Urinary Extracellular Vesicles and Salt-Losing Tubulopathies: A Proteomic Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Patients
2.3. Urine Collection and Exosome Isolation
2.4. Electrophoresis and Western Blotting Analysis
2.4.1. Benzyldimethyl-n-Hexadecylammonium Chloride/Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (16-BAC/SDS-PAGE)
2.4.2. Western Blotting Analysis
2.5. Mass Spectrometry and Protein Identification
2.6. Bioinformatic Analysis
2.7. Statistical Analysis
3. Results
3.1. uEV Characterization
3.2. Two-Dimensional 16-BAC/SDS Polyacrylamide Gel Electrophoresis Separation of uEV Proteins
3.3. Identification of uEV Proteins
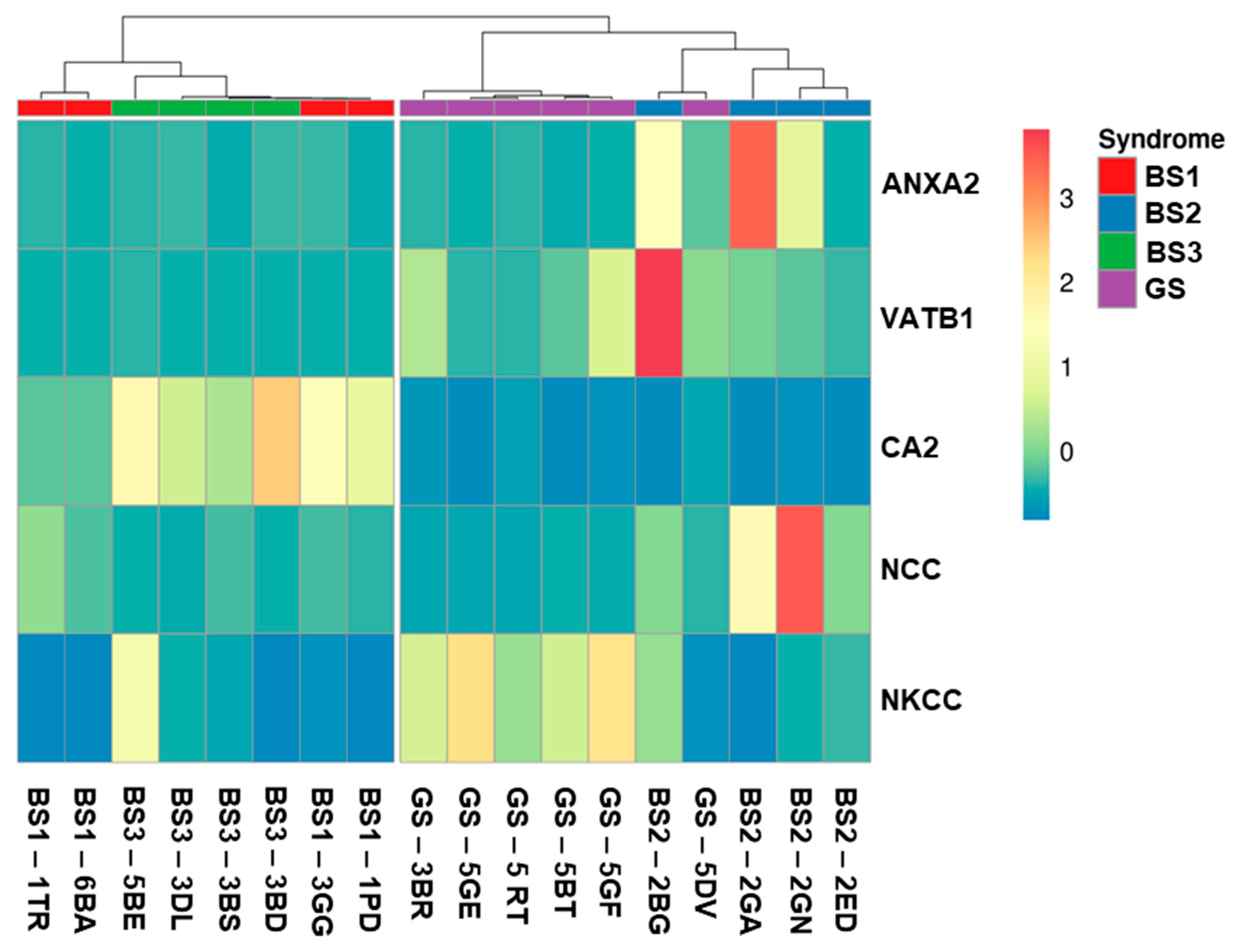
3.4. Western Blotting of Differential uEV Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Seyberth, H.W.; Weber, S.; Kömhoff, M. Bartter’s and Gitelman’s syndrome. Curr. Opin. Pediatr. 2017, 29, 179–186. [Google Scholar] [CrossRef]
- Besouw, M.T.P.; Kleta, R.; Bockenhauer, D. Bartter and Gitelman syndromes: Questions of class. Pediatr. Nephrol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Koulouridis, E.; Koulouridis, I. Molecular pathophysiology of Bartter’s and Gitelman’s syndromes. World J. Pediatr. 2015, 11, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Salih, M.; Bovée, D.M.; van der Lubbe, N.; Danser, A.H.J.; Zietse, R.; Feelders, R.A.; Hoorn, E.J. Increased Urinary Extracellular Vesicle Sodium Transporters in Cushing Syndrome with Hypertension. J. Clin. Endocrinol. Metab. 2018, 103, 2583–2591. [Google Scholar] [CrossRef] [PubMed]
- Riveira-Munoz, E.; Chang, Q.; Godefroid, N.; Hoenderop, J.G.; Bindels, R.J.; Dahan, K.; Devuyst, O. Transcriptional and functional analyses of SLC12A3 mutations: New clues for the pathogenesis of Gitelman syndrome. J. Am. Soc. Nephrol. 2007, 18, 1271–1283. [Google Scholar] [CrossRef]
- Cunha, T.D.S.; Heilberg, I.P. Bartter syndrome: Causes, diagnosis, and treatment. Int. J. Nephrol. Renovasc. Dis. 2018, 11, 291–301. [Google Scholar] [CrossRef]
- Matsunoshita, N.; Nozu, K.; Shono, A.; Nozu, Y.; Fu, X.J.; Morisada, N.; Kamiyoshi, N.; Ohtsubo, H.; Ninchoji, T.; Minamikawa, S.; et al. Differential diagnosis of Bartter syndrome, Gitelman syndrome, and pseudo-Bartter/Gitelman syndrome based on clinical characteristics. Genet. Med. 2016, 18, 180–188. [Google Scholar] [CrossRef]
- Nozu, K.; Yamamura, T.; Horinouchi, T.; Nagano, C.; Sakakibara, N.; Ishikura, K.; Hamada, R.; Morisada, N.; Iijima, K. Inherited salt-losing tubulopathy: An old condition but a new category of tubulopathy. Pediatr. Int. 2019. [Google Scholar] [CrossRef]
- Gollasch, B.; Anistan, Y.M.; Canaan-Kuhl, S.; Gollasch, M. Late-onset Bartter syndrome type II. Clin. Kidney J. 2017, 10, 594–599. [Google Scholar] [CrossRef][Green Version]
- Sharma, A.; Linshaw, M.A. A novel compound heterozygous ROMK mutation presenting as late onset Bartter syndrome associated with nephrocalcinosis and elevated 1,25(OH)(2) vitamin D levels. Clin. Exp. Nephrol. 2011, 15, 572–576. [Google Scholar] [CrossRef]
