Proteomics of Brucella
Abstract
1. Introduction
2. Proteomics Technologies and Their Use for Brucella
3. Proteomics-based Detection of Brucella spp. Immunodominant Proteins
4. Proteomics Strategies in the Study of Brucella-Host Cell Interaction and Stress Response
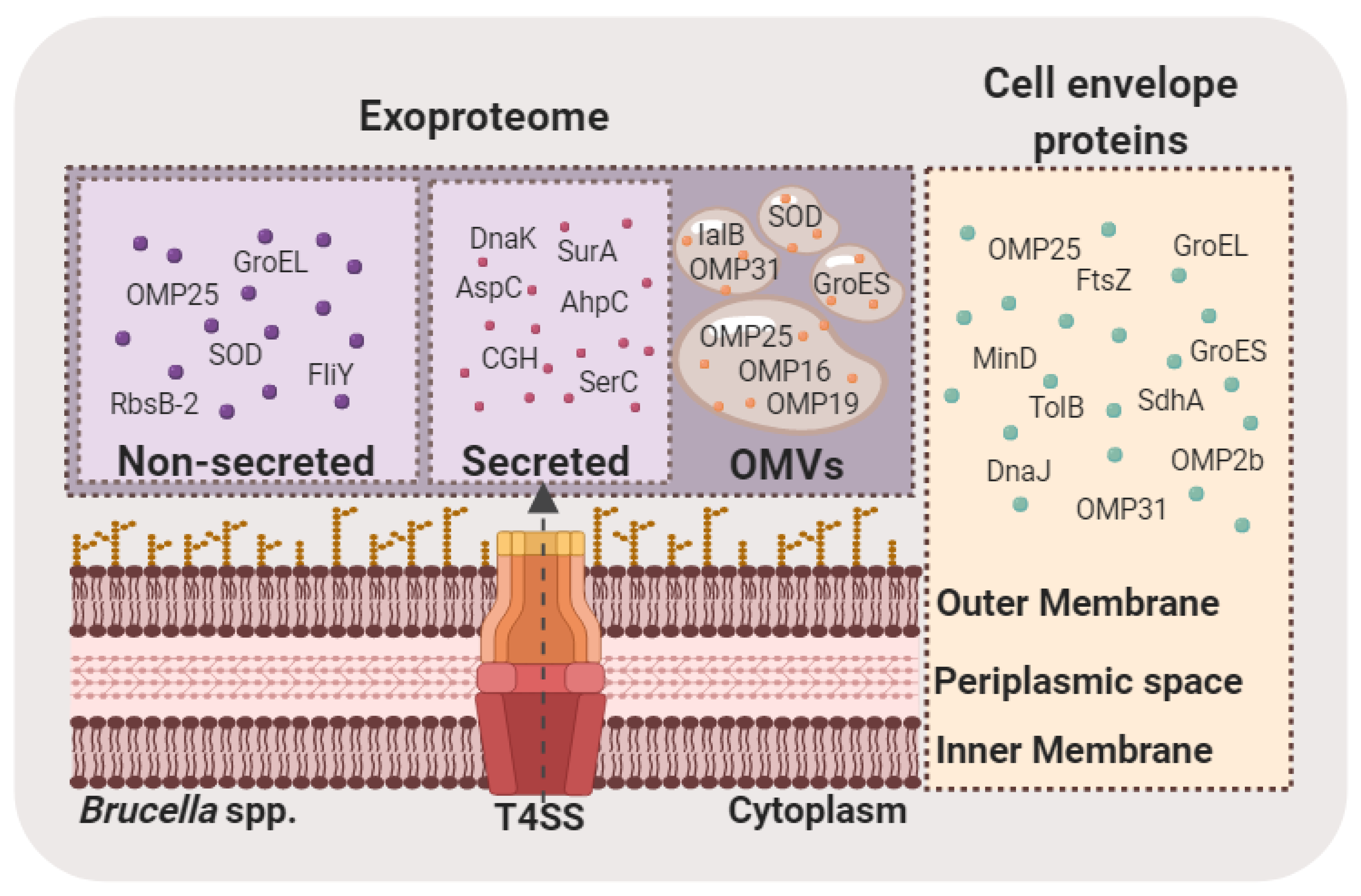
5. Brucella Exoproteome
6. Antibiotic Targets and Resistance
7. Proteogenomics
8. Proteases—A Yet Untouched Topic in Proteomics for Brucella
9. Concluding Remarks
| Reference | Species/Strain | Sample | Experimental Design | MS Method | Immunogenic Proteins Identified |
|---|---|---|---|---|---|
| [45] | B. abortus 1119-3 | Whole cell proteins | Identification of immunogenic proteins by 2-D immunoblots probed with rabbit hyperimmune serum against B. abortus 111-3. | MALDI-MSnLC-ESI-MS/MS | 6 |
| [57] | B. abortus 2308 | Cell envelope proteins | Detection of immunogenic proteins by 2-D Western blotting with human serum from a B. suis-infected patient and with serum from an infected bovine. | MALDI-TOF MSLC-MS/ MS | 18 |
| [58] | B. melitensis M5 | Whole cell and membrane proteins | Identification of immunogenic proteins by 2-D immunoblotting with Brucella-infected bovine sera. | MALDI-TOF MS | 61 |
| [59] | B. melitensis 16M | Whole cell soluble proteins | Identification of antigens recognized by Brucella-infected goat and human sera in 2-D immunoblots. | LC–MS/MS | 11 |
| [60] | B. abortus S19 | Antigen preparation obtained after TX-114 extraction | Detection of infection markers by 1-D and 2-D immunoblots probed with sera from naturally infected or S-19-vaccinated cattle. | LC–MS/MS | 5 |
| [61] | B. abortus 544 | Whole cell proteins | Detection of immunodominant proteins by 2-D immunoblots probed with sera from experimentally infected mice. | MALDI-TOF MS | 17 |
| [62] | B. abortus RB51 | Insoluble proteins | Analysis of immunogenic proteins by 2-DE and Western blot with sera from B. abortus 2308-infected cattle. | MALDI-TOF/TOF MS | 11 |
| [105] | B. abortus and B. melitensis field strains | Whole cell proteins | Detection of antigenic proteins by SDS–PAGE and Western blotting with sera from naturally infected hosts (cows, buffaloes, sheep, and goats). | MALDI-TOF MS | 16 |
| [63] | B. abortus and B. melitensis field strains | Whole cell proteins | Identification of immunodominant proteins by 2-DE and Western blot with sera from naturally infected hosts (cows, buffaloes, sheep, and goats). | MALDI-TOF MS | 36 |
| Reference | Species/Strain | Experimental Design | MS Method | Main Findings |
|---|---|---|---|---|
| [65] | B. suis 1330 | 2-D-DIGE based analysis of intramacrophagic proteome of B. suis at 48 h post-infection, compared to extracellularly grown stationary-phase-bacterial proteome. | MALDI-TOF MS | One hundred and sixty-eight proteins with differential abundance. Most of the proteins identified are involved in metabolic pathways and downregulated intracellularly. |
| [66] | B. abortus 2308 and S19 | Comparison of intramacrophagic proteomes of virulent B. abortus 2308 and the attenuated strain S19 at different times (0, 3, 20 and 44 h post-infection). | LC MS/MS | At early times post-infection, the virulent strain altered its respiration and downregulated the expression of proteins involved in metabolic and biosynthetic pathways. These changes are reverted to pre-infection levels at 44 h post-infection. |
| [68] | B. suis 1330 | 2-D-DIGE based comparative analysis of B. suis proteome under low oxygen conditions (anaerobiosis and microaerobiosis) and control condition (aerobiosis). | MALDI-TOF MS | Upregulated glycolysis and denitrification in microaerobiosis and anaerobiosis. |
| [71] | B. abortus 1119-3 | 2-DE-based proteome analysis of cell envelope proteins of mutant strains defective in internalization into host cells. | LC-ESI-MS | Identification of bacterial loci involved in altered expression of cell envelope proteins such as OMP25, OMP2b and OMP28. |
| [67] | B. abortus 2308 | 2-DE-based comparative proteomic analysis of intracellular and laboratory-grown B. abortus. | MALDI-TOF MS | Two cyclophilins were identified as overexpressed during the intracellular phase. The double mutant strain in the genes coding for these proteins is attenuated in cellular and mice infection models. |
