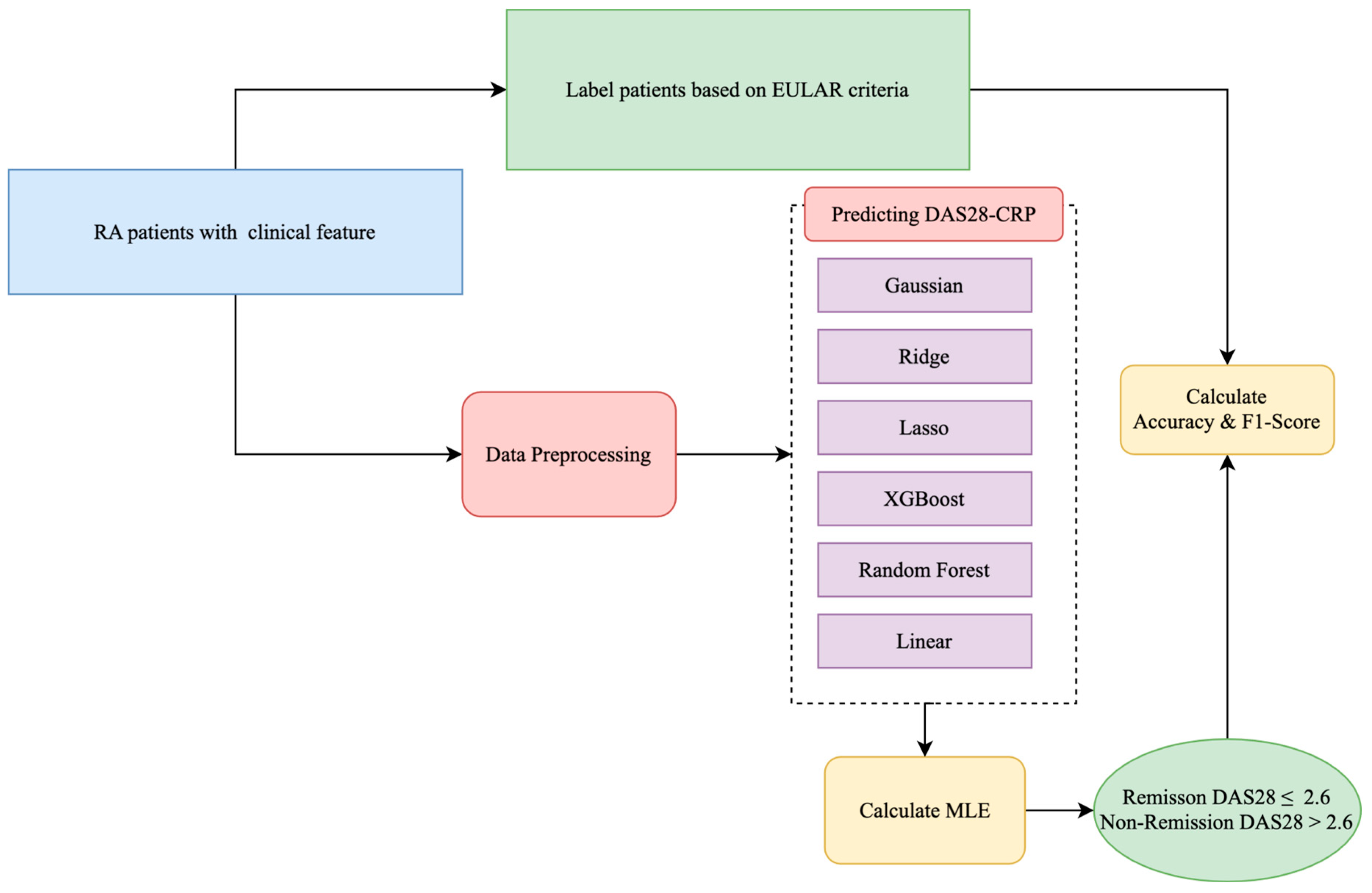
1. Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune disease characterized by persistent joint inflammation, leading to pain, swelling, and, over time, joint damage and disability [
1,
2]. It affects millions worldwide and remains one of the most common causes of chronic joint disease [
3]. Despite advances in treatment, managing RA remains challenging due to the wide variability in individual treatment responses [
4].
