A Microsomal Proteomics View of H2O2- and ABA-Dependent Responses
Abstract
1. Introduction
2. Materials and Methods
2.1. Treatments of Arabidopsis Cell Suspension Culture
2.2. Microsomal Protein Isolation
2.3. Trypsin Digestion and Protein Identification by Tandem Mass Spectrometry
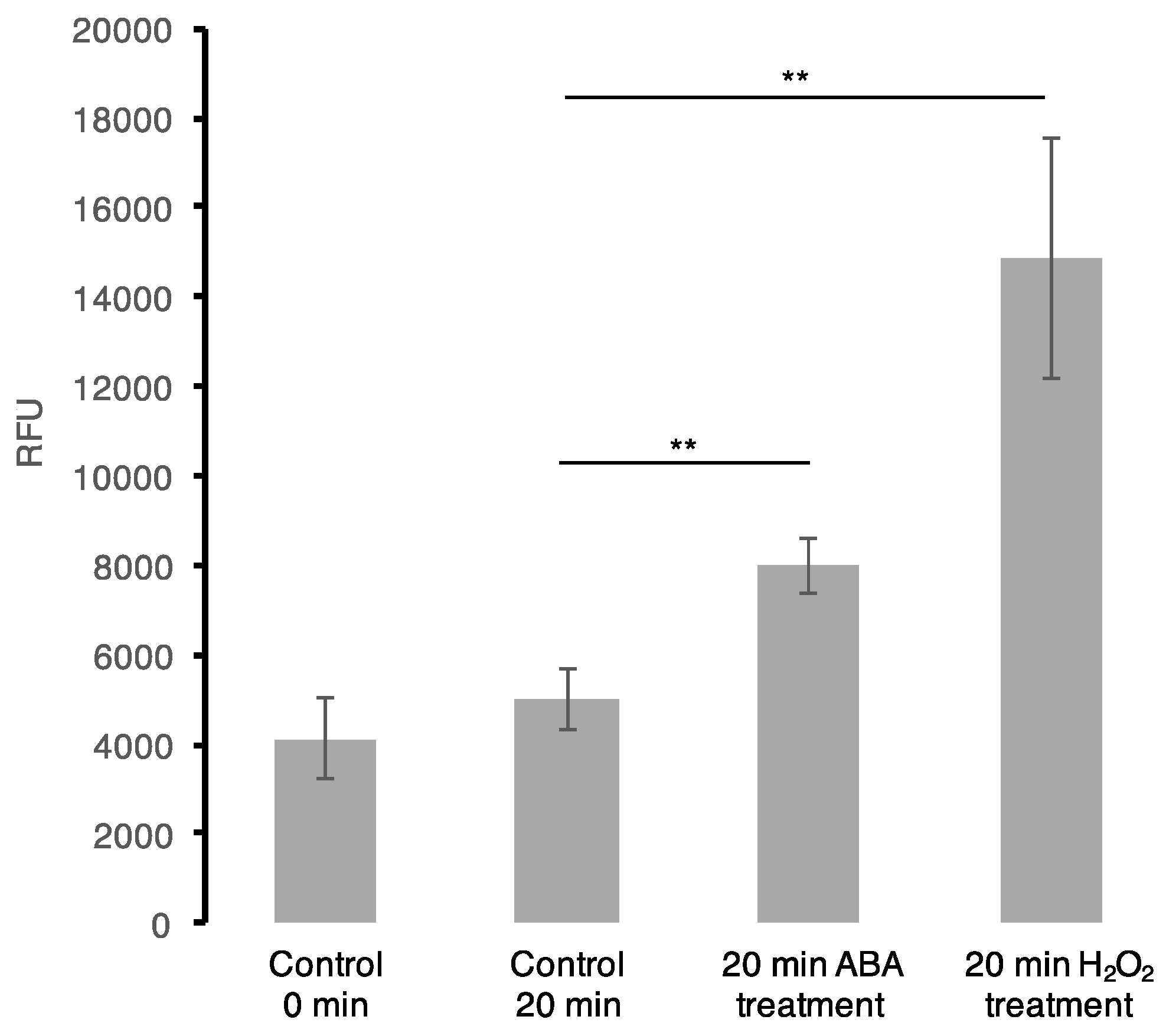
2.4. Intracellular ROS Assay and ABA Measurements
2.5. Bioinformatic Analyses
3. Results
3.1. H2O2-Responsive Microsomal Proteins
3.2. ABA-Responsive Microsomal Proteins
3.3. The Microsomal Proteome of the ABA and H2O2 Responses Show Similarity
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dat, J.; Vandenabeele, S.; Vranova, E.; Van Montagu, M.; Inze, D.; Van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell Mol. Life Sci. 2000, 57, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Luna, C.M.; Pastori, G.M.; Driscoll, S.; Groten, K.; Bernard, S.; Foyer, C.H. Drought controls on H2O2 accumulation, catalase (CAT) activity and CAT gene expression in wheat. J. Exp. Bot. 2005, 56, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, Y.; Yang, Y.; Wu, H.; Wang, D.; Liu, J. Involvement of hydrogen peroxide and nitric oxide in salt resistance in the calluses from Populus euphratica. Plant Cell Environ. 2007, 30, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Herr, E.H.; Orvar, B.L.; van Camp, W.; Willekens, H.; Inze, D.; Ellis, B.E. Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl. Acad. Sci. USA 1999, 96, 14165–14170. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Free radicals, reactive oxygen species and human disease: A critical evaluation with special reference to atherosclerosis. Br. J. Exp. Pathol. 1989, 70, 737–757. [Google Scholar] [PubMed]
- Moller, I.M. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 561–591. [Google Scholar] [CrossRef] [PubMed]
- Millar, H.; Considine, M.J.; Day, D.A.; Whelan, J. Unraveling the role of mitochondria during oxidative stress in plants. IUBMB Life 2001, 51, 201–205. [Google Scholar] [PubMed]
- Schmidtmann, E.; Konig, A.C.; Orwat, A.; Leister, D.; Hartl, M.; Finkemeier, I. Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol. Plant 2014, 7, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Verniquet, F.; Gaillard, J.; Neuburger, M.; Douce, R. Rapid inactivation of plant aconitase by hydrogen peroxide. Biochem. J. 1991, 276, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Leaver, C.J. The cytotoxic lipid peroxidation product, 4-hydroxy-2-nonenal, specifically inhibits decarboxylating dehydrogenases in the matrix of plant mitochondria. FEBS Lett. 2000, 481, 117–121. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Heazlewood, J.L.; Herald, V.; Holtzapffel, R.; Day, D.A.; Leaver, C.J.; Millar, A.H. The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 2002, 32, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Desikan, R.; Reynolds, A.; Hancock, J.T.; Neill, S.J. Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem. J. 1998, 330, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klusener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signaling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.G.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.M.; Zhao, J.; Scandalios, J.G. Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J. 2000, 22, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernandez, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Joshi-Saha, A.; Valon, C.; Leung, J. A brand new START: Abscisic acid perception and transduction in the guard cell. Sci. Signal. 2011, 4, re4. [Google Scholar] [CrossRef] [PubMed]
