Differences in Physiological Responses of Two Oat (Avena nuda L.) Lines to Sodic-Alkalinity in the Vegetative Stage
Abstract
:1. Introduction
2. Results
2.1. Plant Growth
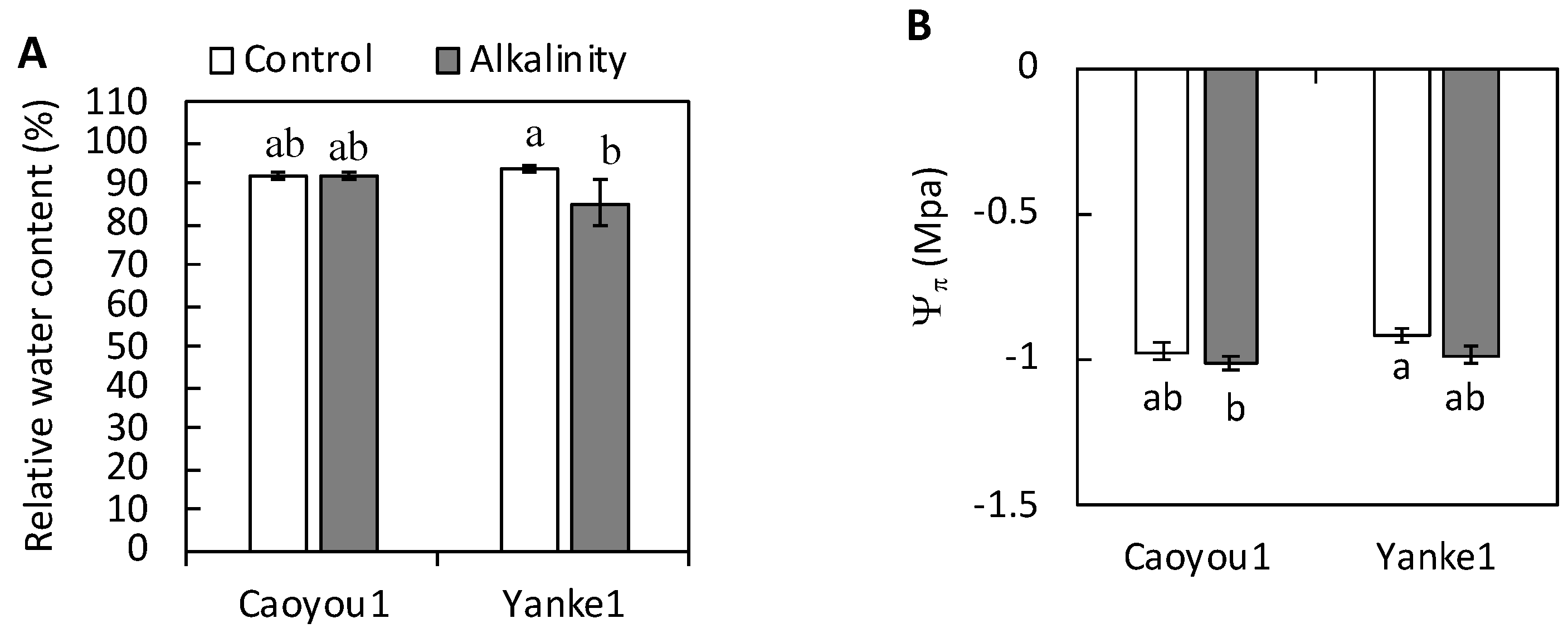
2.2. Leaf Relative Water Content and Leaf Osmotic Potential
2.3. K+, Na+, and K+/Na+ Ratio in Leaf, Sheath, and Root Tissues
2.4. Mg2+ and Total P Concentrations in Leaf Tissue
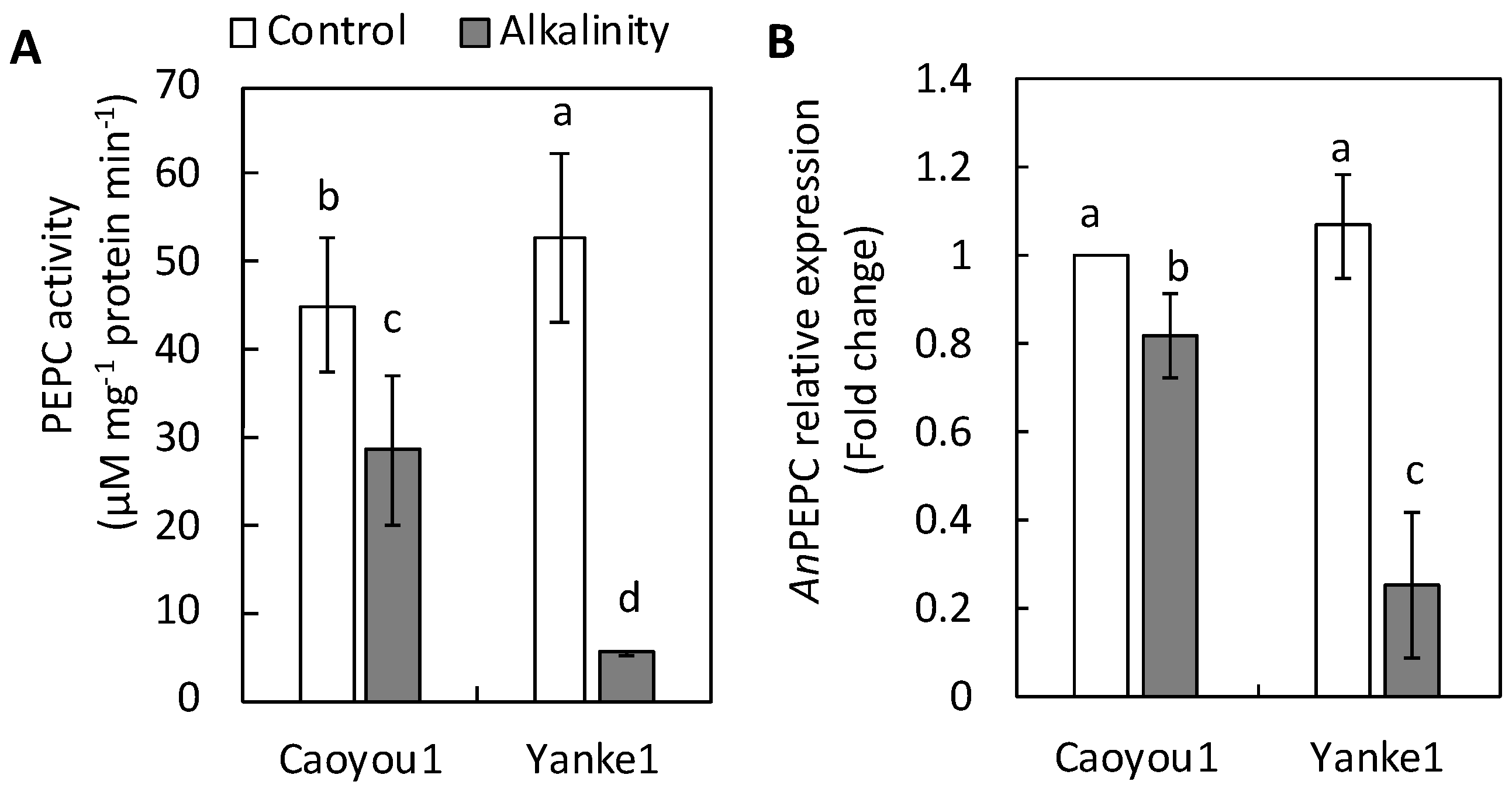
2.5. Phosphoenolpyruvate Carboxylase Activity and Phosphoenolpyruvate Carboxylase-Coding Gene Expression in Leaf Tissue
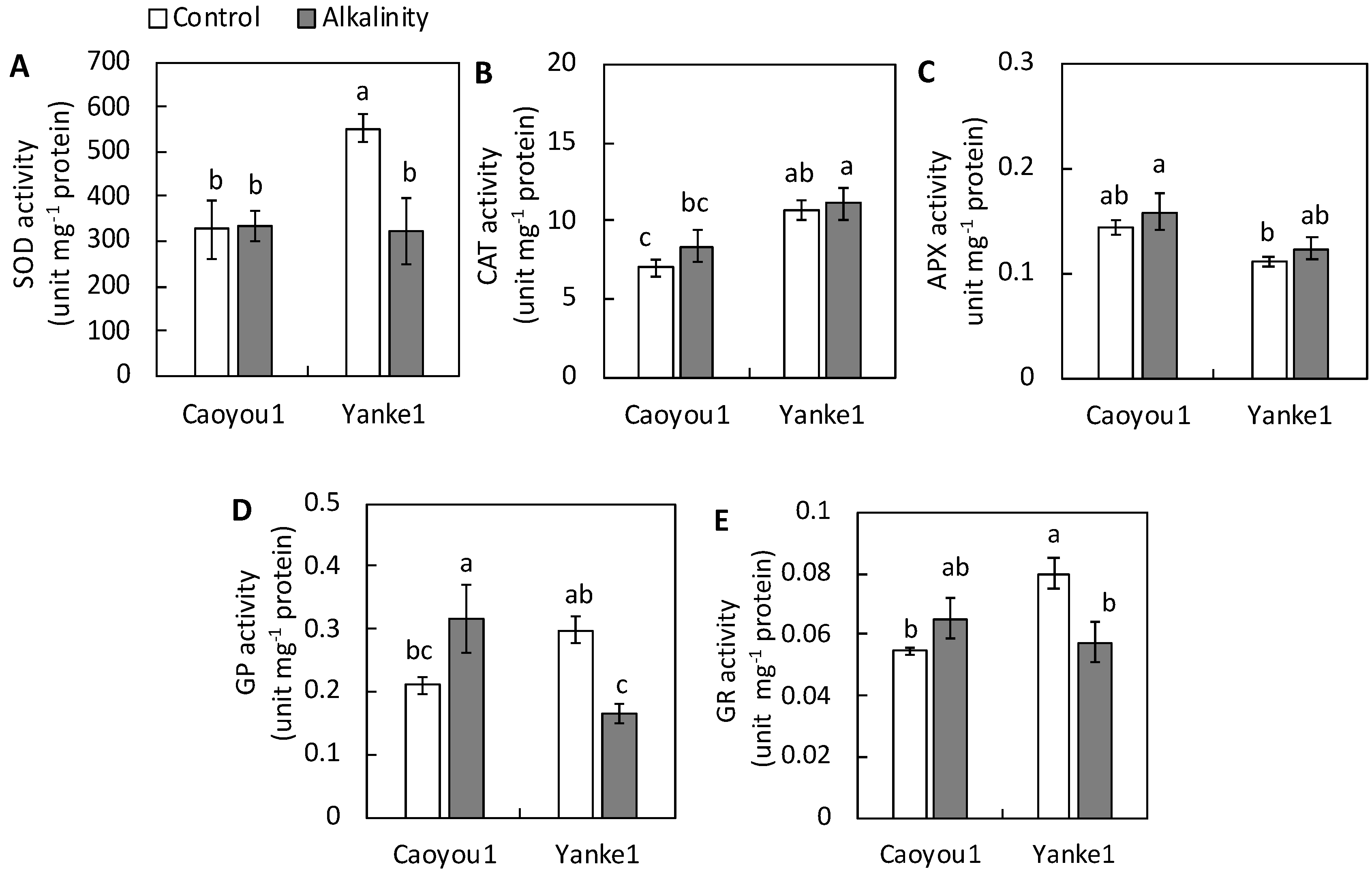
2.6. Antioxidant Enzyme Activity in Leaf Tissue
2.7. Expression Analysis of Antioxidant Enzyme-Coding Genes
3. Discussion
3.1. Water Uptake Capacity in Leaves and K+ Accumulation in Roots
3.2. Total P and Phosphoenolpyruvate Carboxylase Activity in Leaf Tissue
3.3. Differential Antioxidative Capacity in Leaf Tissue
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, Alkalinity Treatment, and Post-Treatment Soil pH
4.2. Measurement of Shoot Height, Total Leaf Area, and Dry Weight
4.3. Measurement of Relative Water Content and Osmotic Potential in Leaf Tissue
4.4. Determination of Na+ and K+ in Different Plant Organs
4.5. Determination of Mg2+ and Total P in Leaf Tissue
4.6. Measurement of Phosphoenolpyruvate Carboxylase Activity in Leaf Tissue
4.7. Measurement of Antioxidant Enzyme Activity in Leaf Tissue
4.8. RNA Isolation and Real-Time Polymerase Chain Reaction
4.9. Data Analysis and Statistics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jin, H.; Kim, H.R.; Plaha, P.; Liu, S.K.; Park, J.Y.; Piao, Y.Z.; Yang, Z.H.; Jiang, G.B.; Kwak, S.S.; An, G.; et al. Expression profiling of the genes induced by Na2CO3 and NaCl stresses in leaves and roots of Leymus chinensis. Plant Sci. 2008, 175, 784–792. [Google Scholar] [CrossRef]
- Song, J.; Fan, H.; Zhao, Y.Y.; Jia, Y.H.; Du, X.H.; Wang, B.S. Effect of salinity on germination, seedling emergence, seedling growth and ion accumulation of a euhalophyte Suaeda salsa in an intertidal zone and on saline inland. Aquat. Bot. 2008, 88, 331–337. [Google Scholar] [CrossRef]