| [25] | B. melitensis 16M | 2-DE-based comparative proteomic analysis of wild type and hfq mutant under stress conditions. | MALDI-TOF/TOF MS | MS identified 55 proteins with differential abundance in the mutant strain. These proteins belong to diverse functional groups including transport and metabolism, outer membrane proteins, post-translational modification and cellular processes. |
| [27] | B. abortus A19 | 2-DE-based comparative proteomic analysis of THP-1-derived macrophages infected or uninfected with B. abortus A19. | MALDI-TOF/TOF MS | MS identified 44 proteins with differential abundance. These proteins were involved in cytoskeleton, signal transduction, energy metabolism, host macromolecular biosynthesis, and stress response. |
| [35] | B. melitensis 16M and VTRM1 | Quantitative proteomic approach to study protein redistribution between membrane domains of monocytes exposed or not exposed to Brucella. | iTRAQ MALDI TOF/TOF | Several proteins were distinctly enriched or depleted in membrane domains upon exposure to rough and smooth B. melitensis strains. |
| [69] | B. abortus2308 | 2-D DIGE based differential proteomic profile of bovine chorioallantoic membrane explants uninfected and at early stages of infection with B. abortus. | MALDI-TOF/TOF MS | Several proteins upregulated during infection are associated with modulation of the innate host immune response to infection with B. abortus, including proteins related to TLR signaling and ROS production, as well as proteins associated with inflammation and intracellular trafficking. |
| [37] | B. melitensis 16M | Comparative proteomics approach to identify Brucella-specific proteins and pathways affected by changes in bacterial c-di-GMP levels. | LC MS/MS | c-di-GMP levels affect multiple processes related to bacterial virulence, such as nutrient acquisition, cell wall formation, and the type IV secretion system. |
| [36] | B. abortus2308 | Comparative proteomic analysis of B. abortus isolated within the host macrophage cell at late post-infection times and in vitro-cultured Brucella. | iTRAQ MALDI TOF/TOF | Identification of 197 differentially modulated proteins in intracellular Brucella. Many of them were related with iron metabolism, known to play a central role in Brucella invasiveness and virulence. |
| [38] | B. abortus104-M | Label-free quantitative proteomic analysis for the identification of proteins involved in stress resistance. | LC MS/MS | Identification of over 1000 differentially abundant proteins under relevant stress conditions. Proteins were included in diverse functional groups such as oxidative phosphorylation, ABC transporters, two-component systems, biosynthesis of secondary metabolites, the citrate cycle, thiamine metabolism, and nitrogen metabolism. |
| [70] | B. melitesis M5 | Comparative 2-DE-based proteomic analysis of lung tissue of BALB/c mice uninfected and infected by exposure to aerosolized bacteria. | MALDI TOF/TOF | Identification of 12 proteins differentially expressed in lung tissue during infection. The proteins with increased expression were related to protein transport, antioxidant function, and antiviral or cell activation. Proteins with decreased expression were related to cytoskeletal structure, enzyme activation, or cell intoxication and transformation. |
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Seleem, M.N.; Boyle, S.M.; Sriranganathan, N. Brucellosis: A re-emerging zoonosis. Vet. Microbiol. 2010, 140, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E. Retrospective and prospective perspectives on zoonotic brucellosis. Front. Microbiol. 2014, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Pappas, G.; Papadimitriou, P.; Akritidis, N.; Christou, L.; Tsianos, E.V. The new global map of human brucellosis. Lancet Infect. Dis. 2006, 6, 91–99. [Google Scholar] [CrossRef]
- Doganay, G.D.; Doganay, M. Brucella as a potential agent of bioterrorism. Recent Pat. Antiinfect. Drug Discov. 2013, 8, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Sieira, R.; Comerci, D.J.; Sánchez, D.O.; Ugalde, R.A. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 2000, 182, 4849–4855. [Google Scholar] [CrossRef] [PubMed]
- Delrue, R.M.; Martinez-Lorenzo, M.; Lestrate, P.; Danese, I.; Bielarz, V.; Mertens, P.; De Bolle, X.; Tibor, A.; Gorvel, J.P.; Letesson, J.J. Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol. 2001, 3, 487–497. [Google Scholar] [CrossRef]
- O’Callaghan, D.; Cazevieille, C.; Allardet-Servent, A.; Boschiroli, M.L.; Bourg, G.; Foulongne, V.; Frutos, P.; Kulakov, Y.; Ramuz, M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 2002, 33, 1210–1220. [Google Scholar] [CrossRef]
