; Puiu, D.; Novoa, B.; Figueras, A.; Posada, D.; Canchaya, C. A First Insight into the Genome of the Filter-Feeder Mussel Mytilus galloprovincialis. PLoS ONE 2016, 11, e0151561. [Google Scholar]
- Lu, J.; Holmgren, A. The Thioredoxin Antioxidant System. Free. Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef]
- Aceto, A.; Amicarelli, F.; Sacchetta, P.; Dragani, B.; Bucciarelli, T.; Masciocco, L.; Miranda, M.; Di Ilio, C. Developmental Aspects of Detoxifying Enzymes in Fish (Salmo Iridaeus). Free Radic. Res. 1994, 21, 285–294. [Google Scholar] [CrossRef]
- Antas, P.; Carneiro, M.; Reis, B.; Castelo-Branco, R.; Azevedo, J.; Urbatzka, R.; Campos, A.; Vasconcelos, V.; Martins, J.C. Gst Transcriptional Changes Induced by a Toxic Microcystis Aeruginosa Strain in Two Bivalve Species During Exposure and Recovery Phases. Ecotoxicology 2018, 27, 1272–1280. [Google Scholar] [CrossRef]
- Trevisan, R.; Mello, D.F.; Uliano-Silva, M.; Delapedra, G.; Arl, M.; Dafre, A.L. The Biological Importance of Glutathione Peroxidase and Peroxiredoxin Backup Systems in Bivalves During Peroxide Exposure. Mar. Environ. Res. 2014, 101, 81–90. [Google Scholar] [CrossRef]
- Gerdol, M.; Moreira, R.; Cruz, F.; Gómez-Garrido, J.; Vlasova, A.; Rosani, U.; Venier, P.; Naranjo-Ortiz, M.A.; Murgarella, M.; Greco, S.; et al. Massive gene presence-absence variation shapes an open pan-genome in the Mediterranean mussel. Genome Biol. 2020, 21, 275. [Google Scholar] [CrossRef]