- Tanji, K.K. Salinity in the soil environment. In Salinity: Environment—Plants—Molecules; Läuchli, A., Lüttge, U., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 21–51. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Beltran, J.; Manzur, C.L. Overview of salinity problems in the world and FAO strategies to address the problem. In Proceedings of the International Salinity Forum, Riverside, CA, USA, 25–27 April 2005; pp. 311–313. [Google Scholar]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Marschner, H. Adaptation of plants to adverse chemical soil conditions. In Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: Cambridge, MA, USA, 1995; 651p, ISBN 978-0-12-473542-2. [Google Scholar]
- Yang, C.W.; Chong, J.N.; Li, C.; Kim, C.; Shi, D.C.; Wang, D.L. Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant Soil 2007, 294, 263–276. [Google Scholar] [CrossRef]
- Rouphael, Y.; Cardarelli, M.; Mattia, E.D.; Tullio, M.; Rea, E.; Colla, G. Enhancement of alkalinity tolerance in two cucumber genotypes inoculated with an arbuscular mycorrhizal biofertilizer containing Glomus intraradices. Biol. Fertil. Soils 2010, 46, 499–509. [Google Scholar] [CrossRef]
- Fredeen, A.L.; Raab, T.K.; Rao, I.M.; Terry, N. Effects of phosphorus nutrition on photosynthesis in Glycine max (L.). Merr. Planta 1990, 181, 399–405. [Google Scholar] [CrossRef]
- Li, R.; Shi, F.; Fukuda, K.; Yang, Y. Effects of salt and alkali stresses on germination, growth, photosynthesis and ion accumulation in alfalfa (Medicago sativa L.). Soil Sci. Plant Nutr. 2010, 56, 725–733. [Google Scholar] [CrossRef]
- Poschenrieder, C.; Fernández, J.A.; Rubio, L.; Pérez, L.; Terés, J.; Barceló, J. Transport and use of bicarbonate in plants: Current knowledge and challenges ahead. Int. J. Mol. Sci. 2018, 19, 1352. [Google Scholar] [CrossRef] [Green Version]
- Karki, S.; Rizal, G.; Quick, W.P. Improvement of photosynthesis in rice (Oryza sativa L.) by inserting the C4 pathway. Rice 2013, 6, 28. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Liu, J.; Wu, J.; Liu, H.; Xu, H. Effects of salt stress on plasmalemma permeability, osmolyte accumulation and protective enzyme activities in oat plants. J. Food Agric. Environ. 2013, 11, 696–701. [Google Scholar]