Achieving remission, where disease activity is minimal or absent, is the primary goal of RA therapy. In clinical practice, remission is typically assessed using composite indices such as the Disease Activity Score for 28 joints (DAS28), which incorporates laboratory markers (e.g., C-reactive protein or CRP), joint counts, and patient-reported symptoms [
5]. Identifying likely non-responders before treatment begins could help to avoid irreversible joint damage and guide earlier intervention.
Biological disease-modifying antirheumatic drugs (bDMARDs) have significantly improved outcomes, especially for patients unresponsive to conventional agents like methotrexate [
6,
7]. However, their effectiveness varies across individuals, and it may take 3 to 6 months to determine treatment response. During this time, non-responders may suffer from side effects, continued inflammation, and financial burdens [
8], creating uncertainty for both clinicians and patients. Early prediction of treatment response could lead to more personalized decision-making, which reduces exposure to ineffective therapies and improves outcomes [
9,
10,
11].
Artificial intelligence (AI) is increasingly transforming medicine by enabling the efficient analysis of large and complex datasets beyond the capacity of traditional statistical methods. Within AI, machine learning (ML) holds particular promise for automating tasks such as diagnosis, risk stratification, and treatment selection by identifying patterns in clinical data. Studies across domains like radiology, cardiology, oncology, and neurology have demonstrated that ML models can match, or even surpass, human performance in specific diagnostic and prognostic tasks [
12]. In the context of treatment planning, ML enables early prediction of patient response, supports risk stratification, and facilitates timely clinical decision-making [
13,
14,
15,
16].
Recent ML studies in RA have used a wide range of input data, ranging from routine clinical features to advanced imaging and molecular profiles [
8,
17,
18,
19,
20]. While non-routine data sources like synovial gene signatures [
19] or multi-omics [
20] can reveal underlying disease mechanisms, their cost and complexity limit clinical integration.
Many existing models use accessible clinical data but often simplify the prediction task by reducing it to binary classification, labeling patients as responders or non-responders based on a fixed remission threshold [
18,
21,
22]. While these studies have identified important clinical predictors, including drug-specific patterns [
8] and sustained versus short-term response [
21], they remain limited by their reliance on dichotomous outcomes. Some recent approaches have introduced risk stratification and model explainability to improve clinical applicability [
21,
22], yet the binary framework remains a central constraint.
Remission criteria vary across guidelines and continue to evolve [
23], making rigid thresholds less reliable in practice. For instance, a patient with a DAS28-CRP score of 2.7 is grouped with someone experiencing much higher disease activity, despite only narrowly missing the remission cutoff of 2.6. Predicting continuous DAS28-CRP scores instead allows for more nuanced and flexible insights that better reflect clinical decision-making.
In addition, a recent systematic review by Mendoza-Pinto et al. [
24] highlighted that many published models have a high or unclear risk of bias and often lack external validation. As a result, their reported performance may not translate reliably across clinical settings, underscoring the need for rigorous external validation.
To address these limitations, we developed a ML framework to predict 12-month DAS28-CRP scores in patients with RA initiating bDMARD therapy, using routinely collected baseline clinical data. Our feature set included joint counts, C-reactive protein (CRP), visual analog scale (VAS) scores, and concurrent medications—variables commonly documented in standard rheumatology practice. This design makes the model practical to implement and broadly applicable across diverse clinical settings.
Unlike many existing studies that frame remission prediction as a binary classification task, our approach models DAS28-CRP as a continuous outcome. This enables more nuanced and generalizable predictions that support flexible, individualized treatment planning.