- García Castaño, A.; Pérez de Nanclares, G.; Madariag, L.; Aguirre, M.; Madrid, Á.; Chocrón, S.; Nadal, I.; Navarro, M.; Lucas, E.; Fijo, J.; et al. Poor phenotype-genotype association in a large series of patients with Type III Bartter syndrome. PLoS ONE 2017, 12, e0173581. [Google Scholar] [CrossRef] [PubMed]
- Corbetta, S.; Raimondo, F.; Tedeschi, S.; Syrèn, M.L.; Rebora, P.; Savoia, A.; Baldi, L.; Bettinelli, A.; Pitto, M. Urinary exosomes in the diagnosis of Gitelman and Bartter syndromes. Nephrol. Dial. Transplant. 2015, 30, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Merchant, M.L.; Rood, I.M.; Deegens, J.K.J.; Klein, J.B. Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nat. Rev. Nephrol. 2017, 13, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, F.; Cerra, D.; Magni, F.; Pitto, M. Urinary proteomics for the study of genetic kidney diseases. Expert Rev. Proteomics 2016, 13, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.L.; Bastos, C.; Faria, N.; Karet Frank, F.E. Making urinary extracellular vesicles a clinically tractable source of biomarkers for inherited tubulopathies using a small volume precipitation method: Proof of concept. J. Nephrol. 2020, 33, 383–386. [Google Scholar] [CrossRef]
- Raimondo, F.; Morosi, L.; Corbetta, S.; Chinello, C.; Brambilla, P.; Della Mina, P.; Villa, A.; Albo, G.; Battaglia, C.; Bosari, S.; et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol. Biosyst. 2013, 9, 1220–1233. [Google Scholar] [CrossRef]
- Hartinger, J.; Stenius, K.; Högemann, D.; Jahn, R. 16-BAC/SDS-PAGE: A two-dimensional gel electrophoresis system suitable for the separation of integral membrane proteins. Anal. Biochem. 1996, 240, 126–133. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Raimondo, F.; Corbetta, S.; Savoia, A.; Chinello, C.; Cazzaniga, M.; Rocco, F.; Bosari, S.; Grasso, M.; Bovo, G.; Magni, F.; et al. Comparative membrane proteomics: A technical advancement in the search of renal cell carcinoma biomarkers. Mol. BioSyst. 2015, 11, 1708–1716. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Dear, J.W.; Street, J.M.; Bailey, M.A. Urinary exosomes: A reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics 2013, 13, 1572–1580. [Google Scholar] [CrossRef] [PubMed]
- Webber, J.; Clayton, A. How pure are your vesicles? J. Extracell Vesicles 2013, 2, 19861. [Google Scholar] [CrossRef] [PubMed]
- Moon, P.G.; You, S.; Lee, J.E.; Hwang, D.; Baek, M.C. Urinary exosomes and proteomics. Mass Spectrom Rev. 2011, 30, 1185–1202. [Google Scholar] [CrossRef]
- Wang, Z.; Hill, S.; Luther, J.M.; Hachey, D.L.; Schey, K.L. Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT). Proteomics 2012, 12, 329–338. [Google Scholar] [CrossRef]
- Wang, S.; Kojima, K.; Mobley, J.A.; West, A.B. Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBio Med. 2019, 45, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Thongboonkerd, V. Roles for Exosome in Various Kidney Diseases and Disorders. Front. Pharmacol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, F. Highlights on the capacities of “gel-based” proteomics. Proteome Sci. 2010, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Santoni, V.; Molloy, M.; Rabilloud, T. Membrane proteins and proteomics: Un amour impossible? Electrophoresis 2000, 21, 1054–1070. [Google Scholar] [CrossRef]