- de Jong, M.F.; Sun, Y.-H.; den Hartigh, A.B.; van Dijl, J.M.; Tsolis, R.M. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol. Microbiol. 2008, 70, 1378–1396. [Google Scholar] [CrossRef]
- de Barsy, M.; Jamet, A.; Filopon, D.; Nicolas, C.; Laloux, G.; Rual, J.-F.; Muller, A.; Twizere, J.-C.; Nkengfac, B.; Vandenhaute, J.; et al. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab. Cell. Microbiol. 2011, 13, 1044–1058. [Google Scholar] [CrossRef]
- Marchesini, M.I.; Herrmann, C.K.; Salcedo, S.P.; Gorvel, J.-P.; Comerci, D.J. In search of Brucella abortus type iv secretion substrates: Screening and identification of four proteins translocated into host cells through virb system. Cell. Microbiol. 2011, 13, 1261–1274. [Google Scholar] [CrossRef]
- Myeni, S.; Child, R.; Ng, T.W.; Kupko, J.J.; Wehrly, T.D.; Porcella, S.F.; Knodler, L.A.; Celli, J. Brucella Modulates Secretory Trafficking via Multiple Type IV Secretion Effector Proteins. PLoS Pathog. 2013, 9, e1003556. [Google Scholar] [CrossRef]
- Salcedo, S.P.S.P.; Marchesini, M.I.M.I.; Degos, C.; Terwagne, M.; Bargen, K.V.; Lepidi, H.; Herrmann, C.K.C.K.C.K.; Santos Lacerda, T.L.T.L.T.L.; Imbert, P.R.C.P.R.C.; Pierre, P.; et al. BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Front. Cell. Infect. Microbiol. 2013, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Döhmer, P.H.; Valguarnera, E.; Czibener, C.; Ugalde, J.E. Identification of a type IV secretion substrate of Brucella abortus that participates in the early stages of intracellular survival. Cell. Microbiol. 2014, 16, 396–410. [Google Scholar] [CrossRef]
- Mahan, M.J.; Slauch, J.M.; Hanna, P.C.; Camilli, A.; Tobias, J.W.; Waldor, M.K.; Mekalanos, J.J. Selection for bacterial genes that are specifically induced in host tissues: The hunt for virulence factors. Infect. Agents Dis. 1993, 2, 263–268. [Google Scholar] [PubMed]
- Hensel, M.; Shea, J.; Gleeson, C.; Jones, M.; Dalton, E.; Holden, D. Simultaneous identification of bacterial virulence genes by negative selection. Science 1995, 269, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.; Völker, U. Proteome analysis of host-pathogen interactions: Investigation of pathogen responses to the host cell environment. Proteomics 2011, 11, 3203–3211. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hu, M.; Yu, K.; Zeng, X.; Liu, X. Mass spectrometry-based proteomic approaches to study pathogenic bacteria-host interactions. Protein Cell 2015, 6, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Llarena, F.J.; Bou, G. Proteomics as a tool for studying bacterial virulence and antimicrobial resistance. Front. Microbiol. 2016, 7, 1–21. [Google Scholar] [CrossRef] [PubMed]
- DelVecchio, V.G.; Kapatral, V.; Redkar, R.J.; Patra, G.; Mujer, C.; Los, T.; Ivanova, N.; Anderson, I.; Bhattacharyya, A.; Lykidis, A.; et al. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 2002, 99, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.A.; Eschenbrenner, M.; Horn, T.A.; Kraycer, J.A.; Mujer, C.V.; Hagius, S.; Elzer, P.; DelVecchio, V.G. Global analysis of the Brucella melitensis proteome: Identification of proteins expressed in laboratory-grown culture. Proteomics 2002, 2, 1047–1060. [Google Scholar] [CrossRef]
- Eschenbrenner, M.; Wagner, M.A.; Horn, T.A.; Kraycer, J.A.; Mujer, C.V.; Hagius, S.; Elzer, P.; DelVecchio, V.G. Comparative Proteome Analysis of Brucella melitensis Vaccine Strain Rev 1 and a Virulent Strain, 16M. J. Bacteriol. 2002, 184, 4962–4970. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Olarte, E.; Guzmán-Verri, C.; Paramithiotis, E.; Moreno, E. What Have We Learned From Brucella Proteomics. In Brucella: Molecular Microbiology and Genomics; Ignacio, L.-G., O’Callaghan, D., Eds.; Caister Academic Press: Cambridge, MA, USA, 2012; pp. 103–132. [Google Scholar]
- Välikangas, T.; Suomi, T.; Elo, L.L. A comprehensive evaluation of popular proteomics software workflows for label-free proteome quantification and imputation. Brief. Bioinform. 2018, 19, 1344–1355. [Google Scholar] [CrossRef]
- Bachor, R.; Waliczek, M.; Stefanowicz, P.; Szewczuk, Z. Trends in the design of new isobaric labeling reagents for quantitative proteomics. Molecules 2019, 24, 701. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, T.; Xu, J.; Ke, Y.; Du, X.; Yuan, X.; Wang, Z.; Gong, C.; Zhuang, Y.; Lei, S.; et al. Impact of Hfq on global gene expression and intracellular survival in Brucella melitensis. PLoS ONE 2013, 8, e71933. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Kumar, S.; Dohre, S.; Afley, P.; Sengupta, N.; Alam, S.I. Identification of a protective protein from stationary-phase exoproteome of Brucella abortus. Pathog. Dis. 2014, 70, 75–83. [Google Scholar] [CrossRef]