We trained and evaluated several regression models—Ridge, Lasso, Support Vector Regression, Random Forest, and XGBoost—on a cohort of 154 RA patients from University Hospital Erlangen. A nested cross-validation scheme was used to ensure robust performance estimation and reduce overfitting. To evaluate generalizability, we applied the models to an independent external dataset from the Austrian BioReg registry, which includes a more diverse, real-world patient population from multiple clinical sites. This external validation provides insights into model robustness across distinct clinical contexts and sets our study apart from similar works that lack validation beyond internal data.
Ultimately, this study presents a practical, data-driven tool to support early RA management and improve treatment outcomes through more personalized care.
3. Results
The internal dataset included 154 patients with rheumatoid arthritis (RA) and 13 routinely collected clinical features (see
Table 1). These included demographics, laboratory markers, joint counts, and composite disease activity scores. We evaluated eight ML models—Ridge, Lasso, SVR, Random Forest, Decision Tree, KNN, AdaBoost, and XGBoost—on two prediction tasks: (1) regression of 12-month DAS28-CRP scores following bDMARD therapy and (2) classification of remission status based on the EULAR threshold. All models were trained on the internal dataset and externally validated using the Austrian BioReg registry. Model performance was assessed using standard metrics and evaluated for fairness across age groups. We also analyzed feature importance stability.
3.1. Regression Performance
Table 3 summarizes the regression performance using MAE, MSE, and R
2 based on nested cross-validation for the internal cohort and direct evaluation on the external cohort. Overall, Ridge regression achieved the best performance across both datasets, with the lowest MAE and MSE and highest R
2 values (MAE = 0.633, R
2 = 0.542 internally; MAE = 0.678, R
2 = 0.491 externally). Lasso and SVR followed closely behind. In contrast, Decision Tree and KNN consistently underperformed, exhibiting higher errors and lower R
2 scores, indicating weaker generalization.
The strong performance of Ridge can be attributed to its regularization, which shrinks but retains all input features, thus reducing overfitting while preserving informative signals, especially in datasets with collinear predictors, such as clinical measurements.
All features were scaled using MinMax normalization, which slightly outperformed standard and robust scaling across key metrics for the Ridge model. It yielded the highest R2, accuracy, and F1 score and the lowest MAE. Given its consistent but modest advantage, MinMax was selected as the preferred scaling method.
For context, the observed DAS28-CRP scores ranged from 0.00 to 8.42, capturing the full spectrum from remission to high disease activity. With this range in mind, an MAE value of 0.63 indicates relatively precise prediction, suitable for stratifying disease progression at the population level.
3.2. Classification Performance
To enable clinical interpretation, continuous DAS28-CRP predictions were binarized using the established EULAR threshold of 2.6 into remission and non-remission categories. Classification performance was evaluated using accuracy, precision, recall, and F1 score (
Table 4).
Ridge, SVR, and Lasso achieved the best classification performances on the internal dataset, with Ridge obtaining the highest accuracy (0.818) and F1 score (0.800). These results are consistent with the regression findings. In contrast, Decision Tree and KNN lagged behind in all metrics.
We then assessed generalizability using the external BioReg dataset. Ridge again emerged as the top-performing model (accuracy = 0.773, F1 = 0.762), followed by Lasso and SVR. Although tree-based models such as AdaBoost and XGBoost demonstrated high recall, their precision was notably lower, leading to reduced F1 scores. This pattern indicates that these models tend to overpredict remission.
To better understand this behavior, we analyzed class distributions. The internal dataset was relatively balanced (46.4% remission), while the external dataset was moderately imbalanced (only 39.8% in remission). This imbalance likely influenced model behavior, particularly precision–recall trade-offs. Models like AdaBoost and SVR achieved high recall by predicting more positive (remission) cases, but this came at the cost of precision. These findings emphasize the importance of considering both precision and recall in imbalanced clinical classification tasks.