- Arve, L.E.; Carvalho, D.R.; Olsen, J.E.; Torre, S. ABA induces H2O2 production in guard cells, but does not close the stomata on Vicia faba leaves developed at high air humidity. Plant Signal. Behav. 2014, 9, e29192. [Google Scholar] [CrossRef] [PubMed]
- Desikan, R.; Cheung, M.K.; Bright, J.; Henson, D.; Hancock, J.T.; Neill, S.J. ABA, hydrogen peroxide and nitric oxide signaling in stomatal guard cells. J. Exp. Bot. 2004, 55, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Pei, Z.M.; Mori, I.C.; Schroeder, J. Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1–1 and abi2–1 protein phosphatase 2C mutants. Plant Cell 2001, 13, 2513–2523. [Google Scholar] [CrossRef] [PubMed]
- Giraudat, J.; Parcy, F.; Bertauche, N.; Gosti, F.; Leung, J.; Morris, P.C.; Bouvier-Durand, M.; Vartanian, N. Current advances in abscisic acid action and signaling. Plant Mol. Biol. 1994, 26, 1557–1577. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, J.; He, J.; Qin, Y.; Hua, D.; Duan, Y.; Chen, Z.; Gong, Z. ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet. 2014, 10, e1004791. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhang, A.; Zhang, J.; Jiang, M. Abscisic acid is a key inducer of hydrogen peroxide production in leaves of maize plants exposed to water stress. Plant Cell Physiol. 2006, 47, 1484–1495. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, J. Involvement of plasma-membrane NADPH oxidase in abscisic acid- and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 2002, 215, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-H.; Chao, Y.-Y.; Kao, C.H. Abscisic acid is an inducer of hydrogen peroxide production in leaves of rice seedlings grown under potassium deficiency. Bot. Stud. 2012, 53, 229–237. [Google Scholar]
- Marondedze, C.; Turek, I.; Parrott, B.; Thomas, L.; Jankovic, B.; Lilley, K.S.; Gehring, C. Structural and functional characteristics of cGMP-dependent methionine oxidation in Arabidopsis thaliana proteins. Cell Commun. Signal. 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Wong, A.; Groen, A.; Serrano, N.; Jankovic, B.; Lilley, K.; Gehring, C.; Thomas, L. Exploring the Arabidopsis proteome: Influence of protein solubilization buffers on proteome coverage. Int. J. Mol. Sci. 2015, 16, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Ordonez, N.M.; Marondedze, C.; Thomas, L.; Pasqualini, S.; Shabala, L.; Shabala, S.; Gehring, C. Cyclic mononucleotides modulate potassium and calcium flux responses to H2O2 in Arabidopsis roots. FEBS Lett. 2014, 588, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.; Marondedze, C.; Ederli, L.; Pasqualini, S.; Gehring, C. Proteomic signatures implicate cAMP in light and temperature responses in Arabidopsis thaliana. J. Proteom. 2013, 83, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Groen, A.; Thomas, L.; Lilley, K.; Marondedze, C. Identification and quantitation of signal molecule-dependent protein phosphorylation. Methods Mol. Biol. 2013, 1016, 121–137. [Google Scholar] [PubMed]
- Bevan, M.; Bancroft, I.; Bent, E.; Love, K.; Goodman, H.; Dean, C.; Bergkamp, R.; Dirkse, W.; van Staveren, M.; Stiekema, W.; et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998, 391, 485–488. [Google Scholar] [CrossRef] [PubMed]
- AgriGO. Available online: http://bioinfo.cau.edu.cn/agriGO/ (accessed on 1 February 2017).