- Sielaff, M.; Kuharev, J.; Bohn, T.; Hahlbrock, J.; Bopp, T.; Tenzer, S.; Distler, U. Evaluation of Fasp, Sp3, and Ist Protocols for Proteomic Sample Preparation in the Low Microgram Range. J. Proteome Res. 2017, 16, 4060–4072. [Google Scholar] [CrossRef] [PubMed]
- Mikulášek, K.; Konečná, H.; Potěšil, D.; Holánková, R.; Havliš, J.; Zdráhal, Z. SP3 Protocol for Proteomic Plant Sample Preparation Prior Lc-Ms/Ms. Front. Plant Sci. 2021, 12, 369. [Google Scholar] [CrossRef]
- Cleland, T.P. Human Bone Paleoproteomics Utilizing the Single-Pot, Solid-Phase-Enhanced Sample Preparation Method to Maximize Detected Proteins and Reduce Humics. J. Proteome Res. 2018, 17, 3976–3983. [Google Scholar] [CrossRef] [PubMed]
- Blankenburg, S.; Hentschker, C.; Nagel, A.; Hildebrandt, P.; Michalik, S.; Dittmar, D.; Surmann, K.; Völker, U. Improving Proteome Coverage for Small Sample Amounts: An Advanced Method for Proteomics Approaches with Low Bacterial Cell Numbers. Proteomics 2019, 19, e1900192. [Google Scholar] [CrossRef] [PubMed]
- Balotf, S.; Wilson, R.; Tegg, R.S.; Nichols, D.S.; Wilson, C.R. Optimisation of Sporosori Purification and Protein Extraction Techniques for the Biotrophic Protozoan Plant Pathogen Spongospora Subterranea. Molecules 2020, 25, 3109. [Google Scholar] [CrossRef]
- Tremblay, T.-L.; Hill, J.J. Adding polyvinylpyrrolidone to low level protein samples significantly improves peptide recovery in FASP digests: An inexpensive and simple modification to the FASP protocol. J. Proteom. 2020, 230, 104000. [Google Scholar] [CrossRef]