3.3. Fairness Across Age Groups
To assess potential bias, we evaluated model accuracy across younger (<55 years) and older (≥55 years) age groups, using 55 as the threshold based on the median age in the dataset.
Table 5 reports the classification accuracy for each subgroup and the performance gap.
Ridge and SVR demonstrated the most balanced performances, with age-based accuracy gaps of only 0.024 and 0.022, respectively. In contrast, tree-based models such as AdaBoost and XGBoost showed wider disparities. These results suggest that regularized linear models are not only more generalizable but also fairer in terms of age subgroup performances.
3.4. Feature Importance and Model Stability
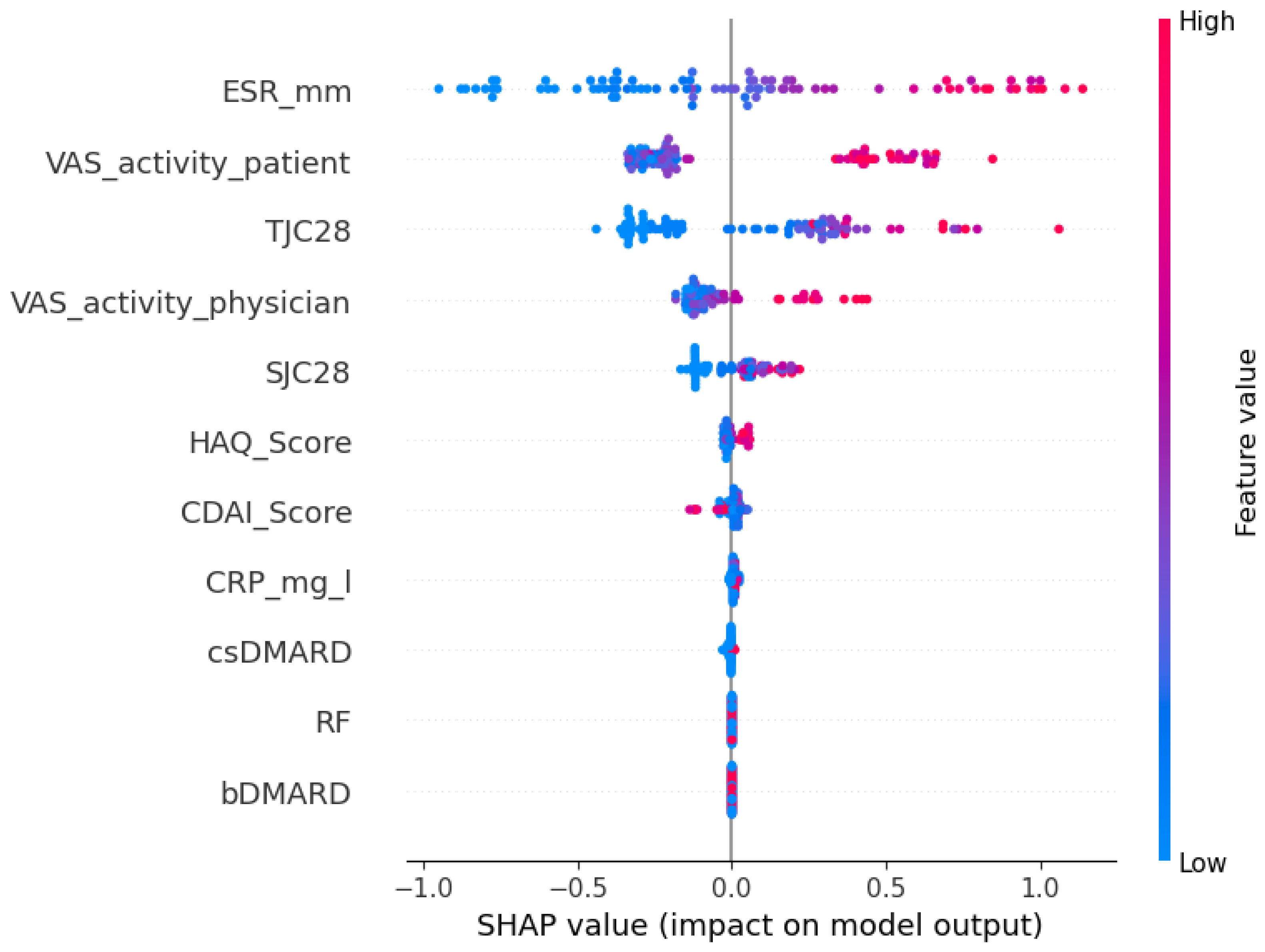
To examine model interpretability and generalizability, we used SHAP (SHapley Additive exPlanations), a game-theoretic approach that quantifies each feature’s contribution to model predictions. The SHAP summary plots rank features by their overall importance across the dataset. Each point represents an individual patient, with the x-axis showing the magnitude and direction of that feature’s impact on the predicted disease activity score. Point color reflects the feature value—red for high and blue for low—enabling intuitive interpretation of how feature levels influence predictions. For instance, in the internal cohort (
Figure 2), where the patient’s global health assessment is the most influential feature, the plot shows that patients with high VAS scores (red points) have positive SHAP values, indicating that a patient’s self-reported disease activity directly contributes to a higher predicted DAS28-CRP score.
As shown in
Figure 2 and
Figure 3, the top five most predictive features were consistent across both cohorts: a patient’s global health assessment (VAS), ESR, TJC28, a physician’s global health assessment (VAS), and SJC28. Although the order of the top features varied slightly, the consistency in key predictors confirms that Ridge captures stable clinical signals transferable across populations.
3.5. Summary
Ridge regression consistently demonstrated the best overall performance across all evaluation criteria. It achieved high predictive accuracy in both regression and classification tasks, generalized effectively to an external cohort, and maintained stable performance across age groups. Furthermore, it relied on consistent and clinically meaningful features, supporting interpretability. These results highlight Ridge regression as a robust, generalizable, and practical model for predicting treatment outcomes in RA, with strong potential for integration into personalized clinical decision-making.