- Moreno-Sanchez, R.; Saavedra, E.; Rodriguez-Enriquez, S.; Gallardo-Perez, J.C.; Quezada, H.; Westerhoff, H.V. Metabolic control analysis indicates a change of strategy in the treatment of cancer. Mitochondrion 2010, 10, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Araujo, W.L.; Nunes-Nesi, A.; Nikoloski, Z.; Sweetlove, L.J.; Fernie, A.R. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 2012, 35, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Tretter, L.; Adam-Vizi, V. Alpha-ketoglutarate dehydrogenase: A target and generator of oxidative stress. Philos. Trans. R. Soc. Lond. Biol. Sci. 2005, 360, 2335–2345. [Google Scholar] [CrossRef] [PubMed]
- Janero, D.R.; Hreniuk, D. Suppression of TCA cycle activity in the cardiac muscle cell by hydroperoxide-induced oxidant stress. Am. J. Physiol. 1996, 270, C1735–C1742. [Google Scholar] [PubMed]
- Moeder, W.; Del Pozo, O.; Navarre, D.A.; Martin, G.B.; Klessig, D.F. Aconitase plays a role in regulating resistance to oxidative stress and cell death in Arabidopsis and Nicotiana benthamiana. Plant Mol. Biol. 2007, 63, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Argos, P. Homology between IRE-BP, a regulatory RNA-binding protein, aconitase, and isopropylmalate isomerase. Nucleic Acids Res. 1991, 19, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Thomas, L.; Serrano, N.L.; Lilley, K.S.; Gehring, C. The RNA-binding protein repertoire of Arabidopsis thaliana. Sci. Rep. 2016, 6, 29766. [Google Scholar] [CrossRef] [PubMed]
- Heyndrickx, K.S.; Vandepoele, K. Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol. 2012, 159, 884–901. [Google Scholar] [CrossRef] [PubMed]
- Martiniere, A.; Li, X.; Runions, J.; Lin, J.; Maurel, C.; Luu, D.T. Salt stress triggers enhanced cycling of Arabidopsis root plasma-membrane aquaporins. Plant Signal. Behav. 2012, 7, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Niittyla, T.; Fuglsang, A.T.; Palmgren, M.G.; Frommer, W.B.; Schulze, W.X. Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell Proteom. 2007, 6, 1711–1726. [Google Scholar] [CrossRef] [PubMed]
- Kammerloher, W.; Fischer, U.; Piechottka, G.P.; Schaffner, A.R. Water channels in the plant plasma membrane cloned by immunoselection from a mammalian expression system. Plant J. 1994, 6, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Dynowski, M.; Schaaf, G.; Loque, D.; Moran, O.; Ludewig, U. Plant plasma membrane water channels conduct the signaling molecule H2O2. Biochem. J. 2008, 414, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Wudick, M.M.; Li, X.; Valentini, V.; Geldner, N.; Chory, J.; Lin, J.; Maurel, C.; Luu, D.T. Subcellular redistribution of root aquaporins induced by hydrogen peroxide. Mol. Plant 2015, 8, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.G.; Barrett-Wilt, G.A.; Sussman, M.R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 15986–15991. [Google Scholar] [CrossRef] [PubMed]
- Grondin, A.; Rodrigues, O.; Verdoucq, L.; Merlot, S.; Leonhardt, N.; Maurel, C. Aquaporins contribute to ABA-Triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 2015, 27, 1945–1954. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.S.; Hwang, B.K. The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 2011, 155, 447–463. [Google Scholar] [CrossRef] [PubMed]