- Ashraf, M. Biotechnological approach of improving plant salt tolerance using antioxidants as marker. Biotechnol. Adv. 2009, 27, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 9. [Google Scholar] [CrossRef]
- Abogadallah, G.M. Insights into the significance of antioxidative defense under salt stress. Plant Signal Behav. 2010, 5, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V. Mechanisms of oxidative stress in plants: From classical chemistry to cell biology. Environ. Exp. Bot. 2015, 109, 212–228. [Google Scholar] [CrossRef]
- Burrows, V.D. Hulless oat development, applications, and opportunities. In Oats; American Association of Cereal Chemists: St. Paul, MN, USA, 2011; pp. 31–50. [Google Scholar]
- Martínez-Villaluenga, C.; Peñas, E. Health benefits of oat: Current evidence and molecular mechanisms. Curr. Opin. Food Sci. 2017, 14, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Bai, J.; Liu, J.; Zhang, N.; Yang, J.; Sa, R.; Wu, L. Effect of alkali stress on soluble sugar, antioxidant enzymes and yield of oat. J. Integr. Agri. 2013, 12, 1441–1449. [Google Scholar] [CrossRef]
- Bai, J.; Yan, W.; Wang, Y.; Yin, Q.; Liu, J.; Wight, C.; Ma, B. Screening oat genotypes for tolerance to salinity and alkalinity. Front. Plant Sci. 2018, 9, 1302. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Wu, N.; Liu, J.L.; Yang, N.N.; Yang, Y.Y. Effects of mild saline-alkali on photosynthetic physiology and yield of different oat cultivars. Southwest China J. Agric. Sci. 2018, 31, 2041–2046. [Google Scholar]
- Islam, M.S.; Akhter, M.M.; El Sabagh, A.; Liu, L.Y.; Nguyen, N.T.; Ueda, A.; Masaoka, Y.; Saneoka, H. Comparative studies on growth and physiological responses to saline and alkaline stresses of Foxtail millet (Setaria italica L.) and Proso millet (Panicum miliaceum L.) Aust. J. Crop Sci. 2011, 5, 1269–1277. [Google Scholar]