4. Discussion
This study evaluated the feasibility of using baseline clinical data to predict 12-month disease activity in patients with RA who were beginning bDMARD therapy. By applying ML models to estimate continuous DAS28-CRP scores and subsequently categorizing patients using a clinically relevant remission threshold, we developed a flexible and interpretable framework that extends beyond binary classification.
Among the eight ML models tested, Ridge regression consistently achieved the strongest performance, demonstrating the highest internal R2, lowest prediction errors, and best classification metrics. Lasso and SVR also performed well, while tree-based models like Decision Tree and AdaBoost underperformed, especially on external validation. These results underscore the suitability of regularized linear models for clinical prediction tasks involving structured, tabular data and relatively small cohorts.
External validation using the Austrian BioReg dataset revealed a modest decline in model performance, as expected due to differences in cohort characteristics and data collection procedures. Notably, Ridge regression maintained solid generalizability, with a classification accuracy of 0.773 and an F1 score of 0.762. In terms of regression, Ridge also yielded the lowest external mean absolute error (MAE = 0.678) and the highest R2 value (0.491) among all tested models. These results highlight Ridge’s ability to retain meaningful predictive signals across clinical settings and confirm its value as a robust choice for generalizable outcome modeling in RA.
The modest decline in performance observed during external validation likely reflects clinical and methodological differences between the Erlangen and BioReg cohorts. As shown in
Table 1, BioReg patients had higher average CRP levels and significantly lower csDMARD co-medication rates, suggesting differences in baseline inflammation and treatment protocols. Additionally, the BioReg cohort was drawn from multiple outpatient clinics and private practices, which may introduce variation into data documentation and patient management compared to the single-center Erlangen cohort. These cohort-specific differences may have contributed to the observed distribution shift, partially explaining the drop in predictive accuracy. Such performance variation is expected when transitioning from a controlled academic setting to a heterogeneous real-world population and highlights the importance of external validation in assessing model generalizability.
