Silicon in Horticultural Crops: Cross-talk, Signaling, and Tolerance Mechanism under Salinity Stress
Abstract
1. Introduction
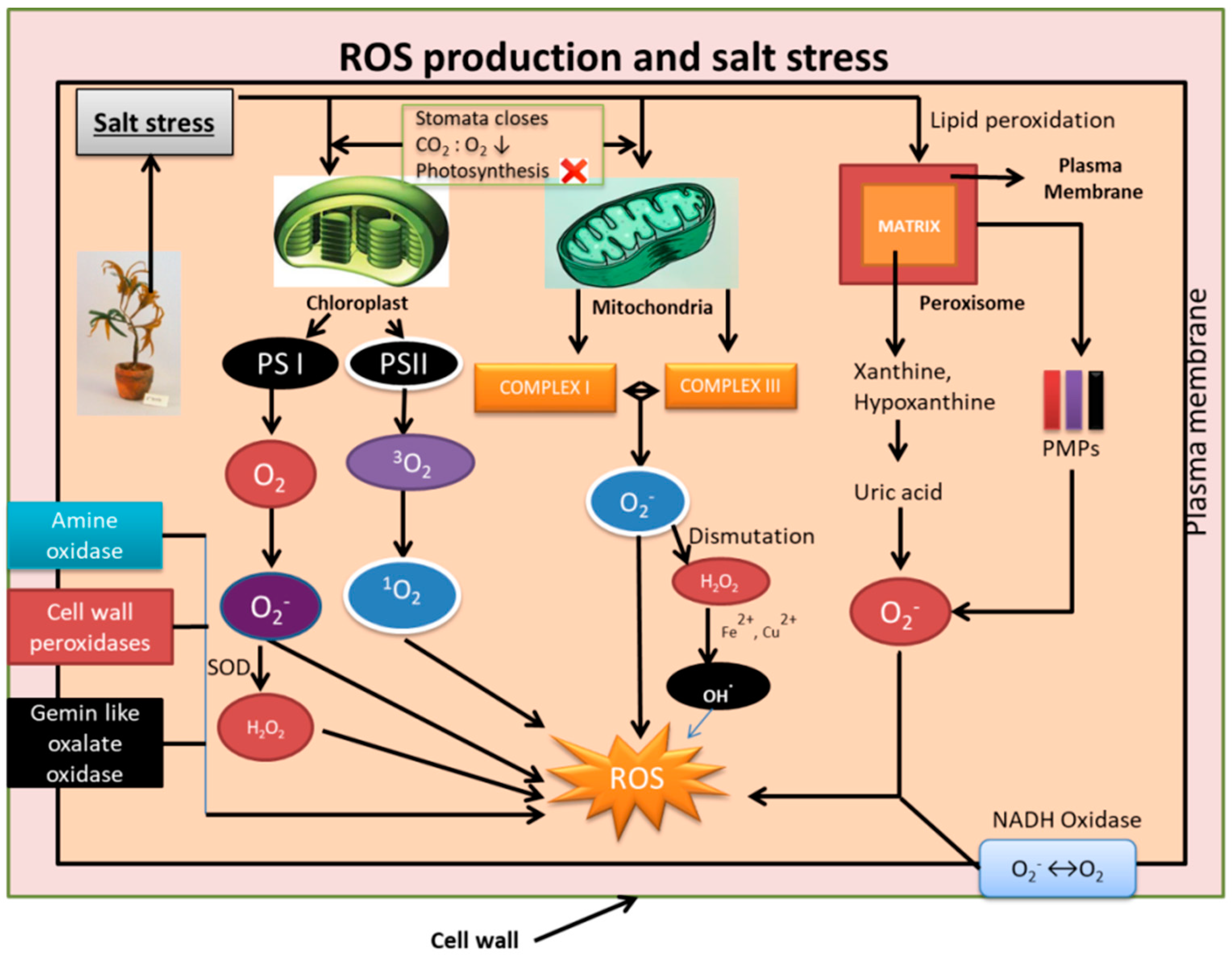
2. Reactive Oxygen Species (ROS) and their Production under Salinity Stress
2.1. In Chloroplasts
2.2. In Mitochondria
2.3. In Peroxisomes
3. ROS Detoxification: A Response towards Salt Stress Tolerance in Plants
3.1. In Chloroplasts
3.2. In Mitochondria
3.3. In Peroxisomes
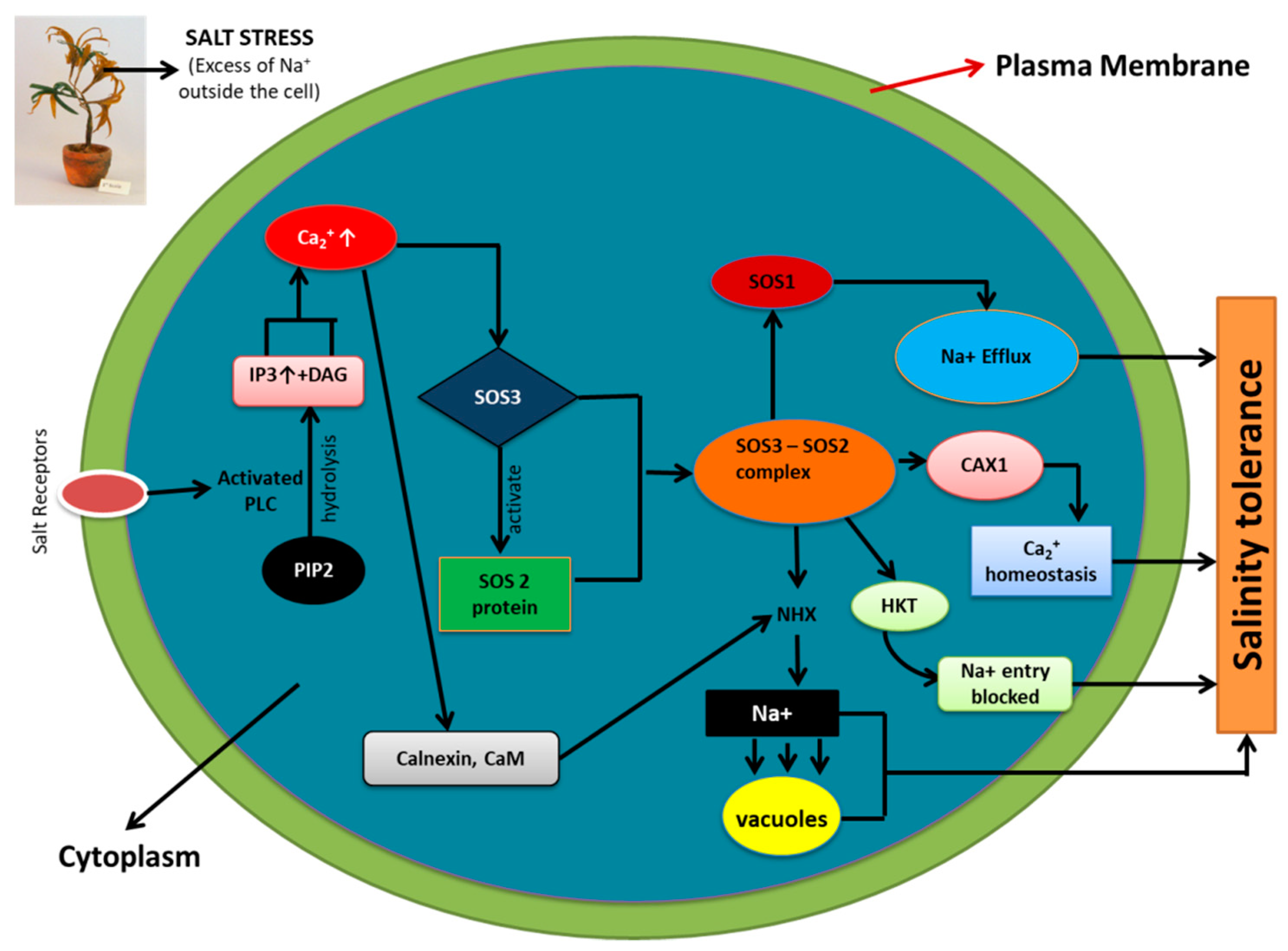
4. Role of Ion Pumps, Calcium, and SOS Pathway in Maintaining Ion Homeostasis during Salinity Stress
5. Role of Phytohormones and Transcription Factors during Salinity Stress
6. Role of Osmoprotectant Osmolytes during Salinity Stress
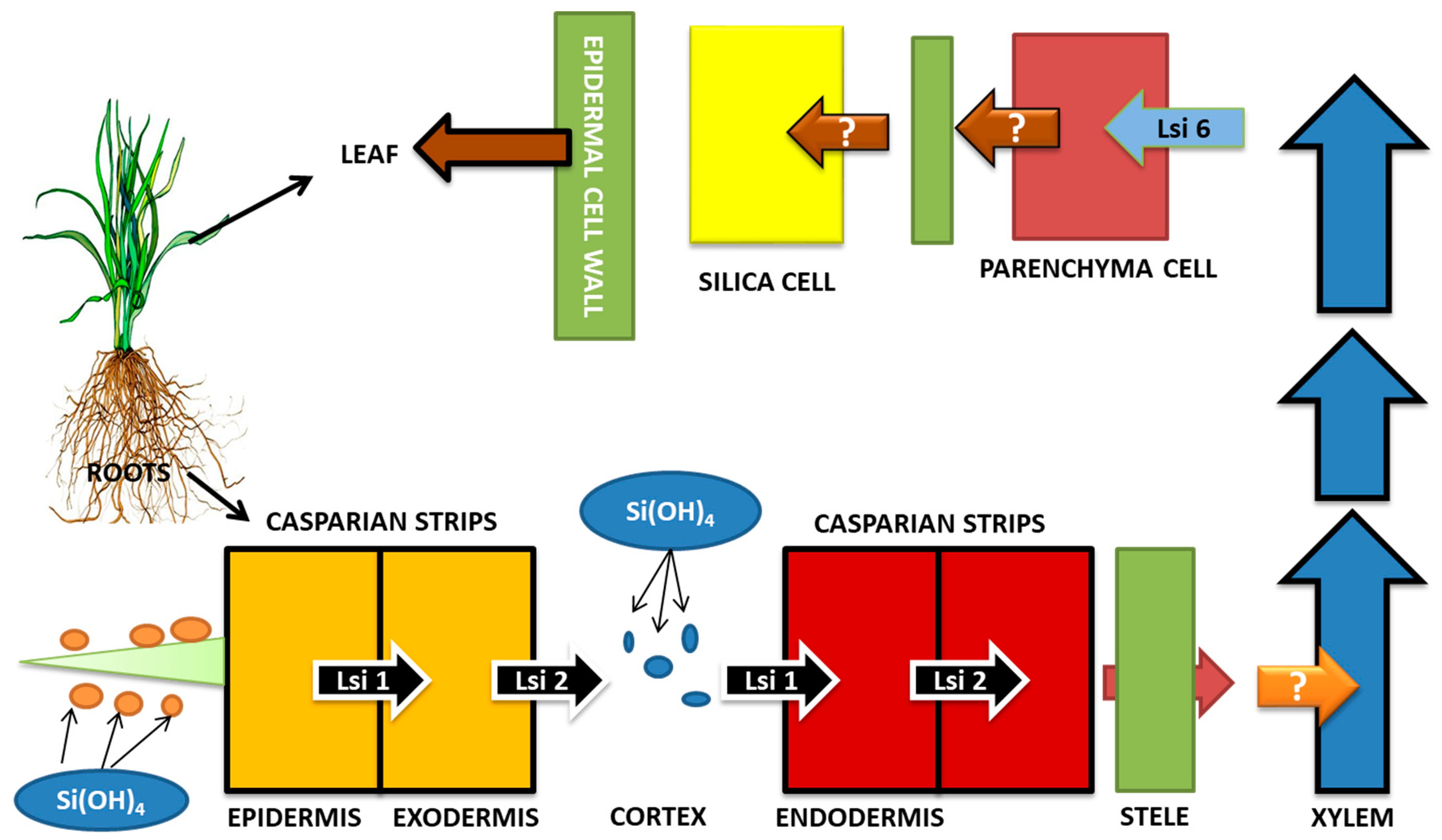
7. Si Uptake, Transport, and Accumulation
8. Si-Mediated Regulation of ROS
9. Si-Mediated Na+ and K+ Homeostasis