- Yoneten, K.K.; Kasap, M.; Akpinar, G.; Kanli, A.; Karaoz, E. Comparative Proteomics Analysis of Four Commonly Used Methods for Identification of Novel Plasma Membrane Proteins. J. Membr. Biol. 2019, 252, 587–608. [Google Scholar] [CrossRef]
- Trevisiol, S.; Ayoub, D.; Lesur, A.; Ancheva, L.; Gallien, S.; Domon, B. The use of proteases complementary to trypsin to probe isoforms and modifications. Proteomics 2016, 16, 715–728. [Google Scholar] [CrossRef]
- Pan, Y.; Mao, J.; Deng, Z.; Dong, M.; Bian, Y.; Ye, M.; Zou, H. The proteomic analysis improved by cleavage kinetics-based fractionation of tryptic peptides. Proteomics 2015, 15, 3613–3616. [Google Scholar] [CrossRef] [PubMed]
- Erozenci, L.A.; Böttger, F.; Bijnsdorp, I.V.; Jimenez, C.R. Urinary exosomal proteins as (pan-)cancer biomarkers: Insights from the proteome. FEBS Lett. 2019, 593, 1580–1597. [Google Scholar] [CrossRef] [PubMed]
- Lonnerholm, G.; Ridderstrale, Y. Intracellular distribution of carbonic anhydrase in the rat kidney. Kidney Int. 1980, 17, 162–174. [Google Scholar] [CrossRef]
- Xu, J.; Barone, S.; Brooks, M.B.; Soleimani, M. Double knockout of carbonic anhydrase II (CAII) and Na(+)-Cl(-) cotransporter (NCC) causes salt wasting and volume depletion. Cell Physiol. Biochem. 2013, 32, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.A.; Imenez Silva, P.H.; Bourgeois, S. Molecular Pathophysiology of Acid-Base Disorders. Semin. Nephrol. 2019, 39, 340–352. [Google Scholar] [CrossRef]
- Mazaheri, M.; Assadi, F.; Sadeghi-Bojd, S. Adjunctive acetazolamide therapy for the treatment of Bartter syndrome. Int. Urol. Nephrol. 2020, 52, 121–128. [Google Scholar] [CrossRef]
- Katoh, M. Functional proteomics, human genetics and cancer biology of GIPC family members. Exp. Mol. Med. 2013, 7, 45. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Pal, K.; Sharma, A.K.; Dutta, S.K.; Lau, J.S.; Yan, I.K.; Wang, E.; Elkhanany, A.; Alkharfy, K.M.; Sanyal, A.; et al. GAIP interacting protein C-terminus regulates autophagy and exosome biogenesis of pancreatic cancer through metabolic pathways. PLoS ONE 2014, 9, e114409. [Google Scholar] [CrossRef]
- Lou, X.; McQuistan, T.; Orlando, R.A.; Farquhar, M.G.J. GAIP, GIPC and Galphai3 are concentrated in endocytic compartments of proximal tubule cells: Putative role in regulating megalin’s function. Am. Soc. Nephro. 2002, 13, 918–927. [Google Scholar]
- Harris, A.N.; Grimm, P.R.; Lee, H.W.; Delpire, E.; Fang, L.; Verlander, J.W.; Welling, P.A.; Weiner, I.D. Mechanism of Hyperkalemia-Induced Metabolic Acidosis. J. Am. Soc. Nephrol. 2018, 29, 1411–1425. [Google Scholar] [CrossRef]
- Frische, S.; Chambrey, R.; Trepiccione, F.; Zamani, R.; Marcussen, N.; Alexander, R.T.; Skjødt, K.; Svenningsen, P.; Dimke, H. H+-ATPase B1 subunit localizes to thick ascending limb and distal convoluted tubule of rodent and human kidney. Am. J. Physiol. Renal Physiol. 2018, 315, F429–F444. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T. Improving outcomes for patients with distal renal tubular acidosis: Recent advances and challenges ahead. Pediatric Health Med. Ther. 2018, 9, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Naveen, P.S.; Srikanth, L.; Venkatesh, K.; Sarma, P.V.; Sridhar, N.; Krishnakishore, C.; Sandeep, Y.; Manjusha, Y.; Sivakumar, V. Distal renal tubular acidosis with nerve deafness secondary to ATP6B1 gene mutation. Saudi J. Kidney Dis. Transpl. 2015, 26, 119–121. [Google Scholar]