To further investigate model interpretability and generalizability, we examined feature importance using the SHAP values for the best-performing Ridge model. Across both the internal and external cohorts, the top five most influential features were largely consistent: the patient’s global health assessment (VAS), the erythrocyte sedimentation rate (ESR), the tender joint count (TJC28), the physician’s global assessment (VAS), and the swollen joint count (SJC28). While the exact ordering of feature importance varied slightly between datasets, the stability of these core predictors suggests that Ridge regression captures clinically meaningful signals that generalize across populations. This interpretability is especially important in clinical settings, where understanding the drivers of model predictions can support clinician confidence and informed decision-making.
By modeling DAS28-CRP as a continuous outcome rather than a binary label, our approach allows clinicians to interpret disease activity more flexibly. Remission criteria vary between guidelines, such as those from EULAR and ACR, and have evolved over time. Predicting a continuous disease activity score enables clinicians to apply whichever threshold is appropriate for their specific clinical context, offering greater utility than a fixed classification scheme. In practice, treatment decisions are often guided not only by whether a patient crosses a defined cutoff (e.g., DAS28-CRP < 2.6) but also by how close they are to it. For instance, a predicted score of 2.7 might prompt continued monitoring or minor treatment adjustments, whereas a value of 4.5 would suggest a clearly insufficient response. By providing a full-spectrum estimate of disease activity, the model supports more nuanced and individualized care while remaining adaptable to different remission definitions or local standards.
In clinical practice, the model output could help to identify patients less likely to respond to the selected bDMARD. This information could support more frequent monitoring, early treatment escalation, or the selection of an alternative bDMARD class. Such individualized guidance could help to avoid prolonged ineffective treatment and enable faster achievement of disease control.
Few prior studies have modeled DAS28-CRP as a continuous outcome, limiting opportunities for the direct comparison of regression results. To enable performance benchmarking, we additionally converted our regression outputs into binary remission classifications using the standard DAS28-CRP threshold. This allowed us to assess classification metrics commonly reported in the literature. Our models achieved over 80% accuracy and F1 score on internal data and over 75% on the external dataset—performance levels that are comparable to or exceed those typically reported for similar clinical prediction tasks. These results support the validity and robustness of our regression framework, even when viewed through the lens of conventional binary evaluation metrics.
Despite these encouraging results, several limitations should be considered. The sample size, particularly in the external validation cohort, was relatively modest, which may affect generalizability and model stability. The models were trained exclusively on baseline clinical features available at the initiation of bDMARD therapy. While this supports early risk stratification, it does not capture subsequent changes in disease activity or short-term treatment response. Incorporating early follow-up data, such as 3-month DAS28-CRP scores, could enhance predictive accuracy and enable more adaptive treatment planning. Additionally, our reliance on routinely collected clinical variables means that potentially important but unmeasured confounders, such as smoking status, physical activity, or socioeconomic factors, were not included. The model also did not differentiate between bDMARD classes, such as TNF inhibitors and JAK inhibitors, which may vary in effectiveness and response patterns. Future work could explore stratified modeling approaches to better account for this therapeutic heterogeneity.
While the model demonstrated strong performance in retrospective evaluation, prospective studies are needed to validate its clinical utility and robustness in real-world settings. Future research should also examine the integration of early treatment response signals and evaluate performance across more diverse populations.
Overall, this study demonstrates that ML models trained on routine clinical data can support remission prediction in RA. The ability to identify patients who are less likely to respond to bDMARDs early in the treatment process could reduce unnecessary delays, improve patient outcomes, and inform more personalized treatment strategies. With further validation and refinement, these models have the potential to become valuable tools in clinical rheumatology.