- Liu, L.; El-Shemy, H.A.; Saneoka, H. Effects of 5-aminolevulinic acid on water uptake, ionic toxicity, and antioxidant capacity of Swiss chard (Beta vulgaris L.) under sodic-alkaline conditions. J. Plant Nutr. Soil Sci. 2017, 180, 535–543. [Google Scholar] [CrossRef]
- Sun, J.; He, L.; Li, T. Response of seedling growth and physiology of Sorghum bicolor (L.) Moench to saline-alkali stress. PLoS ONE 2019, 14, e0220340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Zhang, S.; Xu, X.; Korpelainen, H.; Li, C. Effects of increased alkalinity on Na+ and K+ contents, lipid peroxidation and antioxidative enzymes in two populations of Populus cathayana. Biol. Plant. 2009, 53, 597–600. [Google Scholar] [CrossRef]
- Wang, J.; Liu, W.Z.; Mu, H.F.; Dand, T.H. Inorganic phosphorus fractions and phosphorus availability in a calcareous soil receiving 21-year superphosphate application. Pedosphere 2010, 20, 304–310. [Google Scholar] [CrossRef]
- Plaxton, W.C.; Shane, M.W. The role of post-translational enzyme modifications in the metabolic adaptations of phosphorus-deprived plants. Annu. Plant Rev. 2015, 48, 99–124. [Google Scholar]
- Zhang, H.F.; Xu, W.G.; Wang, H.W.; Hu, L.; Li, Y.; Qi, X.L.; Zhang, L.; Li, C.X.; Hua, X. Pyramiding expression of maize genes encoding phosphoenolpyruvate carboxylase (PEPC) and pyruvate orthophosphate dikinase (PPDK) synergistically improve the photosynthetic characteristics of transgenic wheat. Protoplasma 2014, 251, 1163–1173. [Google Scholar] [CrossRef]
- Elsawy, H.I.A.; Mekawy, A.M.M.; Elhity, M.A.; Abdel-dayem, S.M.; Abdelaziz, M.N.; Assaha, D.V.M.; Ueda, A.; Saneoka, H. Differential responses of two Egyptian barley (Hordeum vulgare L.) cultivars to salt stress. Plant Physiol. Biochem. 2018, 127, 425–435. [Google Scholar] [CrossRef]
- Kong, L.; Huo, H.; Mao, P. Antioxidant response and related gene expression in aged oat seed. Front. Plant Sci. 2015, 6, 158. [Google Scholar] [CrossRef]