- Norgett, E.E.; Golder, Z.J.; Lorente-Cánovas, B.; Ingham, N.; Steel, K.P.; Karet Frank, F.E. Atp6v0a4 knockout mouse is a model of distal renal tubular acidosis with hearing loss, with additional extrarenal phenotype. Proc. Natl. Acad. Sci. USA 2012, 109, 13775–13780. [Google Scholar] [CrossRef] [PubMed]
- Dathe, C.; Daigeler, A.L.; Seifert, W.; Jankowski, V.; Mrowka, R.; Kalis, R.; Wanker, E.; Mutig, K.; Bachmann, S.; Paliege, A. Annexin A2 mediates apical trafficking of renal Na+-K+-2Cl− cotransporter. J. Biol. Chem. 2014, 289, 9983–9997. [Google Scholar] [CrossRef]
- Sabaratnam, R.; Geertsen, L.; Skjødt, K.; Højlund, K.; Dimke, H.; Lund, L.; Svenningsen, P. In human nephrectomy specimens, the kidney level of tubular transport proteins does not correlate with their abundance in urinary extracellular vesicles. Am. J. Physiol. Renal Physiol. 2019, 317, F560–F571. [Google Scholar] [CrossRef]
- Blijdorp, C.J.; Hoorn, E.J. Urinary extracellular vesicles: The mothership connection. Am. J. Physiol. Renal Physiol. 2019, 317, F648–F649. [Google Scholar] [CrossRef]
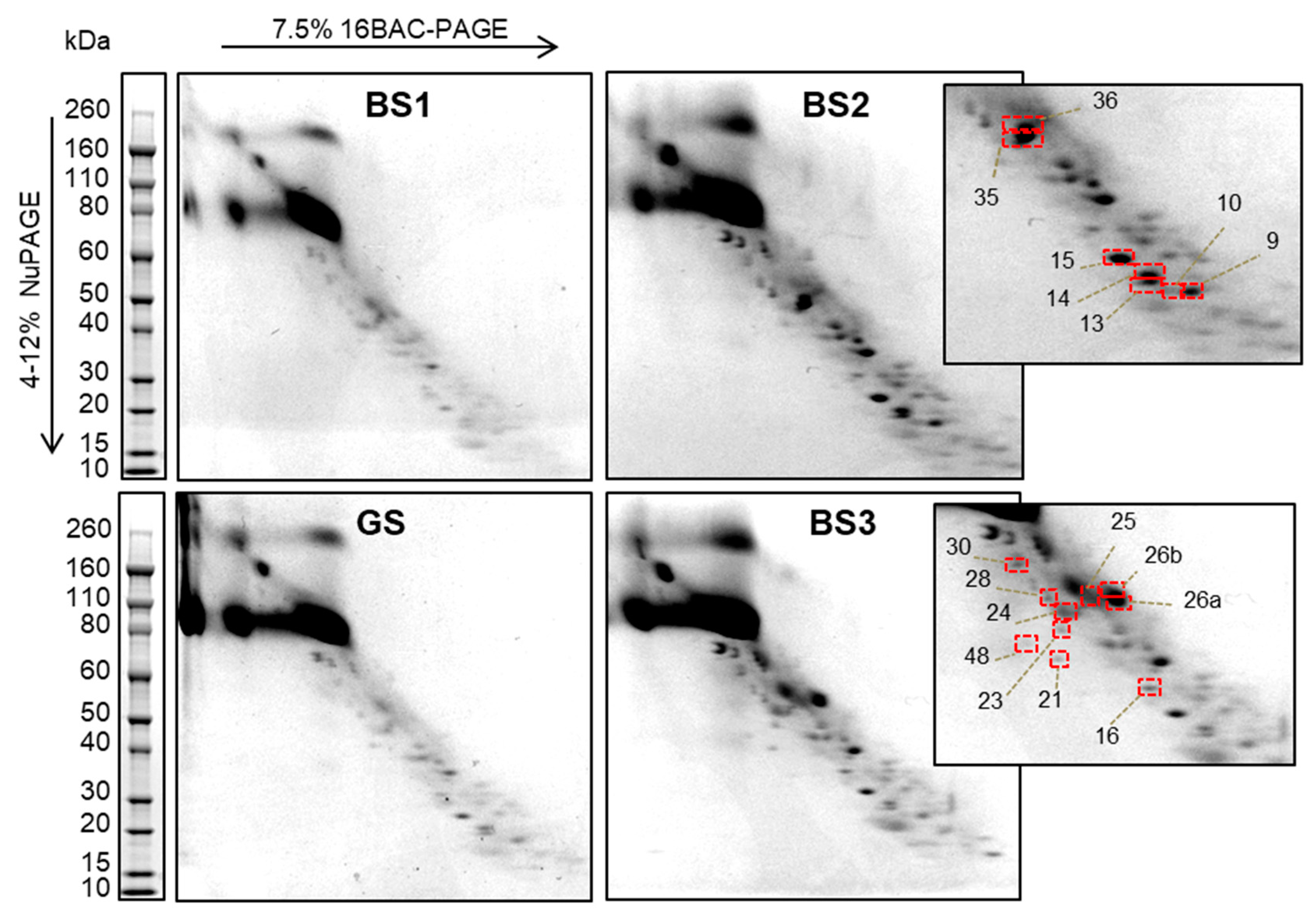
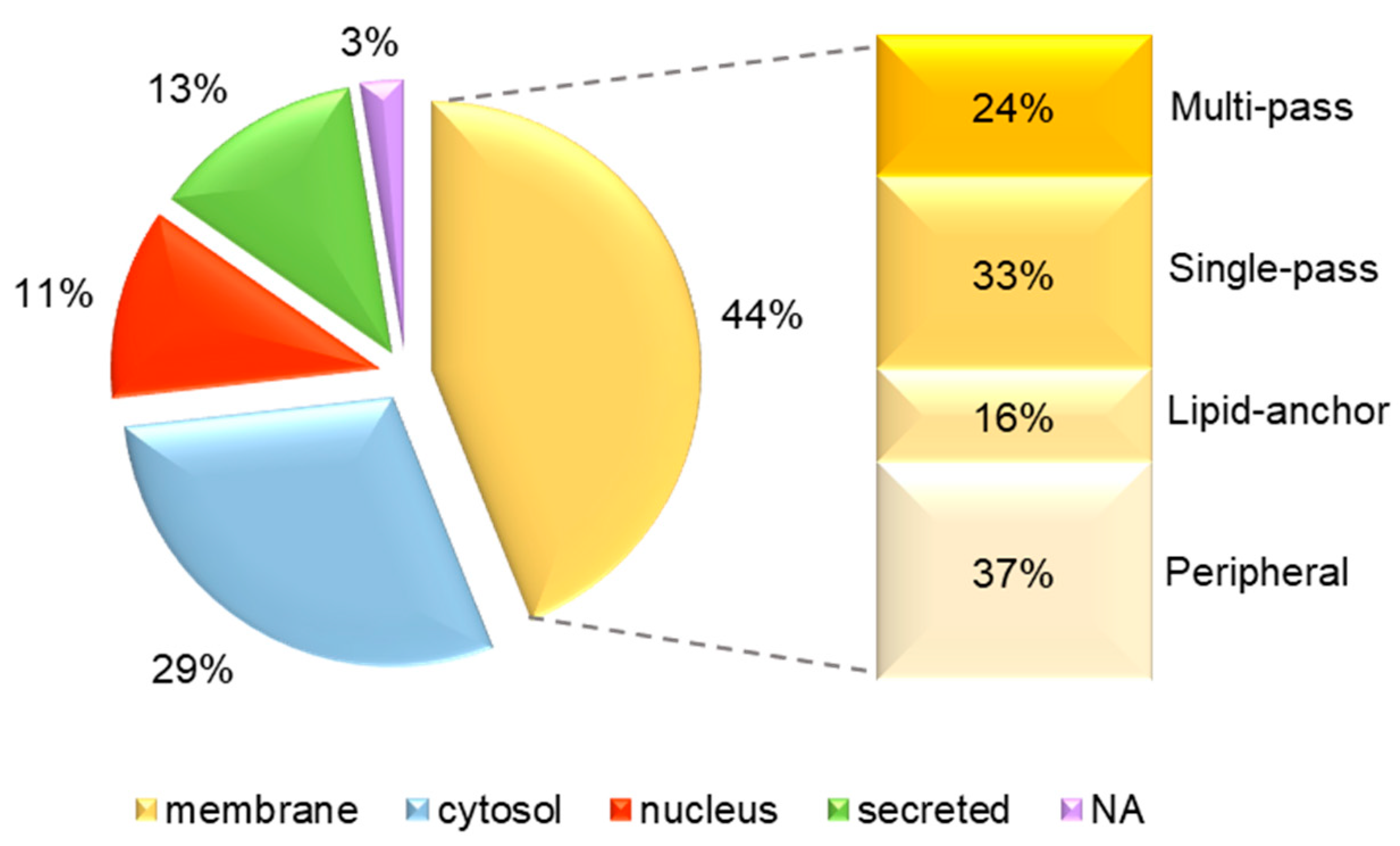
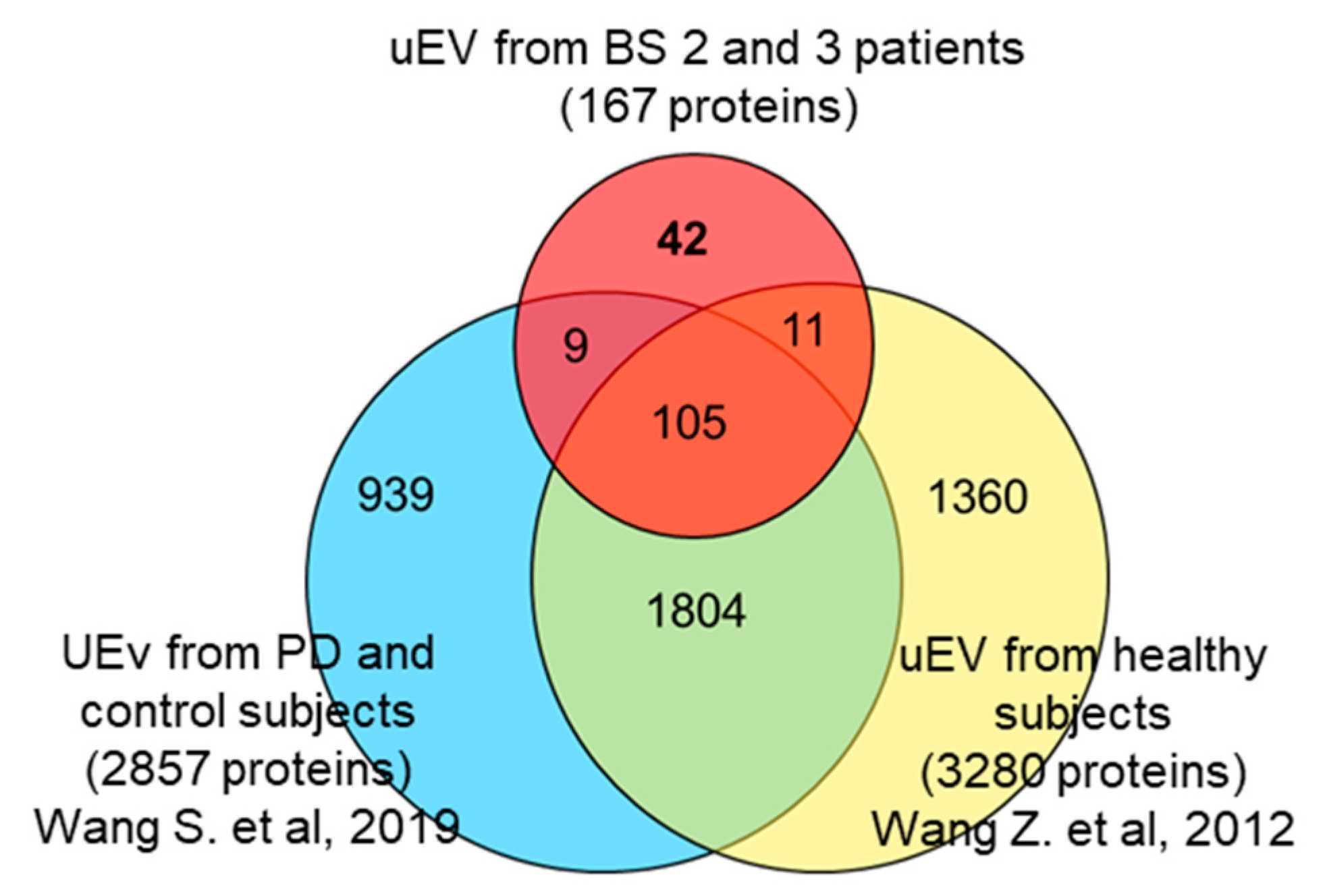
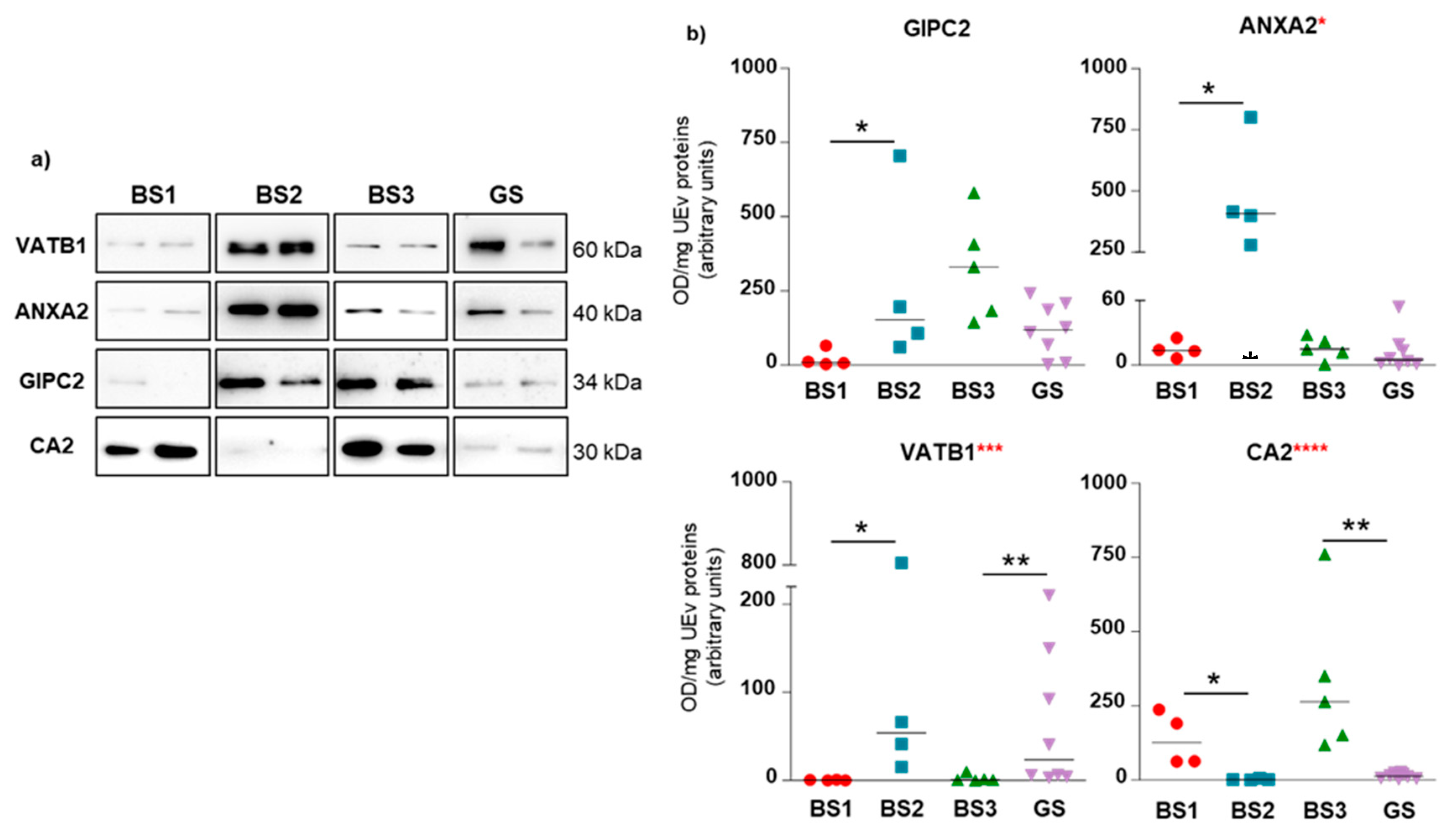
| Patient Code | Age | Sex | Gene Affected | Mutation at DNA Level * | Changes at protein Level | Mutation Type | 16-BAC | WB |
|---|---|---|---|---|---|---|---|---|
| Gitelman Syndrome | ||||||||
| 3BR04 | 7 | M | SLC12A3 | c.[2981G>A] | p.[Cys994Tyr] | M | X | |
| c.[506-?_741+?del] | p.[Val169_Gln247del] | LD | ||||||
| 5BL94 | 16 | M | SLC12A3 | c.[2029G>A] | p.[Val677Met] | M | X | |
| c.[506-?_741+?del] | p.[Val169_Gln247del] | LD | ||||||
| 5BT06 | 5 | M | SLC12A3 | c.[1924C>G] | p.[Arg642Gly] | M | X | |
| c.[2981G>A] | p.[Cys994Tyr] | M | ||||||
| 5CM97 | 14 | M | SLC12A3 | c.[2981G>A] | p.[Cys994Tyr] | M | X | |
| c.[2981G>A] | p.[Cys994Tyr] | M | ||||||
| 5CE94 | 17 | F | SLC12A3 | c.[1844C>T] | p.[Ser615Leu] | M | X | X |
| c.[1925G>A] | p.[Arg642His] | M | ||||||
| 5DV85 | 26 | M | SLC12A3 | c.[2899A>G] | p.[Arg967Gly] | M | X | X |
| c.[2899A>G] | p.[Arg967Gly] | M | ||||||
| 5GE01^ | 10 | M | SLC12A3 | c.[1196_1202dup7bp] | p.[Ser402*] | N | X | |
| c.[1424C>G] | p.[Ser475Cys] | M | ||||||
| 5GF01^ | 10 | M | SLC12A3 | c.[1196_1202dup7bp] | p.[Ser402*] | N | X | |
| c.[1424C>G] | p.[Ser475Cys] | M | ||||||
| 5MR86 | 25 | F | SLC12A3 | c.[2560delC]; | p.[Leu854fs] | F | X | |
| c.[1180+1G>T] | p.[Gly394fs] | F | ||||||
| 5RT94 | 18 | M | SLC12A3 | c.[238dupCC] | p.[Arg80fs] | F | X | |
| c.[2952-?_3090+?del] | p.[Ile984_Gln1030del] | LD | ||||||
| Bartter Type 1 Syndrome | ||||||||
| 1PD90 | 21 | M | SLC12A1 | c.[347G>A] | p.[Arg116His] | M | X | X |
| c.[1954G>A] | p.[Gly652Ser] | M | ||||||
| 1TR02 | 9 | M | SLC12A1 | c.[551T>A] | p.[Leu184Gln] | M | X | X |
| c.[611T>C] | p.[Val204Ala] | M | ||||||