- Leonhardt, N.; Kwak, J.M.; Robert, N.; Waner, D.; Leonhardt, G.; Schroeder, J.I. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 2004, 16, 596–615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.L.; Peumans, W.J.; Barre, A.; Astoul, C.H.; Rovira, P.; Rouge, P.; Proost, P.; Truffa-Bachi, P.; Jalali, A.A.H.; Van Damme, E.J.M. Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta 2000, 210, 970–978. [Google Scholar] [PubMed]
- Feng, H.; Xu, W.Z.; Lin, H.H.; Chong, K. Transcriptional regulation of wheat VER2 promoter in rice in response to abscisic acid, jasmonate, and light. J. Genet. Genom. 2009, 36, 371–377. [Google Scholar] [CrossRef]
- Fouquaert, E.; Van Damme, E.J. Promiscuity of the euonymus carbohydrate-binding domain. Biomolecules 2012, 2, 415–434. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Xu, W.Q.; Xiang, Y.; Jia, H.Y.; Zhang, L.X.; Ma, Z.Q. Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol. Biol. 2014, 84, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Le, M.H.; Cao, Y.; Zhang, X.C.; Stacey, G. LIK1, a CERK1-interacting kinase, regulates plant immune responses in Arabidopsis. PLoS ONE 2014, 9, e102245. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Zhang, X.C.; Neece, D.; Ramonell, K.M.; Clough, S.; Kim, S.Y.; Stacey, M.G.; Stacey, G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008, 20, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Kabbage, M.; Kim, H.J.; Britt, R.; Dickman, M.B. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 2011, 7, e1002107. [Google Scholar] [CrossRef] [PubMed]
- Groenendyk, J.; Peng, Z.; Dudek, E.; Fan, X.; Mizianty, M.J.; Dufey, E.; Urra, H.; Sepulveda, D.; Rojas-Rivera, D.; Lim, Y.; et al. Interplay between the oxidoreductase PDIA6 and microRNA-322 controls the response to disrupted endoplasmic reticulum calcium homeostasis. Sci. Signal. 2014, 7, ra54. [Google Scholar] [CrossRef] [PubMed]
- Borgese, N.; Aggujaro, D.; Carrera, P.; Pietrini, G.; Bassetti, M. A role for N-myristoylation in protein targeting: NADH-cytochrome b5 reductase requires myristic acid for association with outer mitochondrial but not ER membranes. J. Cell Biol. 1996, 135, 1501–1513. [Google Scholar] [CrossRef] [PubMed]
- Fukuchi-Mizutani, M.; Mizutani, M.; Tanaka, Y.; Kusumi, T.; Ohta, D. Microsomal electron transfer in higher plants: cloning and heterologous expression of NADH-cytochrome b5 reductase from Arabidopsis. Plant Physiol. 1999, 119, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Bello, R.I.; Alcain, F.J.; Gomez-Diaz, C.; Lopez-Lluch, G.; Navas, P.; Villalba, J.M. Hydrogen peroxide- and cell-density-regulated expression of NADH-cytochrome b5 reductase in HeLa cells. J. Bioenerg. Biomembr. 2003, 35, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Hashimoto, T. Altered microtubule dynamics by expression of modified alpha-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. Plant J. 2005, 43, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, B.; Harris, N.S.; Deyholos, M.K. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 2007, 58, 3591–3607. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bian, Y.; Cheng, K.; Zou, H.; Sun, S.S.; He, J.X. A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. J. Proteom. Res. 2012, 11, 2301–2315. [Google Scholar] [CrossRef] [PubMed]