- Pirog, A.; Faktor, J.; Urban-Wojciuk, Z.; Kote, S.; Chruściel, E.; Arcimowicz, Ł.; Marek-Trzonkowska, N.; Vojtesek, B.; Hupp, T.R.; Shboul, S.A.; et al. Comparison of Different Digestion Methods for Proteomic Analysis of Isolated Cells and Ffpe Tissue Samples. Talanta 2021, 233, 122568. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Dubiak, K.M.; Huber, P.W.; Dovichi, N.J. Miniaturized Filter-Aided Sample Preparation (MICRO-FASP) Method for High Throughput, Ultrasensitive Proteomics Sample Preparation Reveals Proteome Asymmetry in Xenopus laevis Embryos. Anal. Chem. 2020, 92, 5554–5560. [Google Scholar] [CrossRef]
- Gonzalez-Lozano, M.A.; Koopmans, F.; Paliukhovich, I.; Smit, A.B.; Li, K.W. A Fast and Economical Sample Preparation Protocol for Interaction Proteomics Analysis. Proteomics 2019, 19, e1900027. [Google Scholar] [CrossRef]
- Robledo, D.; Palaiokostas, C.; Bargelloni, L.; Martínez, P.; Houston, R. Applications of genotyping by sequencing in aquaculture breeding and genetics. Rev. Aquac. 2018, 10, 670–682. [Google Scholar] [CrossRef] [PubMed]
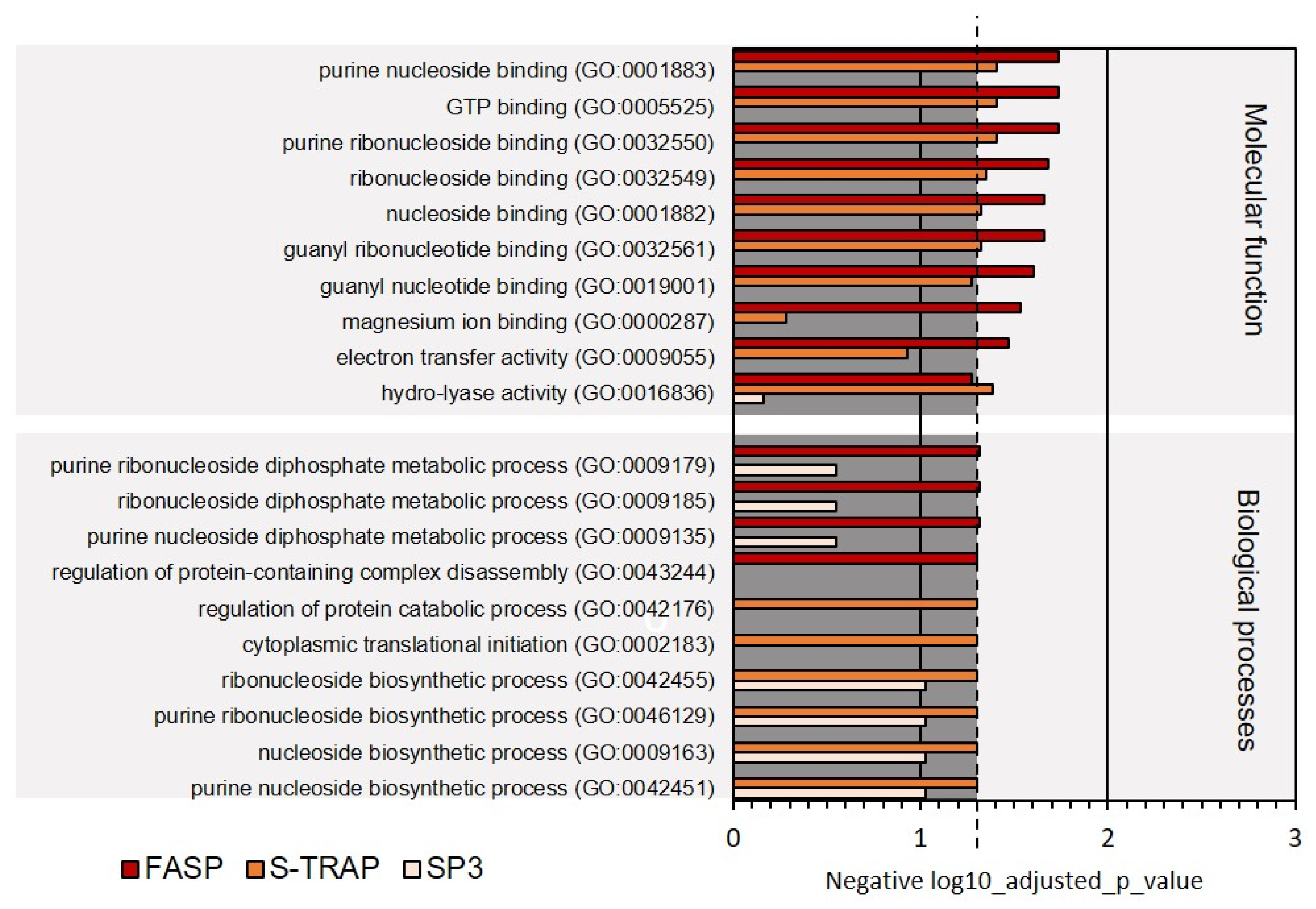
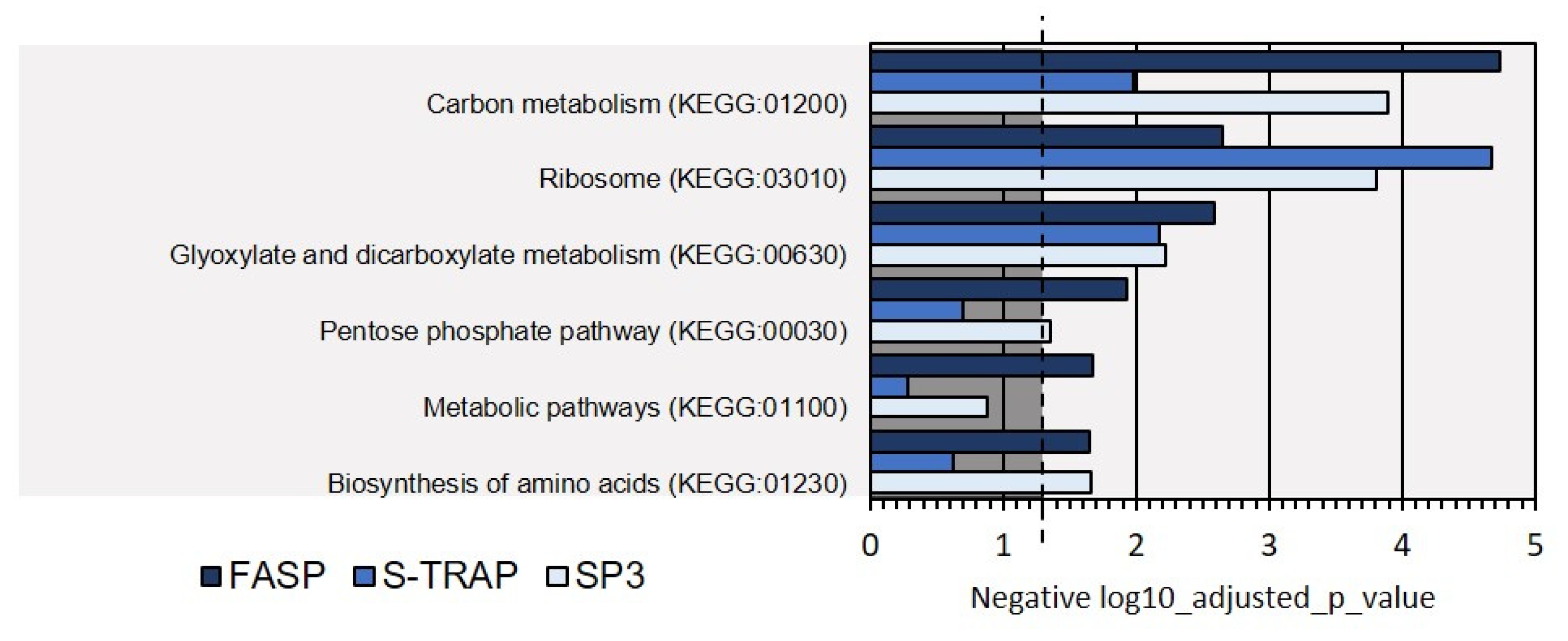
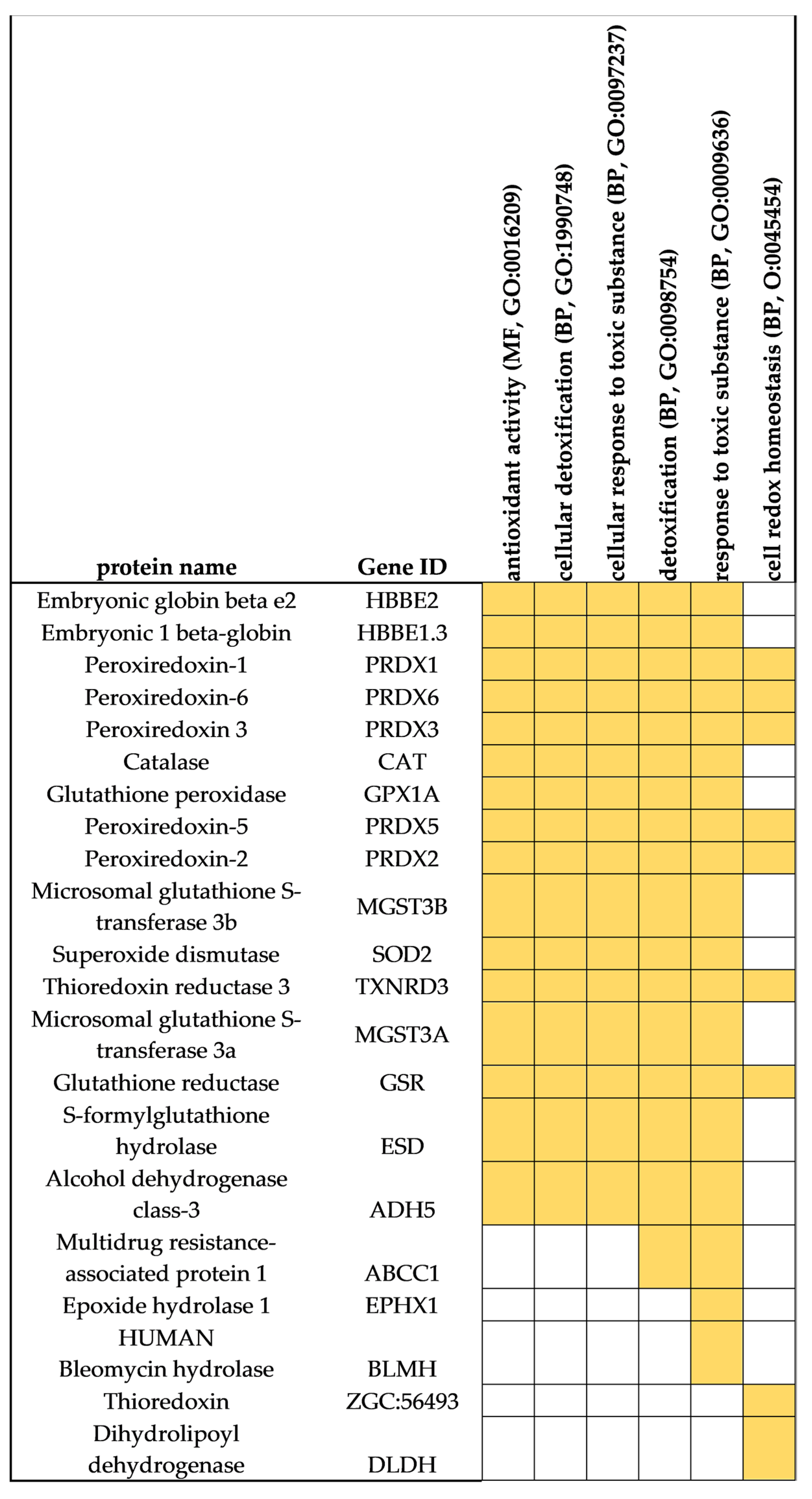
| GO Categories | FASP | S-TRAP | SP3 |
|---|---|---|---|
| Molecular Function | 76 | 78 | 77 |