| 3GG07 | 4 | M | SLC12A1 | c.[730dupG] | p.[Ala244fs] | F | X | X |
| c.[1432G>A] | p.[Gly478Arg] | M | ||||||
| 6BA09 | 5 | F | SLC12A1 | c.[572T>A] | p.[Ile191Asn] | M | X | X |
| c.[1493C>T] | p.[Ala498Val] | M | ||||||
| Bartter Type 2 Syndrome | ||||||||
| 2BG03 | 8 | F | KCNJ1 | c.[256A>G] | p.[Thr86Ala] | M | X | |
| c.[=] | p.[=] | -/- | ||||||
| 2ED04 | 3 | M | KCNJ1 | c.[133A>G] | p.[Lys45Glu] | M | X | X |
| c.[?] | p.[Glu334fs] | F | ||||||
| 2GA04^ | 6 | M | KCNJ1 | c.[572C>T] | p.[Thr191Ile] | M | X | X |
| c.[572C>T] | p.[Thr191Ile] | M | ||||||
| 2GN03^ | 7 | M | KCNJ1 | c.[572C>T] | p.[Thr191Ile] | M | X | X |
| c.[572C>T] | p.[Thr191Ile] | M | ||||||
| Bartter Type 3 Syndrome | ||||||||
| 3BD82 | 29 | M | CLCNKB | c.[371C>T] | p.[Pro124Leu] | M | X | X |
| c.[371C>T] | p.[Pro124Leu] | M | ||||||
| 3BS07 | 4 | F | CLCNKB | c.[725C>A] | p.[Ala242Glu] | M | X | |
| c.[1-?_2064+?del] | p.[0] | LD | ||||||
| 3DL91 | 20 | F | CLCNKB | c.[1-?_2064+?del] | p.[0] | LD | X | X |
| c.[1-?_2064+?del] | p.[0] | LD | ||||||
| 3SS82 | 29 | M | CLCNKB | c.[1-?_2064+?del] | p.[0] | LD | X | X |
| c.[1-?_2064+?del] | p.[0] | LD | ||||||
| 5BE99 | 12 | F | CLCNKB | c.[1101G>A] | p.[Trp367*] | N | X | X |
| c.[1?_2064+?del] | p.[0] | LD | ||||||
| Uniprot ID | Protein Name | Funct | Sub Loc | Mbr Inter | Kidney Expression Score |
|---|---|---|---|---|---|
| Proteins Identified Only in BS2 Spots | |||||
| P41181 | Aquaporin-2 | transp | mbr | M-P | medium |
| P08183 | P-glycoprotein 1 | transp | mbr | M-P | low |
| Q8IZF0 | Na-leak channel non-selective protein * | transp | mbr | M-P | low |
| Q9NZM6 | Polycystin-L2 * | transp | mbr | M-P | medium |
| Q9UBD6 | Ammonium transporter Rh type C | transp | mbr | M-P | medium |
| P55017 | Na-Cl cotransporter (NCC) | transp | mbr | M-P | medium |
| Q8NG04 | Solute carrier family 26 member 10 * | transp | mbr | M-P | NA |
| P58743 | Prestin * | transp | mbr | M-P | NA |
| Q9C0H2 | Protein tweety homolog 3 | transp | mbr | M-P | medium |
| Q9Y2B5 | VPS9 domain-containing protein 1 | transp | cyt | medium | |
| P60709 | Actin, cytoplasmic 1 | struct | cyt | medium | |
| Q9H6S3 | EPS8-like protein 2 | struct | cyt | high | |
| Q96T17 | MAP7 domain-containing protein 2 * | struct | nuc | low | |
| Q96QZ7 | Atrophin-1-interacting protein 3 | struct | mbr | Per | medium |
| Q969L2 | Protein MAL2 | struct | mbr | M-P | low |
| Q92859 | Neogenin | rec | mbr | S-P | low |
| P14384 | Carboxypeptidase M | prot | mbr | L-A | high |
| P08246 | Neutrophil elastase | prot | cyt | NA | |
| P60900 | Proteasome subunit α type-6 | prot | cyt | medium | |
| K7EIQ3 | ZNF561 antisense gene protein 1 * | NA | NA | NA | |
| Q9H579 | Protein MROH8 * | NA | nuc | low | |
| P0CG20 | Proline-rich protein 35 * | NA | NA | NA | |
| O94933 | SLIT and NTRK-like protein 3 * | NA | mbr | S-P | NA |
| Q9Y4F4 | Crescerin-1 | NA | cyt | medium | |
| O95336 | 6-phosphogluconolactonase | enz | cyt | high | |
| O95573 | Arachidonate-CoA ligase | enz | mbr | S-P | high |
| Q9BUT1 | 3-hydroxybutyrate DH type 2 | enz | cyt | high | |
| P00918 | Carbonic anhydrase 2 | enz | mbr | L-A | high |