- Wu, B.; Munkhtuya, Y.; Li, J.; Hu, Y.; Zhang, Q.; Zhang, Z. Comparative transcriptional profiling and physiological responses of two contrasting oat genotypes under salt stress. Sci. Rep. 2018, 8, 16248. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Rahman, M.A.; Rathinasabapathi, B.; Babar, M.A. UPLC-HRMS-based untargeted metabolic profiling reveals changed in chickpea (Cicer arietinum) metabolome following long-term grought stress. Plant Cell Environ. 2019, 42, 115–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mekawy, A.M.M.; Assaha, D.V.M.; Yahagi, H.; Tada, Y.; Ueda, A.; Saneoka, H. Growth, physiological adaptation, and gene expression analysis of two Egyptian rice cultivars under salt stress. Plant Physiol. Biochem. 2015, 87, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.S.; Toribara, T.Y.; Warner, H. Microdetermination of phosphorus. Anal. Chem. 1956, 28, 1756–1758. [Google Scholar] [CrossRef]
- Shi, J.; Yi, K.; Liu, Y.; Xie, L.; Zhou, Z.; Chen, Y.; Hu, Z.; Zheng, T.; Liu, R.; Chen, Y.; et al. Phosphoenolpyruvate carboxylase in arabidopsis leaves plays a crucial role in carbon and nitrogen metabolism. Plant Physiol. 2015, 167, 671–681. [Google Scholar] [CrossRef] [Green Version]
- Dhindsa, R.S.; Plumb-Dhindsa, P.; Thorpe, T.A. Leaf senescence: Correlated with increased permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. Methods Enzymol. 1984, 105, 121–126. [Google Scholar] [PubMed]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidases in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Foyer, C.H.; Halliwell, B. The presence of glutathione and glutathione reductase in chloroplasts: A proposed role in ascorbic acid metabolism. Planta 1976, 133, 21–25. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Ueda, A.; Yahagi, H.; Fujikawa, Y.; Nagaoka, T.; Esaka, M.; Calcaño, M.; González, M.M.; Martich, J.D.H.; Saneoka, H. Comparative physiological analysis of salinity tolerance in rice. Soil Sci. Plant Nutr. 2013, 59, 896–903. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Measurements | Tissues | Caoyou1 | Yanke1 | ||
|---|---|---|---|---|---|
| Control | Sodic-Alkalinity | Control | Sodic-Alkalinity | ||
| Na+ (mg g−1 FW) | Leaf | 0.02 ± 0.00 b | 2.95 ± 0.07 a | 0.01 ± 0.00 b | 2.93 ± 0.10 a |
| Sheath | 0.02 ± 0.00 b | 2.59 ± 0.12 a | 0.02 ± 0.00 b | 2.44 ± 0.07 a | |