- Wu, Y.; Jin, Y.; Pan, W.; Ye, C.; Sun, X.; Sun, Y.; Hu, B.; Zhou, J. Comparative proteomics analysis of host cells infected with Brucella abortus A. Electrophoresis 2014, 35, 1130–1143. [Google Scholar] [CrossRef]
- Lee, J.J.; Lim, J.J.; Kim, D.G.D.H.H.; Simborio, H.L.; Kim, D.G.D.H.H.; Reyes, A.W.B.; Min, W.G.; Lee, H.J.; Kim, D.G.D.H.H.; Chang, H.H.; et al. Characterization of culture supernatant proteins from Brucella abortus and its protection effects against murine brucellosis. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 221–228. [Google Scholar] [CrossRef]
- Wareth, G.; Melzer, F.; Weise, C.; Neubauer, H.; Roesler, U.; Murugaiyan, J. Proteomics-based identification of immunodominant proteins of Brucellae using sera from infected hosts points towards enhanced pathogen survival during the infection. Biochem. Biophys. Res. Commun. 2015, 456, 202–206. [Google Scholar] [CrossRef]
- Tang, T.; Chen, G.; Guo, A.; Xu, Y.; Zhao, L.; Wang, M.; Lu, C.; Jiang, Y.; Zhang, C. Comparative proteomic and genomic analyses of Brucella abortusbiofilm and planktonic cells. Mol. Med. Rep. 2020, 21, 731–743. [Google Scholar]
- Al Dahouk, S.; Jubier-Maurin, V.; Neubauer, H.; Köhler, S. Quantitative analysis of the Brucella suis proteome reveals metabolic adaptation to long-term nutrient starvation. BMC Microbiol. 2013, 13, 199. [Google Scholar] [CrossRef]
- Abdou, E.; de Bagüés, M.P.J.; Martínez-Abadía, I.; Ouahrani-Bettache, S.; Pantesco, V.; Occhialini, A.; Al Dahouk, S.; Köhler, S.; Jubier-Maurin, V. RegA plays a key role in oxygen-dependent establishment of persistence and in isocitrate lyase activity, a critical determinant of in vivo Brucella suis pathogenicity. Front. Cell. Infect. Microbiol. 2017, 7, 185. [Google Scholar] [CrossRef] [PubMed]
- Overlöper, A.; Kraus, A.; Gurski, R.; Wright, P.R.; Georg, J.; Hess, W.R.; Narberhaus, F. Two separate modules of the conserved regulatory RNA AbcR1 address multiple target mRNAs in and outside of the translation initiation region. RNA Biol. 2014, 11. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.F.; Zhang, Y.; Zhao, X.X. Comparative Analysis of Serum Proteome in Healthy and Brucella abortus-Infected Dairy CowsNo Title. Philipp. Agric. Sci. 2014, 97, 122–130. [Google Scholar]
- Lauer, S.A.; Iyer, S.; Sanchez, T.; Forst, C.V.; Bowden, B.; Carlson, K.; Sriranganathan, N.; Boyle, S.M. Proteomic analysis of detergent resistant membrane domains during early interaction of macrophages with rough and smooth brucella melitensis. PLoS ONE 2014, 9, 624–640. [Google Scholar] [CrossRef]
- Roset, M.S.; Alefantis, T.G.; Delvecchio, V.G.; Briones, G. Iron-dependent reconfiguration of the proteome underlies the intracellular lifestyle of Brucella abortus. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Khan, M.; Harms, J.S.; Marim, F.M.; Armon, L.; Hall, C.L.; Liu, Y.-P.P.; Banai, M.; Oliveira, S.C.; Splitter, G.A.; Smith, J.A. The bacterial second messenger cyclic di-GMP regulates Brucella pathogenesis and leads to altered host immune response. Infect. Immun. 2016, 84, 3458–3470. [Google Scholar] [CrossRef]
- Zai, X.; Yang, Q.; Yin, Y.; Li, R.; Qian, M.; Zhao, T.; Li, Y.; Zhang, J.; Fu, L.; Xu, J.; et al. Relative quantitative proteomic analysis of Brucella abortus reveals metabolic adaptation to multiple environmental stresses. Front. Microbiol. 2017, 8, 8. [Google Scholar] [CrossRef]
- Greco, E.; El-Aguizy, O.; Ali, M.F.; Foti, S.; Cunsolo, V.; Saletti, R.; Ciliberto, E. Proteomic Analyses on an Ancient Egyptian Cheese and Biomolecular Evidence of Brucellosis. Anal. Chem. 2018, 90, 9673–9676. [Google Scholar] [CrossRef]
- Bialer, M.G.; Ruiz-Ranwez, V.; Sycz, G.; Estein, S.M.; Russo, D.M.; Altabe, S.; Sieira, R.; Zorreguieta, A. MapB, the Brucella suis TamB homologue, is involved in cell envelope biogenesis, cell division and virulence. Sci. Rep. 2019, 9, 2158. [Google Scholar] [CrossRef]
- Li, P.; Tian, M.; Hu, H.; Yin, Y.; Guan, X.; Ding, C.; Wang, S.; Yu, S. Lable-free based comparative proteomic analysis of secretory proteins of rough Brucella mutants. J. Proteomics 2019, 195, 66–75. [Google Scholar] [CrossRef]