| Q12873 | ATP-dependent helicase CHD3 | enz | nuc | low | |
| Q13237 | cGMP-dependent protein kinase 2 | enz | mbr | L-A | medium |
| Q9BYJ1 | Epidermis-type lipoxygenase 3 * | enz | cyt | NA | |
| Q96RQ9 | l-amino-acid oxidase * | enz | org | NA | |
| Q8IY17 | Neuropathy target esterase * | enz | mbr | S-P | high |
| P60174 | Triosephosphate isomerase | enz | cyt | medium | |
| P02768 | Serum albumin | bind | secr | NA | |
| P05090 | Apolipoprotein D | bind | secr | high | |
| P05026 | Na/K-transporting ATPase sub β-1 | bind | mbr | S-P | medium |
| Q16854 | Deoxyguanosine kinase, mitochondrial * | bind | org | medium | |
| Q0VDD8 | Dynein heavy chain 14, axonemal * | bind | cyt | low | |
| P01133 | Pro-epidermal growth factor | bind | mbr | S-P | NA |
| O75955 | Flotillin-1 | bind | mbr | Per | high |
| Q86YZ3 | Hornerin | bind | cyt | NA | |
| Q8N1G4 | Leu-rich repeat-containing protein 47 | bind | cyt | high | |
| Q8WV92 | MIT domain-containing protein 1 | bind | mbr | Per | high |
| P15941 | Mucin-1 | bind | mbr | S-P | medium |
| Q8WXI7 | Mucin-16 | bind | mbr | S-P | NA |
| P49321 | Nuclear autoantigenic sperm protein | bind | cyt | low | |
| O60422 | One cut domain family member 3 * | bind | nuc | NA | |
| Q9Y5G0 | Protocadherin γ-B5 | bind | mbr | S-P | NA |
| P05164 | Myeloperoxidase | bind | org | NA | |
| Q01970 | Phospholipase C-β-3 | bind | mbr | Per | low |
| P62191 | 26S protease regulatory sub 4 | bind | cyt | nd | |
| Q8NFJ5 | Retinoic acid-induced protein 3 | bind | mbr | M-P | low |
| P62263 | 40S ribosomal protein S14 | bind | nuc | medium | |
| P02743 | Serum amyloid P-component | bind | secr | NA | |
| P12931 | Proto-oncogene c-Src | bind | mbr | Per | medium |
| Q16851 | UDP-glucose pyrophosphorylase | bind | cyt | low | |
| Q15904 | V-type proton ATPase subunit S1 | bind | mbr | S-P | high |
| Q9UN37 | Vacuolar protein sorting-associated protein 4A | bind | mbr | Per | medium |
| O75351 | Vacuolar protein sorting-associated protein 4B | bind | mbr | Per | medium |
| Q86YA3 | protein ZGRF1 * | bind | mbr | S-P | medium |
| Q8IYB9 | Zinc finger protein 595 * | bind | nuc | NA | |
| B4DX44 | Zinc finger protein 736 * | bind | nuc | medium | |
| Proteins Identified in Both BS2 and BS3 Spots | |||||
| P53990 | IST1 homolog (hIST1) | bind | cyt | high | |
| P21281 | V-ATPase subunit B 2 | transp | mbr | Per | high |
| O43508 | TNF ligand superfamily member 12 * | rec | mbr | S-P | medium |
| P01008 | Antithrombin-III | prot inh | secr | medium | |
| P01042 | Kininogen-1 | prot inh | secr | high | |
| P16444 | Dipeptidase 1 | prot | mbr | L-A | high |
| P55786 | Puromycin-sensitive aminopeptidase | prot | cyt | medium | |
| Q9Y6R7 | IgGFc-binding protein | na | secr | NA | |
| P07948 | Tyr-protein kinase Lyn | enz | mbr | Per | ND |
| P33908 | Mannosidase α class 1A member 1 | enz | mbr | S-P | high |
| P50995 | Annexin A11 | bind | cyt | medium | |
| Q01518 | Adenylyl cyclase-associated protein 1 | bind | mbr | Per | medium |
| O75131 | Copine-3 | bind | nuc | low | |
| P68104 | Elongation factor 1-α 1 | bind | cyt | medium | |
| P02675 | Fibrinogen β chain | bind | secr | NA | |