- Boldogh, I.R.; Pon, L.A. Interactions of mitochondria with the actin cytoskeleton. Biochim. Biophys. Acta 2006, 1763, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Bright, J.; Desikan, R.; Hancock, J.T.; Weir, I.S.; Neill, S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006, 45, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Dong, F.; Gao, J.; Galbraith, D.W.; Song, C.P. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 2001, 126, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Penson, S.P.; Schuurink, R.C.; Fath, A.; Gubler, F.; Jacobsen, J.V.; Jones, R.L. cGMP is required for gibberellic acid-induced gene expression in barley aleurone. Plant Cell 1996, 8, 2325–2333. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Groen, A.J.; Thomas, L.; Lilley, K.S.; Gehring, C. A Quantitative phosphoproteome analysis of cGMP-dependent cellular responses in Arabidopsis thaliana. Mol. Plant 2016, 9, 621–623. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
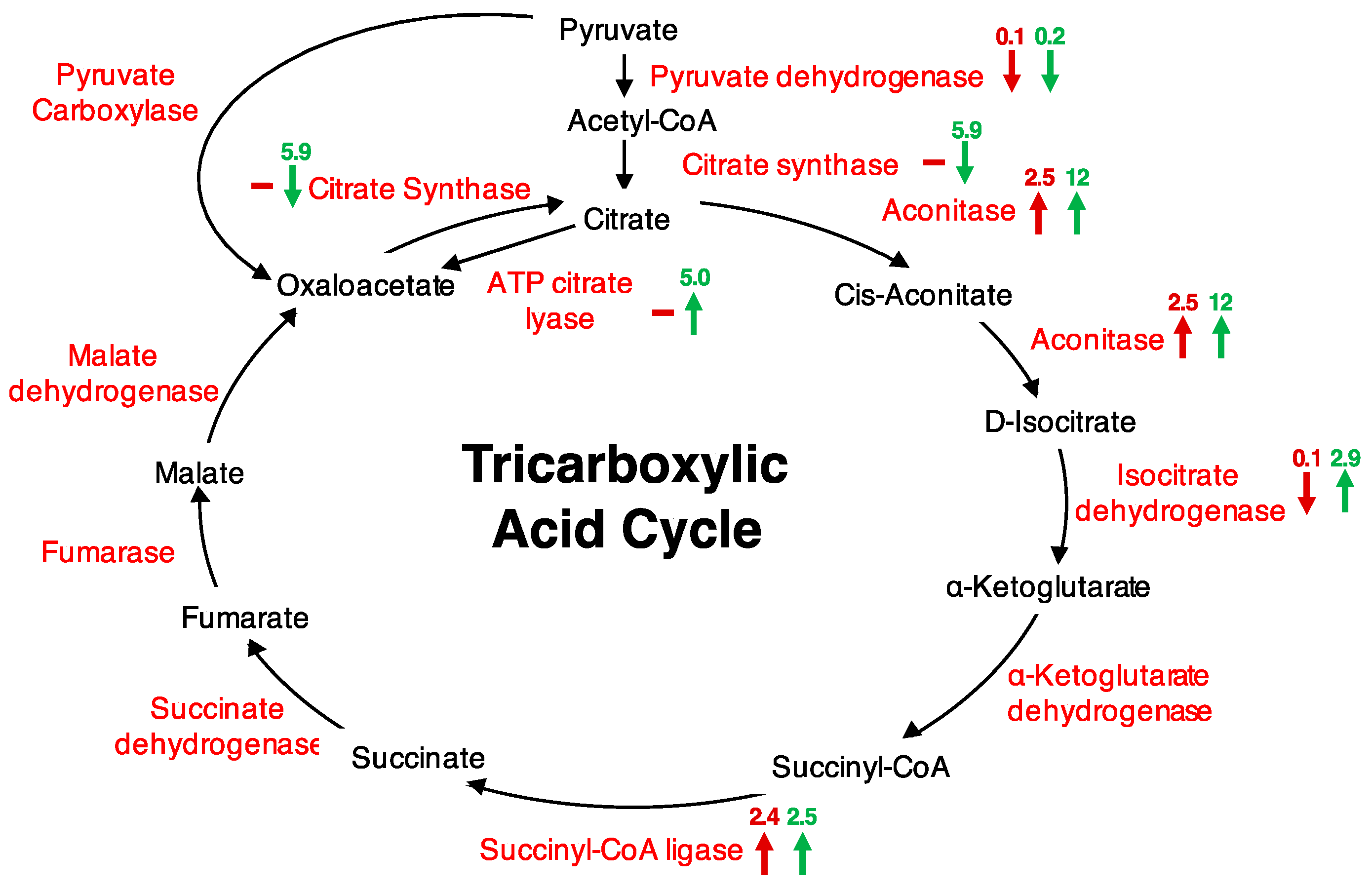
| Accession Number | Protein Name | ANOVA (p-Value) | FC 5 min | FC 20 min |
|---|---|---|---|---|
| 1. Metabolism | ||||
| AT1G17745 | D-3-phosphoglycerate dehydrogenase 2 | 0.0089 | 0.1 | 0.1 |
| AT5G26780 | Serine hydroxymethyltransferase 2 | 0.036 | 2.1 | 13 |
| AT3G61440 | Cysteine synthase C1 | 0.00074 | ns | 5 |
| AT4G14880 | O-acetylserine (thiol) lyase isoform A1 | 0.015 | 0.1 | 7.9 |
| AT5G23300 | Pyrimidine d | 0.013 | 0.2 | ns |
| AT3G09820 | Adenosine kinase 1 | 0.0042 | 0.1 | 14.0 |
| AT5G17770 | NADH:cytochrome B5 reductase 1 | 0.017 | 0.4 | 2.5 |
| AT1G74790 | Catalytics | 0.045 | 5.7 | 4.6 |
| AT4G23850 | AMP-dep. synthetase and ligase protein | 0.017 | 0.5 | ns |
| AT1G65290 | Mitochondrial acyl carrier protein 2 | 0.044 | 2.9 | ns |
| AT3G47930 | L-galactono-1,4-lactone dehydrogenase | 0.019 | 0.2 | ns |
| 2. Energy | ||||
| AT1G79550 | Phosphoglycerate kinase | 0.0069 | 0.2 | 2.0 |
| AT1G24180 | Pyruvate dehydrogenase E1 comp. α-2 | 0.014 | 0.1 | 0.2 |
| AT3G48000 | Aldehyde dehydrogenase 2B4 | 0.022 | 4.3 | 7.1 |
| AT3G60750 | Transketolase | 0.0087 | 0.5 | 4.5 |
| AT2G05710 | Aconitase 3 | 0.0002 | 2.5 | 12.0 |
| AT2G20420 | ATP citrate lyase | 0.022 | ns | 5.0 |
| AT5G08300 | Succinyl-CoA ligase, alpha subunit | 0.006 | 2.4 | 2.5 |
| AT2G44350 | Citrate synthase family protein | 0.026 | ns | 5.9 |
| AT1G65930 | Cyt. NADP+-dep. isocitrate dehydrogenase | 0.033 | 0.1 | 2.9 |
| AT1G15120 | Ubiquinol-cytochrome C reductase hinge | 0.016 | 19 | 20 |
| AT3G03100 | NADH:ubiquinone oxidoreductase,17.2kDa | 0.0029 | 0.5 | ns |
| AT5G13430 | Ubiquinol-cytochrome C reductase, Fe-S | 0.02 | 0.2 | ns |
| AT3G14610 | Cytochrome P450, 72A, polypeptide 7 | 0.0026 | 0.1 | 5.4 |
| 3. Cell growth/division | ||||
| AT5G52240 | Membrane steroid binding protein 1 | 0.02 | 0.1 | 7.5 |
| AT1G10930 | DNA helicase (RECQl4A) | 0.0036 | 4.6 | 0.1 |
| AT3G44310 | Nitrilase 1 | 0.023 | 2.9 | ns |
| 4. Transcription | ||||
| AT1G14850 | Nucleoporin 155 | 0.00094 | ns | 0.3 |
| 5. Protein synthesis | ||||
| AT1G01100 | 60S acidic ribosomal | 0.022 | ns | 0.4 |
| AT3G48930 | Nucleic acid-binding, OB-fold-like protein | 0.024 | 2.1 | ns |
| AT3G10090 | Nucleic acid-binding, OB-fold-like protein | 0.027 | ns | 0.3 |