- Zai, X.; Yang, Q.; Liu, K.; Li, R.; Qian, M.; Zhao, T.; Li, Y.; Yin, Y.; Dong, D.; Fu, L.; et al. A comprehensive proteogenomic study of the human Brucella vaccine strain 104 M. BMC Genomics 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Araiza-Villanueva, M.; Avila-Calderón, E.D.; Flores-Romo, L.; Calderón-Amador, J.; Sriranganathan, N.; Qublan, H.A.; Witonsky, S.; Aguilera-Arreola, M.G.; Ruiz-Palma, M.D.; Ruiz, E.A.; et al. Proteomic Analysis of Membrane Blebs of Brucella abortus 2308 and RB51 and Their Evaluation as an Acellular Vaccine. Front. Microbiol. 2019, 10, 2714. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, P.M.; Marín, C.M.; Monreal, D.; González, D.; Garin-Bastuji, B.; Díaz, R.; Mainar-Jaime, R.C.; Moriyón, I.; Blasco, J.M. Efficacy of several serological tests and antigens for diagnosis of bovine brucellosis in the presence of false-positive serological results due to Yersinia enterocolitica O: 9. Clin. Diagn. Lab. Immunol. 2005, 12, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Al Dahouk, S.; Nöckler, K.; Scholz, H.C.; Tomaso, H.; Bogumil, R.; Neubauer, H. Immunoproteomic characterization of Brucella abortus 1119-3 preparations used for the serodiagnosis of Brucella infections. J. Immunol. Methods 2006, 309, 34–47. [Google Scholar] [CrossRef]
- Ko, K.Y.; Kim, J.-W.; Her, M.; Kang, S.-I.; Jung, S.C.; Cho, D.H.; Kim, J.-Y. Immunogenic proteins of Brucella abortus to minimize cross reactions in brucellosis diagnosis. Vet. Microbiol. 2012, 156, 374–380. [Google Scholar] [CrossRef]
- Caroff, M.; Bundle, D.R.; Perry, M.B. Structure of the O-chain of the phenol-phase soluble cellular lipopolysaccharide of Yersinia enterocolitica serotype O: 9. Eur. J. Biochem. 1984, 139, 195–200. [Google Scholar]
- Caroff, M.; Bundle, D.R.; Perry, M.B.; Cherwonogrodzky, J.W.; Duncan, J.R. Antigenic S-type lipopolysaccharide of Brucella abortus 1119-3. Infect. Immun. 1984, 46, 384–388. [Google Scholar]
- Corbel, M.J.; Stuart, F.A.; Brewer, R.A. Observations on serological cross-reactions between smooth Brucella species and organisms of other genera. Dev. Biol. Stand. 1984, 56, 341–348. [Google Scholar]
- Cotton, W.E.; Buck, J.M.; Smith, H.E. Efficacy and safety of abortion vaccines prepared from Brucella abortus strains of different degrees of virulence. J. Agric. Res. 1933, 46, 291–314. [Google Scholar]
- Elberg, S.S.; Faunce, K. Immunization against Brucella infection. VI. Immunity conferred on goats by a nondependent mutant from a streptomycin-dependent mutant strain of Brucella melitensis. J. Bacteriol. 1957, 73, 211–217. [Google Scholar] [CrossRef]
- Schurig, G.G.; Roop, R.M.; Bagchi, T.; Boyle, S.; Buhrman, D.; Sriranganathan, N. Biological properties of RB51; a stable rough strain of Brucella abortus. Vet. Microbiol. 1991, 28, 171–188. [Google Scholar] [CrossRef]
- Tabatabai, L.B.; Pugh, G.W. Modulation of immune responses in Balb/c mice vaccinated with Brucella abortus Cu-Zn superoxide dismutase synthetic peptide vaccine. Vaccine 1994, 12, 919–924. [Google Scholar] [CrossRef]
- Al-Mariri, A.; Tibor, A.; Mertens, P.; De Bolle, X.; Michel, P.; Godfroid, J.; Walravens, K.; Letesson, J.J. Induction of immune response in BALB/c mice with a DNA vaccine encoding bacterioferritin or P39 of Brucella spp. Infect. Immun. 2001, 69, 6264–6270. [Google Scholar] [CrossRef]
- Al-Mariri, A.; Tibor, A.; Mertens, P.; De Bolle, X.; Michel, P.; Godefroid, J.; Walravens, K.; Letesson, J.J. Protection of BALB/c mice against Brucella abortus 544 challenge by vaccination with bacterioferritin or P39 recombinant proteins with CpG oligodeoxynucleotides as adjuvant. Infect. Immun. 2001, 69, 4816–4822. [Google Scholar] [CrossRef] [PubMed]
- Velikovsky, C.A.; Cassataro, J.; Giambartolomei, G.H.; Goldbaum, F.A.; Estein, S.; Bowden, R.A.; Bruno, L.; Fossati, C.A.; Spitz, M. A DNA vaccine encoding lumazine synthase from Brucella abortus induces protective immunity in BALB/c mice. Infect. Immun. 2002, 70, 2507–2511. [Google Scholar] [CrossRef] [PubMed]
- Connolly, J.P.; Comerci, D.; Alefantis, T.G.; Walz, A.; Quan, M.; Chafin, R.; Grewal, P.; Mujer, C.V.; Ugalde, R.A.; DelVecchio, V.G. Proteomic analysis of Brucella abortus cell envelope and identification of immunogenic candidate proteins for vaccine development. Proteomics 2006, 6, 3767–3780. [Google Scholar] [CrossRef]