| Q99816 | Tumor susceptibility gene 101 protein | bind | mbr | Per | high |
| P0CG47 | Polyubiquitin-B | bind | cyt | medium | |
| P42685 | FYN-related kinase | rec | cyt | medium | |
| P19440 | γ-glutamyltransferase 1 | enz | mbr | S-P | high |
| Proteins Identified Only in BS3 Spots | |||||
| P01009 | α-1-antitrypsin | prot inh | secr | NA | |
| P01011 | α-1-antichymotrypsin | prot inh | secr | low | |
| P05154 | Serpin A5 | prot inh | secr | NA | |
| P26038 | Moesin | struct | mbr | Per | medium |
| Q8N957 | ANKF1 * | na | nuc | NA | |
| P04406 | Glyceraldehyde-3-phosphate DH | enz | cyt | medium | |
| P80723 | Brain acid soluble protein 1 | bind | mbr | L-A | ND |
| Q8TF65 | PDZ domain-containing protein GIPC2 | bind | cyt | high | |
| P15313 | V-ATPase subunit B 1 | transp | mbr | Per | high |
| P04075 | Fructose-bisphosphate aldolase A | struct | cyt | medium | |
| P05062 | Fructose-bisphosphate aldolase B | struct | cyt | high | |
| Q9UHR4 | BAI1-associated protein 2-like protein 1 | struct | cyt | low | |
| Q9UJU6 | Drebrin-like protein | struct | cyt | medium | |
| P15311 | Ezrin | struct | mbr | Per | high |
| Q5S007 | Leu-rich repeat Ser/Thr-protein kinase 2 | struct | cyt | high | |
| P22105 | Tenascin-X | struct | secr | NA | |
| P28223 | Serotonin receptor 2A * | rec | mbr | M-P | NA |
| P46098 | Serotonin receptor 3A * | rec | mbr | M-P | NA |
| P15328 | Folate receptor α | rec | mbr | L-A | NA |
| Q9NQ84 | Glycerol kinase | rec | mbr | M-P | high |
| Q08380 | Galectin-3-binding protein | rec | secr | low | |
| O14745 | Na/H exchanger regulatory factor 1 | rec | mbr | Per | high |
| Q6TCH4 | Membrane progesterone P4 receptor δ * | rec | mbr | M-P | NA |
| O43653 | Prostate stem cell antigen | rec | mbr | L-A | low |
| Q6UXT9 | Protein ABHD15 * | prot inh | secr | medium | |
| P05543 | Thyroxine-binding globulin | prot inh | secr | NA | |
| P28838 | Cytosol aminopeptidase | prot | cyt | high | |
| Q9Y646 | Carboxypeptidase Q | prot | org | low | |
| P81605 | Dermcidin | prot | secr | NA | |
| Q9UHL4 | Dipeptidyl peptidase 2 | prot | org | medium | |
| Q6P179 | Endoplasmic reticulum aminopeptidase 2 | prot | mbr | S-P | medium |
| P12955 | Xaa-Pro dipeptidase | prot | cyt | high | |
| Q8IYX3 | Coiled-coil domain-containing protein 116 * | na | cyt | NA | |
| Q8NCU4 | Coiled-coil domain-containing protein 191 * | na | org | medium | |
| Q8WW52 | Protein FAM151A | na | mbr | S-P | high |
| A8MUH7 | PDZ domain-containing 1 pseudogene 1 * | na | NA | NA | |
| Q9BW04 | Specifically androgen-regulated gene protein | na | cyt | medium | |
| P11766 | Alcohol DH class-3 | enz | cyt | high | |
| P00352 | Retinal DH 1 | enz | cyt | low | |
| P49419 | α-aminoadipic semialdehyde DH | enz | cyt | high | |
| Q9H2A2 | Aldehyde DH family 8 member A1 | enz | cyt | medium | |
| P15121 | Aldose reductase | enz | cyt | NA | |
| Q92485 | ASM-like phosphodiesterase 3b | enz | sec | NA | |
| P16278 | β-galactosidase | enz | org | medium | |
| Q00796 | Sorbitol DH | enz | mbr | Per | medium |
| Q9H9B1 | Eu-HMTase1 * | enz | nuc | NA | |
| Q6UWR7 | GPC-Cpde ENPP6 | enz | mbr | L-A | medium |
| P06744 | Glucose-6-phosphate isomerase | enz | cyt | medium | |