| Root | 0.10 ± 0.00 b | 2.42 ± 0.10 a | 0.10 ± 0.01 b | 2.29 ± 0.11 a | |
| K+ (mg g−1 FW) | Leaf | 6.72 ± 0.17 a | 3.20 ± 0.07 b | 7.19 ± 0.54 a | 3.27 ± 0.19 b |
| Sheath | 6.41 ± 0.09 a | 3.24 ± 0.11 b | 6.30 ± 0.04 a | 3.17 ± 0.16 b | |
| Root | 3.06 ± 0.14 a | 2.48 ± 0.08 b | 2.67 ± 0.09 ab | 1.86 ± 0.22 c | |
| K+/Na+ | Leaf | 416.69 ± 39.9 b | 1.08 ± 0.01 c | 574.45 ± 51.02 a | 1.12 ± 0.07 c |
| Sheath | 314.60 ±1 0.23 b | 1.26 ± 0.06 c | 396.84 ± 10.22 a | 1.31 ± 0.10 c | |
| Root | 30.65 ± 1.60 a | 1.03 ± 0.06 b | 29.74 ± 2.26 a | 0.80 ± 0.07 b |
| Lines | Treatments | Soil pH |
|---|---|---|
| Coyou1 | Control | 6.42 ± 0.02 d |
| Sodic-alkalinity | 8.31 ± 0.02 b | |
| Yanke1 | Control | 6.53 ± 0.02 c |
| Sodic-alkalinity | 8.48 ± 0.01 a |
| Genes | Forward Primers (5′–3′) | Reverse Primers (5′–3′) |
|---|---|---|
| AnSOD1 | CACAAGCACTTCACAGGAACAGT | TGCCACTCTGAACATTTCATCAC |
| AnCAT1 | CAGGCTGGCGAGAGATTCC | AGCATCCGTGAGTGCATCAA |
| AnAPX1 | GCTCCGTGAAGTAAGTGTTATCAAAC | CCTGGGAAGGTGCCACAA |
| AnPEPC | AAGCTGCTGGGTCTCTTCAT | AGATGTACAACGAGTGGCCA |
| Anactin | CCAGAGTCTAAGACGCAACC | CGTGAAAGAATGACCCAAAT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Petchphankul, N.; Ueda, A.; Saneoka, H. Differences in Physiological Responses of Two Oat (Avena nuda L.) Lines to Sodic-Alkalinity in the Vegetative Stage. Plants 2020, 9, 1188. https://doi.org/10.3390/plants9091188
Liu L, Petchphankul N, Ueda A, Saneoka H. Differences in Physiological Responses of Two Oat (Avena nuda L.) Lines to Sodic-Alkalinity in the Vegetative Stage. Plants. 2020; 9(9):1188. https://doi.org/10.3390/plants9091188
Chicago/Turabian StyleLiu, Liyun, Nateetorn Petchphankul, Akihiro Ueda, and Hirofumi Saneoka. 2020. "Differences in Physiological Responses of Two Oat (Avena nuda L.) Lines to Sodic-Alkalinity in the Vegetative Stage" Plants 9, no. 9: 1188. https://doi.org/10.3390/plants9091188
APA StyleLiu, L., Petchphankul, N., Ueda, A., & Saneoka, H. (2020). Differences in Physiological Responses of Two Oat (Avena nuda L.) Lines to Sodic-Alkalinity in the Vegetative Stage. Plants, 9(9), 1188. https://doi.org/10.3390/plants9091188