- Zhao, Z.P.; Yan, F.; Ji, W.H.; Luo, D.Y.; Liu, X.; Xing, L.; Duan, Y.Q.; Yang, P.H.; Shi, X.M.; Li, Z.; et al. Identification of immunoreactive proteins of Brucella melitensis by immunoproteomics. Sci. China Life Sci. 2011, 54, 880–887. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Yin, J.; Wang, X.; Cheng, S.; Lang, X.; Wang, X.; Qu, H.; Sun, C.; Wang, J.; et al. Immunoproteomic analysis of Brucella melitensis and identification of a new immunogenic candidate protein for the development of brucellosis subunit vaccine. Mol. Immunol. 2011, 49, 175–184. [Google Scholar] [CrossRef]
- Pajuaba, A.C.A.M.; Silva, D.A.O.; Almeida, K.C.; Cunha-Junior, J.P.; Pirovani, C.P.; Camillo, L.R.; Mineo, J.R. Immunoproteomics of Brucella abortus reveals differential antibody profiles between S19-vaccinated and naturally infected cattle. Proteomics 2012, 12, 820–831. [Google Scholar] [CrossRef]
- Lee, J.J.; Simborio, H.L.; Reyes, A.W.B.; Kim, D.G.; Hop, H.T.; Min, W.; Her, M.; Jung, S.C.; Yoo, H.S.; Kim, S. Proteomic analyses of the time course responses of mice infected with Brucella abortus 544 reveal immunogenic antigens. FEMS Microbiol. Lett. 2014, 357, 164–174. [Google Scholar]
- Kim, J.Y.; Sung, S.R.; Lee, K.; Lee, H.K.; Kang, S., II; Lee, J.J.; Jung, S.C.; Park, Y.H.; Her, M. Immunoproteomics of Brucella abortus RB51 as candidate antigens in serological diagnosis of brucellosis. Vet. Immunol. Immunopathol. 2014, 160, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Eravci, M.; Weise, C.; Roesler, U.; Melzer, F.; Sprague, L.D.; Neubauer, H.; Murugaiyan, J. Comprehensive identification of immunodominant proteins of Brucella abortus and Brucella melitensis using antibodies in the sera from naturally infected hosts. Int. J. Mol. Sci. 2016, 17, 695. [Google Scholar] [CrossRef] [PubMed]
- Semanjski, M.; Macek, B. Shotgun proteomics of bacterial pathogens: Advances, challenges and clinical implications. Expert Rev. Proteomics 2016, 13, 139–156. [Google Scholar] [CrossRef] [PubMed]
- Al Dahouk, S.; Jubier-Maurin, V.; Scholz, H.C.; Tomaso, H.; Karges, W.; Neubauer, H.; Köhler, S. Quantitative analysis of the intramacrophagic Brucella suis proteome reveals metabolic adaptation to late stage of cellular infection. Proteomics 2008, 8, 3862–3870. [Google Scholar] [CrossRef]
- Lamontagne, J.; Forest, A.; Marazzo, E.; Denis, F.; Butler, H.; Michaud, J.-F.; Boucher, L.; Pedro, I.; Villeneuve, A.; Sitnikov, D.; et al. Intracellular adaptation of Brucella abortus. J. Proteome Res. 2009, 8, 1594–1609. [Google Scholar] [CrossRef]
- Roset, M.S.; Fernández, L.G.; DelVecchio, V.G.; Briones, G. Intracellularly induced cyclophilins play an important role in stress adaptation and virulence of brucella abortus. Infect. Immun. 2013, 81, 521–530. [Google Scholar] [CrossRef]
- Al Dahouk, S.; Loisel-Meyer, S.; Scholz, H.C.; Tomaso, H.; Kersten, M.; Harder, A.; Neubauer, H.; Köhler, S.; Jubier-Maurin, V. Proteomic analysis of Brucella suis under oxygen deficiency reveals flexibility in adaptive expression of various pathways. Proteomics 2009, 9, 3011–3021. [Google Scholar] [CrossRef]
- Mol, J.P.S.; Pires, S.F.; Chapeaurouge, A.D.; Perales, J.; Santos, R.L.; Andrade, H.M.; Lage, A.P. Proteomic profile of brucella abortus-Infected bovine chorioallantoic membrane explants. PLoS ONE 2016, 11, 1–16. [Google Scholar] [CrossRef][Green Version]
- Fu, Y.; Wang, Z.; Lu, B.; Zhao, S.; Zhang, Y.; Zhao, Z.; Zhang, C.; Li, J.; Zhou, B.; Guo, Z.; et al. Immune response and differentially expressed proteins in the lung tissue of BALB/c mice challenged by aerosolized Brucella melitensis. J. Int. Med. Res. 2018, 46, 4740–4752. [Google Scholar] [CrossRef]
- Cha, S.B.; Rayamajhi, N.; Lee, W.J.; Shin, M.K.; Jung, M.H.; Shin, S.W.; Kim, J.W.; Yoo, H.S. Generation and envelope protein analysis of internalization defective Brucella abortus mutants in professional phagocytes, RAW 264.7. FEMS Immunol. Med. Microbiol. 2012, 64, 244–254. [Google Scholar] [CrossRef]