| AT3G09200 | Ribosomal protein L10 | 0.012 | 2.9 | ns |
| AT3G04400 | Ribosomal protein L14p/L23e | 0.0095 | 3.8 | 3.0 |
| AT1G23290 | Ribosomal protein L18e/L15 | 0.017 | ns | 0.5 |
| AT4G02230 | Ribosomal protein L19e | 0.0089 | ns | 0.3 |
| AT2G44120 | Ribosomal protein L30/L7 | 0.027 | ns | 0.5 |
| AT5G56710 | Ribosomal protein L31e | 0.013 | 4.6 | ns |
| AT3G45030 | Ribosomal protein S10p/S20e | 0.03 | ns | 2.2 |
| AT5G02960 | Ribosomal protein S12/S23 | 0.016 | 2.3 | ns |
| AT3G60245 | Zinc-binding ribosomal protein | 0.036 | 2.4 | ns |
| 6. Protein destination and storage | ||||
| AT1G14980 | Chaperonin 10 | 0.022 | ns | 2.9 |
| AT3G02530 | Chaperonin 60 family protein | 0.045 | 2.2 | 0.4 |
| AT3G03960 | Chaperonin 60 family protein | 0.041 | 2.1 | ns |
| AT5G56030 | Heat shock protein 81-2 | 0.012 | 0.4 | ns |
| AT3G52140 | Tetratricopeptide repeat-containing protein | 0.04 | 5.3 | ns |
| 7. Transporters | ||||
| AT4G38580 | Farnesylated protein 6 | 0.017 | 0.4 | ns |
| AT3G15660 | Glutaredoxin 4 | 0.011 | ns | 6.0 |
| AT1G27950 | Glycosylphosphatidylinositol-anchored lipid protein transfer 1 | 0.027 | 16.0 | 44.0 |
| AT1G07670 | Endomembrane-type CA-ATPase 4 | 0.0026 | 0.1 | 5.0 |
| AT4G27500 | Proton pump interactor 1 | 0.006 | ns | 2.4 |
| AT4G39080 | Vacuolar proton ATPase A3 | 0.0041 | 0.5 | ns |
| AT3G58730 | Vacuolar proton pump D subunit | 0.044 | 0.4 | ns |
| AT1G15500 | TLC ATP/ADP transporter | 0.0099 | 0.3 | ns |
| AT4G28390 | ADP/ATP carrier 3 | 0.014 | 0.1 | ns |
| AT5G60460 | Preprotein translocase Sec, Sec61-β subunit | 0.0081 | 4.7 | 4 |
| AT3G51890 | Clathrin light chain protein | 0.019 | ns | 0.5 |
| AT3G08530 | Clathrin, heavy chain | 0.0021 | 0.5 | 0.2 |
| AT3G11130 | Clathrin, heavy chain | 0.00044 | 0.5 | 0.4 |
| AT5G19760 | Mitochondrial substrate carrier protein | 0.038 | ns | 2.2 |
| AT5G40810 | Cytochrome C1 | 0.021 | 0.3 | ns |
| 8. Intracellular traffic | ||||
| AT3G49560 | Mit. import inner membrane translocase Tim17 | 0.0031 | 0.1 | ns |
| AT1G61570 | Mit. import inner membrane translocase 13 | 0.0044 | 17 | 23 |
| AT1G27390 | Translocase outer membrane 20-2 | 0.0015 | 0.1 | 2.1 |
| AT3G60600 | Vesicle associated protein | 0.051 | ns | 3 |
| AT4G34660 | SH3 domain-containing protein | 0.039 | 4.7 | ns |
| 9. Cell structure | ||||
| AT5G09810 | Actin 7 | 0.0038 | 0.5 | ns |
| AT5G19770 | Tubulin alpha-3 | 0.016 | 0.1 | ns |
| AT5G62690 | Tubulin beta chain 2 | 0.006 | 0.4 | 0.4 |
| AT2G29550 | Tubulin beta-7 chain | 0.0034 | 0.3 | 0.4 |
| AT4G14960 | Tubulin/FtsZ family protein | 0.036 | 0.4 | 0.5 |
| AT3G08950 | Electron transport SCO1/SenC protein | 0.045 | 0.1 | 4.3 |
| 10. Signal transduction | ||||
| AT3G14840 | LRR transmembrane protein kinase | 0.025 | ns | 11 |
| AT5G59840 | Ras-related small GTP-binding protein | 0.0084 | 0.1 | 2.7 |
| AT3G51800 | Metallopeptidase M24 | 0.0015 | 5.1 | ns |
| 11. Disease/Defence | ||||
| AT3G57280 | Transmembrane proteins 14C | 0.014 | 0.1 | ns |
| AT3G12500 | Basic chitinase | 0.035 | ns | 2.1 |
| AT3G32980 | Peroxidase superfamily protein | 0.012 | ns | 3 |
| AT4G36430 | Peroxidase superfamily protein | 0.015 | 0.1 | ns |
| 12. Unclassified | ||||
| AT2G40765 | Unknown protein | 0.019 | 2 | ns |
| AT2G46540 | Unknown protein | 0.033 | ns | 3.5 |