| P32189 | Glycerol kinase | enz | mbr | Per | medium |
| P15309 | Prostatic acid phosphatase | enz | mbr | S-P | NA |
| O43175 | d-3-phosphoglycerate DH | enz | cyt | high | |
| Q6PHR2 | Ser/Thr-protein kinase ULK3 | enz | cyt | high | |
| Q96BJ3 | AIDA protein | bind | cyt | low | |
| P20073 | Annexin A7 | bind | nuc | medium | |
| P06727 | Apolipoprot A-IV | bind | secr | medium | |
| P02649 | Apolipoprot E | bind | secr | low | |
| Q9UQB8 | BAI-associated protein 2 | bind | mbr | NA | high |
| Q9NRL2 | BAZ protein 1A * | bind | nuc | low | |
| Q8TDL5 | BPI fold-containing family B member 1 | bind | secr | NA | |
| P55286 | Cadherin-8 | bind | mbr | S-P | NA |
| P08571 | Monocyte differentiation antigen CD14 | bind | mbr | L-A | NA |
| P10909 | Clusterin | bind | secr | NA | |
| Q14117 | Dihydropyrimidinase | bind | cyt | high | |
| Q96JB3 | Hypermethylated in cancer 2 protein (Hic2) * | bind | nuc | low | |
| P14618 | Pyruvate kinase PKM | bind | cyt | medium | |
| P30613 | Pyruvate kinase PKLR | bind | cyt | high | |
| Q96JY0 | Protein maelstrom homolog * | bind | cyt | NA | |
| Q86W25 | Ntd-binding oligomerization domain protein 14 * | bind | NA | NA | |
| Q14596 | Next to BRCA1 gene 1 protein * | bind | cyt | medium | |
| O94818 | Nucleolar protein 4 * | bind | nuc | NA | |
| Q6V1P9 | Protocadherin-23 * | bind | mbr | S-P | NA |
| Q12913 | Receptor-type Tyr-protein phosphatase η | bind | mbr | S-P | low |
| P35241 | Radixin | bind | mbr | Per | high |
| Q9UPX8 | Cortactin-binding protein 1 | bind | mbr | NA | low |
| O95343 | Homeobox protein SIX3 * | bind | nuc | NA | |
| O60284 | Suppression of tumorigenicity 18 protein * | bind | nuc | Per | high |
| Q9Y6E0 | Ser/Thr-protein kinase 24 | bind | cyt | medium | |
| Q9BXI6 | TBC1 domain family member 10A | bind | cyt | high | |
| Q7Z2Z1 | Treslin * | bind | nuc | NA | |
| O75674 | TOM1-like protein 1 | bind | mbr | Per | medium |
| O60784 | Target of Myb protein 1 | bind | mbr | NA | medium |
| P59923 | Zinc finger protein 445 * | bind | nuc | NA | |
| Q92629 | δ-sarcoglycan * | NA | mbr | S-P | NA |
| Q9Y6Q2 | Stonin-1 * | NA | cyt | medium | |
| Q8N1K5 | Protein THEMIS * | NA | cyt | NA | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raimondo, F.; Chinello, C.; Porcaro, L.; Magni, F.; Pitto, M. Urinary Extracellular Vesicles and Salt-Losing Tubulopathies: A Proteomic Approach. Proteomes 2020, 8, 9. https://doi.org/10.3390/proteomes8020009
Raimondo F, Chinello C, Porcaro L, Magni F, Pitto M. Urinary Extracellular Vesicles and Salt-Losing Tubulopathies: A Proteomic Approach. Proteomes. 2020; 8(2):9. https://doi.org/10.3390/proteomes8020009
Chicago/Turabian StyleRaimondo, Francesca, Clizia Chinello, Luigi Porcaro, Fulvio Magni, and Marina Pitto. 2020. "Urinary Extracellular Vesicles and Salt-Losing Tubulopathies: A Proteomic Approach" Proteomes 8, no. 2: 9. https://doi.org/10.3390/proteomes8020009
APA StyleRaimondo, F., Chinello, C., Porcaro, L., Magni, F., & Pitto, M. (2020). Urinary Extracellular Vesicles and Salt-Losing Tubulopathies: A Proteomic Approach. Proteomes, 8(2), 9. https://doi.org/10.3390/proteomes8020009