- Jan, A.T. Outer Membrane Vesicles (OMVs) of gram-negative bacteria: A perspective update. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Boigegrain, R.; Salhi, I.; Machold, J.; Fedon, Y.; Arpagaus, M.; Weise, C.; Rittig, M.; Rouot, B. Release of Periplasmic Proteins of Brucella suis upon Acidic Shock Involves the Outer Membrane Protein Omp. Infect. Immun. 2004, 72, 5693–5703. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gamazo, C.; Moriyón, I. Release of outer membrane fragments by exponentially growing Brucella melitensis cells. Infect. Immun. 1987, 55, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Pollak, C.N.; Delpino, M.V.; Fossati, C.A.; Baldi, P.C. Outer Membrane Vesicles from Brucella abortus Promote Bacterial Internalization by Human Monocytes and Modulate Their Innate Immune Response. PLoS ONE 2012, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Avila-Caldern, E.D.; Lopez-Merino, A.; Jain, N.; Peralta, H.; Lpez-Villegas, E.O.; Sriranganathan, N.; Boyle, S.M.; Witonsky, S.; Contreras-Rodríguez, A. Characterization of outer membrane vesicles from Brucella melitensis and protection induced in mice. Clin. Dev. Immunol. 2012, 2012. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z.; Qiao, F.; Ying, T.; Yuan, J.; Zhong, Z.; Zhou, L.; Du, X.; Wang, Z.; Zhao, J.; et al. Comparative proteomics analyses reveal the virB of B. melitensis affects expression of intracellular survival related proteins. PLoS ONE 2009, 4, e5368. [Google Scholar] [CrossRef]
- Delpino, M.V.; Comerci, D.J.; Wagner, M.A.; Eschenbrenner, M.; Mujer, C.V.; Ugalde, R.A.; Fossati, C.A.; Baldi, P.C.; DelVecchio, V.G. Differential composition of culture supernatants from wild-type Brucella abortus and its isogenic virB mutants. Arch. Microbiol. 2009, 191, 571–581. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z.; Qiao, F.; Zhong, Z.; Xu, J.; Wang, Z.; Du, X.; Qu, Q.; Yuan, J.; Jia, L.; et al. and the outer membrane properties of Brucella melitensis. FEMS Microbiol. Lett. 2010, 303, 92–100. [Google Scholar] [CrossRef]
- Paredes-Cervantes, V.; Flores-Mejía, R.; Moreno-Lafont, M.C.; Lanz-Mendoza, H.; Tello-López, Á.T.; Castillo-Vera, J.; Pando-Robles, V.; Hurtado-Sil, G.; González-González, E.; Rodríguez-Cortés, O.; et al. Comparative proteome analysis of Brucella abortus 2308 and its virB type IV secretion system mutant reveals new T4SS-related candidate proteins. J. Proteomics 2011, 74, 2959–2971. [Google Scholar] [CrossRef]
- Hayat, Z.; Khan, H.; Ahmad, I.; Habib, H.; Hayat, K. Antibiotics in the management of brucellosis. Gomal J. Med. Sci. 2018, 16, 114–116. [Google Scholar]
- Marianelli, C.; Ciuchini, F.; Tarantino, M.; Pasquali, P.; Adone, R. Genetic bases of the rifampin resistance phenotype in Brucella spp. J. Clin. Microbiol. 2004, 42, 5439–5443. [Google Scholar] [CrossRef] [PubMed]
- Sandalakis, V.; Psaroulaki, A.; De Bock, P.J.; Christidou, A.; Gevaert, K.; Tsiotis, G.; Tselentis, Y. Investigation of rifampicin resistance mechanisms in brucella abortus using MS-driven comparative proteomics. J. Proteome Res. 2012, 11, 2374–2385. [Google Scholar] [CrossRef]
- Smolkina, T.V.; Zebrev, A.I.; Nikitin, A.V. Effects of rifampicin and doxycycline on the production of hydrogen peroxide by macrophages. Antibiot. Khimioterapiia Antibiot. Chemoterapy 1992, 37, 17–19. [Google Scholar]
- Mahmud, A.; Khan, M.T.; Iqbal, A. Identification of novel drug targets for humans and potential vaccine targets for cattle by subtractive genomic analysis of Brucella abortus strain. Microb. Pathog. 2019, 137. [Google Scholar]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Eschenbrenner, M.; Horn, T.A.; Wagner, M.A.; Mujer, C.V.; Miller-Scandle, T.L.; DelVecchio, V.G. Comparative proteome analysis of laboratory grown Brucella abortus 2308 and Brucella melitensis 16M. J. Proteome Res. 2006, 5, 1731–1740. [Google Scholar] [CrossRef]
- Wheeler, D.L.; Church, D.M.; Federhen, S.; Lash, A.E.; Madden, T.L.; Pontius, J.U.; Schuler, G.D.; Schriml, L.M.; Sequeira, E.; Tatusova, T.A.; et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003, 31, 28–33. [Google Scholar] [CrossRef]
- Cui, G.; Wei, P.; Zhao, Y.; Guan, Z.; Yang, L.; Sun, W.; Wang, S.; Peng, Q. Brucella infection inhibits macrophages apoptosis via Nedd4-dependent degradation of calpain. Vet. Microbiol. 2014, 174, 195–205. [Google Scholar]
- Jakka, P.; Namani, S.; Murugan, S.; Rai, N.; Radhakrishnan, G. The Brucella effector protein TcpB induces degradation of inflammatory caspases and thereby subverts Non-canonical inflammasome activation in macrophages. J. Biol. Chem. 2017, 292, 20613–20627. [Google Scholar] [CrossRef]
- Pasquevich, K.A.; Carabajal, M.V.; Guaimas, F.F.; Bruno, L.; Roset, M.S.; Coria, L.M.; Rey Serrantes, D.A.; Comerci, D.J.; Cassataro, J. Omp19 Enables Brucella abortus to Evade the Antimicrobial Activity from Host’s Proteolytic Defense System. Front. Immunol. 2019, 10, 1436. [Google Scholar] [CrossRef]