| AT4G12590 | Transmembrane protein, DUF106 | 0.033 | 0.3 | ns |
| AT1G08480 | Unknown protein | 0.048 | 0.2 | 0.4 |
| AT3G20370 | TRAF-like family protein | 0.0058 | 0.2 | 3 |
| AT3G49720 | Unknown protein | 0.016 | 0.1 | 0.3 |
| AT4G24330 | DUF1682, unknown protein | 0.0077 | 0.1 | 2.2 |
| AT2G33585 | Unknown protein | 0.019 | 0.1 | 13 |
| Accession Number | Protein Name | ANOVA (p-Value) | FC 5 min | FC 20 min |
|---|---|---|---|---|
| 1. Metabolism | ||||
| AT2G26400 | Acireductone dioxygenase 3 | 0.035 | 3.9 | 6.8 |
| AT4G34200 | D-3-phosphoglycerate dehydrogenase 1 | 0.017 | 0.5 | ns |
| AT1G17745 | D-3-phosphoglycerate dehydrogenase 2 | 0.029 | 0.1 | 0.2 |
| AT4G13940 | S-adenosyl-L-homocysteine hydrolase | 0.028 | 2.5 | 2.0 |
| AT3G17820 | Glutamine synthetase 1.3 | 0.023 | 3.7 | 3.5 |
| AT5G17770 | NADH cytochrome B5 reductase 1 | 0.046 | 2.3 | 2.3 |
| AT3G15730 | Phospholipase D α 1 | 0.014 | 3.2 | 3.8 |
| 2. Energy | ||||
| AT2G01140 | Aldolase superfamily protein | 0.0075 | 5.0 | 5.1 |
| AT5G43940 | GroES-like zinc-binding dehydrogenase | 0.014 | 4.5 | 5.2 |
| AT4G34870 | Rotamase cyclophilin 5 | 0.0015 | 3.1 | 2.7 |
| AT2G05710 | Aconitase 3 | 0.035 | 8.0 | 9.9 |
| AT1G15120 | Ubiquinol-cytochrome C reductase hinge | 0.041 | 15.0 | 11.0 |
| AT2G33220 | GRIM-19 protein | 0.017 | 2.9 | 7.1 |
| 3. Cell growth/division | ||||
| AT5G43070 | WPP domain protein 1 | 0.042 | 0.4 | 0.2 |
| 4. Transcription | ||||
| AT2G18740 | Small nuclear ribonucleoprotein | 0.02 | 7.7 | 4.3 |
| AT5G40480 | Embryo defective 3012 | 0.0084 | 5.6 | 7.9 |
| AT1G14850 | Nucleoporin 155 | 0.0058 | 0.1 | 0.2 |
| 5. Protein synthesis | ||||
| AT1G08360 | Ribosomal protein L1p/L10e | 0.037 | 3.1 | ns |
| AT4G10450 | Ribosomal protein L6 | 0.00092 | 12.0 | >0.1 |
| AT5G08180 | Ribosomal protein L7Ae | 0.022 | 11.0 | 8.7 |
| AT3G04400 | Ribosomal protein L14p | 0.016 | 3.5 | 2.6 |
| AT3G14600 | Ribosomal protein L18ae | 0.033 | 3.4 | 3.3 |
| AT3G45030 | Ribosomal protein S10p/S20e | 0.0041 | 2.3 | ns |
| AT5G47930 | Zinc-binding ribosomal protein | 0.027 | 8.6 | 7.0 |
| AT4G00810 | 60S acidic ribosomal protein family | 0.042 | 0.4 | 0.4 |
| AT1G01100 | 60S acidic ribosomal protein family | 0.045 | 0.4 | 0.4 |
| AT5G47700 | 60S acidic ribosomal protein family | 0.0085 | 0.3 | 0.4 |
| 6. Protein destination and storage | ||||
| AT1G14980 | Chaperonin 10 | 0.0061 | 2.5 | 3.0 |
| AT3G16420 | PYK10-binding protein 1 | 0.017 | 0.3 | 0.5 |
| AT5G56030 | Heat shock protein 81-2 | 0.0038 | 0.4 | 0.5 |
| AT1G03220 | Eukaryotic aspartyl protease | 0.0045 | 2.3 | 2.5 |
| 7. Transporters | ||||
| AT3G53420 | Plasma membrane intrinsic protein 2A | 0.031 | 0.3 | ns |
| AT3G42050 | Vacuolar ATP synthase subunit H | 0.045 | 4.4 | 4.2 |
| AT4G27500 | Proton pump interactor 1 | 0.0048 | ns | 2.3 |
| AT2G34250 | SecY protein transport | 0.011 | 2.5 | 3.9 |
| AT3G08530 | Clathrin, heavy chain | 0.0019 | 0.2 | 0.4 |
| AT3G11130 | Clathrin, heavy chain | 0.00054 | 0.3 | 0.4 |
| 8. Intracellular traffic | ||||
| AT2G29530 | TIM10 zinc finger protein | 0.021 | 6.0 | 5.0 |
| AT1G61570 | Mit. import inner membrane 13 translocase | 0.042 | 19.0 | 11.0 |
| 9. Cell structure | ||||
| AT4G30270 | Xyloglucan endotransglucosylase | 0.045 | ns | 3.0 |
| AT3G18780 | Actin 2 | 0.016 | 0.3 | ns |