| Biological Processes | 219 | 215 | 213 |
| Total | 295 | 293 | 290 |
| GO Categories | FASP | S-TRAP | SP3 |
|---|---|---|---|
| Molecular Function | 63 | 58 | 32 |
| Biological Processes | 95 | 90 | 75 |
| Total | 158 | 148 | 107 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araújo, M.J.; Sousa, M.L.; Felpeto, A.B.; Turkina, M.V.; Fonseca, E.; Martins, J.C.; Vasconcelos, V.; Campos, A. Comparison of Sample Preparation Methods for Shotgun Proteomic Studies in Aquaculture Species. Proteomes 2021, 9, 46. https://doi.org/10.3390/proteomes9040046
Araújo MJ, Sousa ML, Felpeto AB, Turkina MV, Fonseca E, Martins JC, Vasconcelos V, Campos A. Comparison of Sample Preparation Methods for Shotgun Proteomic Studies in Aquaculture Species. Proteomes. 2021; 9(4):46. https://doi.org/10.3390/proteomes9040046
Chicago/Turabian StyleAraújo, Mário Jorge, Maria Lígia Sousa, Aldo Barreiro Felpeto, Maria V. Turkina, Elza Fonseca, José Carlos Martins, Vítor Vasconcelos, and Alexandre Campos. 2021. "Comparison of Sample Preparation Methods for Shotgun Proteomic Studies in Aquaculture Species" Proteomes 9, no. 4: 46. https://doi.org/10.3390/proteomes9040046
APA StyleAraújo, M. J., Sousa, M. L., Felpeto, A. B., Turkina, M. V., Fonseca, E., Martins, J. C., Vasconcelos, V., & Campos, A. (2021). Comparison of Sample Preparation Methods for Shotgun Proteomic Studies in Aquaculture Species. Proteomes, 9(4), 46. https://doi.org/10.3390/proteomes9040046