- Liu, Y.; Dong, H.; Peng, X.; Gao, Q.; Jiang, H.; Xu, G.; Qin, Y.; Niu, J.; Sun, S.; Li, P.; et al. RNA-seq reveals the critical role of Lon protease in stress response and Brucella virulence. Microb. Pathog. 2019, 130, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.T.; Kovach, M.E.; Allen, C.A.; Ficht, T.A.; Roop, R.M. The Brucella abortus Lon functions as a generalized stress response protease and is required for wild-type virulence in BALB/c mice. Mol. Microbiol. 2000, 35, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.W.; Roop, R.M. Brucella abortus HtrA functions as an authentic stress response protease but is not required for wild-type virulence in BALB/c mice. Infect. Immun. 2001, 69, 5911–5913. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martin Roop, R.; Phillips, R.W.; Hagius, S.; Walker, J.V.; Booth, N.J.; Todd Fulton, W.; Edmonds, M.D.; Elzer, P.H. Re-examination of the role of the Brucella melitensis HtrA stress response protease in virulence in pregnant goats. Vet. Microbiol. 2001, 82, 91–95. [Google Scholar] [CrossRef]
- Ekaza, E.; Teyssier, J.; Ouahrani-Bettache, S.; Liautard, J.P.; Köhler, S. Characterization of Brucella suis clpB and clpAB mutants and participation of the genes in stress responses. J. Bacteriol. 2001, 183, 2677–2681. [Google Scholar] [CrossRef]
- Bandara, A.B.; Sriranganathan, N.; Schurig, G.G.; Boyle, S.M. Carboxyl-terminal protease regulates Brucella suis morphology in culture and persistence in macrophages and mice. J. Bacteriol. 2005, 187, 5767–5775. [Google Scholar] [CrossRef] [PubMed]
- Weeks, J.N.; Galindo, C.L.; Drake, K.L.; Adams, G.L.; Garner, H.R.; Ficht, T.A. Brucella melitensis VjbR and C12-HSL regulons: Contributions of the N-dodecanoyl homoserine lactone signaling molecule and LuxR homologue VjbR to gene expression. BMC Microbiol. 2010, 10, 167. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Caswell, C.C.; Foreman, R.; Roop, R.M.; Crosson, S. The Brucella abortus general stress response system regulates chronic mammalian infection and is controlled by phosphorylation and proteolysis. J. Biol. Chem. 2013, 288, 13906–13916. [Google Scholar] [CrossRef]
- Arriola Benitez, P.C.; Rey Serantes, D.; Herrmann, C.K.; Pesce Viglietti, A.I.; Vanzulli, S.; Giambartolomei, G.H.; Comerci, D.J.; Delpino, M.V. The Effector Protein BPE005 from Brucella abortus Induces Collagen Deposition and Matrix Metalloproteinase 9 Downmodulation via Transforming Growth Factor β1 in Hepatic Stellate Cells. Infect. Immun. 2016, 84, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Choi, Y.S.; Park, S.H.; Kim, Y.R.; Chu, H.; Hwang, K.J.; Park, M.Y. Lon Mutant of Brucella abortus Induces Tumor Necrosis Factor-Alpha in Murine J774.A1 Macrophage. Osong Public Heal. Res. Perspect. 2013, 4, 301–307. [Google Scholar] [CrossRef]
- Cerletti, M.; Paggi, R.A.; Guevara, C.R.; Poetsch, A.; De Castro, R.E. Global role of the membrane protease LonB in Archaea: Potential protease targets revealed by quantitative proteome analysis of a lonB mutant in Haloferax volcanii. J. Proteomics 2015, 121, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cerletti, M.; Paggi, R.; Troetschel, C.; Ferrari, M.C.; Guevara, C.R.; Albaum, S.; Poetsch, A.; De Castro, R. LonB Protease Is a Novel Regulator of Carotenogenesis Controlling Degradation of Phytoene Synthase in Haloferax volcanii. J. Proteome Res. 2018, 17, 1158–1171. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.I.; Cerletti, M.; Paggi, R.A.; Trötschel, C.; De Castro, R.E.; Poetsch, A.; Giménez, M.I. Haloferax volcanii Proteome Response to Deletion of a Rhomboid Protease Gene. J. Proteome Res. 2018, 17, 961–977. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Melzer, F.; Weise, C.; Neubauer, H.; Roesler, U.; Murugaiyan, J. Mass spectrometry data from proteomics-based screening of immunoreactive proteins of fully virulent Brucella strains using sera from naturally infected animals. Data Br. 2015, 4, 587–590. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poetsch, A.; Marchesini, M.I. Proteomics of Brucella. Proteomes 2020, 8, 8. https://doi.org/10.3390/proteomes8020008
Poetsch A, Marchesini MI. Proteomics of Brucella. Proteomes. 2020; 8(2):8. https://doi.org/10.3390/proteomes8020008
Chicago/Turabian StylePoetsch, Ansgar, and María Inés Marchesini. 2020. "Proteomics of Brucella" Proteomes 8, no. 2: 8. https://doi.org/10.3390/proteomes8020008
APA StylePoetsch, A., & Marchesini, M. I. (2020). Proteomics of Brucella. Proteomes, 8(2), 8. https://doi.org/10.3390/proteomes8020008