| AT5G09810 | Actin 7 | 0.00073 | 0.5 | 0.5 |
| AT5G19770 | Tubulin alpha-3 | 0.044 | 0.5 | 0.3 |
| AT5G62690 | Tubulin beta chain 2 | 0.003 | 0.3 | 0.4 |
| AT2G29550 | Tubulin beta-7 chain | 0.0072 | 0.3 | 0.4 |
| AT4G14960 | Tubulin/FtsZ family protein | 0.019 | 0.4 | 0.3 |
| 10. Signal transduction | ||||
| AT3G14840 | LRR transmembrane protein kinase | 0.014 | 8.3 | 14.0 |
| AT3G51800 | Metallopeptidase M24 | 0.027 | 3.4 | ns |
| 11. Disease/Defence | ||||
| AT1G78850 | D-mannose binding lectin protein | 0.0013 | 14.0 | 18.0 |
| AT2G43610 | Chitinase family protein | 0.05 | ns | 0.3 |
| AT4G38740 | Rotamase CYP 1 | 0.02 | 2.6 | ns |
| AT1G20620 | Catalase 3 | 0.0064 | 3.5 | 2.1 |
| Accession Number | Protein Name | H2O2 5 min | H2O2 20 min | ABA 5 min | ABA 20 min |
|---|---|---|---|---|---|
| 1. Metabolism | |||||
| AT1G17745 | D-3-phosphoglycerate dehydrogenase 2 | 0.1 | 0.1 | 0.1 | 0.2 |
| AT5G17770 | NADH:cytochrome B5 reductase 1 | 0.4 | 2.5 | 2.3 | 2.3 |
| 2. Energy | |||||
| AT2G05710 | Aconitase 3 | 2.5 | 12 | 8 | 9.9 |
| AT1G15120 | Ubiquinol-cytochrome C reductase hinge | 19 | 20 | 15 | 11 |
| 3. Transcription | |||||
| AT1G14850 | Nucleoporin 155 | ns | 0.3 | 0.1 | 0.2 |
| 4. Protein synthesis | |||||
| AT3G04400 | Ribosomal protein L14p/L23e | 3.8 | 3 | 3.5 | 2.6 |
| AT3G45030 | Ribosomal protein S10p/S20e | ns | 2.2 | 2.3 | ns |
| AT1G01100 | 60S acidic ribosomal protein family | ns | 0.4 | 0.4 | 0.4 |
| 5. Protein destination and storage | |||||
| AT1G14980 | Chaperonin 10 | ns | 2.9 | 2.5 | 3 |
| AT5G56030 | Heat shock protein 81-2 | 0.4 | ns | 0.4 | 0.5 |
| 6. Transporters | |||||
| AT4G27500 | Proton pump interactor 1 | ns | 2.4 | ns | 2.3 |
| AT3G08530 | Clathrin, heavy chain | 0.5 | 0.2 | 0.2 | 0.4 |
| AT3G11130 | Clathrin, heavy chain | 0.5 | 0.4 | 0.3 | 0.4 |
| 7. Intracellular traffic | |||||
| AT1G61570 | Mit. import inner membrane 13 translocase | 17 | 23 | 19 | 11 |
| 8. Cell structure | |||||
| AT5G09810 | Actin 7 | 0.5 | ns | 0.5 | 0.5 |
| AT4G14960 | Tubulin/FtsZ family protein | 0.4 | 0.5 | 0.4 | 0.3 |
| AT5G62690 | Tubulin beta chain 2 | 0.4 | 0.4 | 0.3 | 0.4 |
| AT2G29550 | Tubulin beta-7 chain | 0.3 | 0.4 | 0.3 | 0.4 |
| AT5G19770 | Tubulin alpha-3 | 0.1 | ns | 0.5 | 0.3 |
| 9. Signal transduction | |||||
| AT3G51800 | Metallopeptidase M24 family protein | 5.1 | ns | 3.4 | ns |
| AT3G14840 | LRR transmembrane protein kinase | ns | 11 | 8.3 | 14 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alqurashi, M.; Thomas, L.; Gehring, C.; Marondedze, C. A Microsomal Proteomics View of H2O2- and ABA-Dependent Responses. Proteomes 2017, 5, 22. https://doi.org/10.3390/proteomes5030022
Alqurashi M, Thomas L, Gehring C, Marondedze C. A Microsomal Proteomics View of H2O2- and ABA-Dependent Responses. Proteomes. 2017; 5(3):22. https://doi.org/10.3390/proteomes5030022
Chicago/Turabian StyleAlqurashi, May, Ludivine Thomas, Chris Gehring, and Claudius Marondedze. 2017. "A Microsomal Proteomics View of H2O2- and ABA-Dependent Responses" Proteomes 5, no. 3: 22. https://doi.org/10.3390/proteomes5030022
APA StyleAlqurashi, M., Thomas, L., Gehring, C., & Marondedze, C. (2017). A Microsomal Proteomics View of H2O2- and ABA-Dependent Responses. Proteomes, 5(3), 22. https://doi.org/10.3390/proteomes5030022