10. Si-Mediated Biosynthesis of Compatible Solutes and Phytohormone
11. Si Efficiency in Salinity-Stressed Horticultural Crops Employing Proteomic Approaches
12. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hu, Y.; Schmidhalter, U. Limitation of salt stress to plant growth. In Plant Toxicology; Hock, B., Elstner, C.F., Eds.; Marcel Dekker Inc.: New York, NY, USA, 2002; pp. 91–224. [Google Scholar]
- Rasool, S.; Hameed, A.; Azooz, M.M.; Muneeb-u-Rehman; Siddiqi, T.O.; Ahmad, P. Salt stress: Causes, types and responses of plants. In Ecophysiol and Responses of Plants under Salt Stress; Ahmad, P., Azooz, M.M., Prasad, M.N.V., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–24. [Google Scholar]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene expression profiling of plants under salt stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Ouhibi, C.; Attia, H.; Rebah, F.; Msilini, N.; Chebbi, M.; Aarrouf, J.; Urban, L.; Lachaal, M. Salt stress mitigation by seed priming with UV-C in lettuce plants: Growth, antioxidant activity and phenolic compounds. Plant Physiol. Biochem. 2014, 83, 126–133. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Shah, F.; Wu, W. Soil and crop management strategies to ensure higher crop productivity within sustainable environments. Sustainability 2019, 11, 1485. [Google Scholar] [CrossRef]
- Ismail, A.; Takeda, S.; Nick, P. Life and death under salt stress: Same players, different timing? J. Exp. Bot. 2014, 65, 2963–2979. [Google Scholar] [CrossRef] [PubMed]
- Akbarimoghaddam, H.; Galavi, M.; Ghanbari, A.; Panjehkeh, N. Salinity effects on seed germination and seedling growth of bread wheat cultivars. Trakia J. Sci. 2011, 9, 43–50. [Google Scholar]
- Singh, K.N.; Chatrath, R. Salinity tolerance. In Application of Physiology in Wheat Breeding; Reynolds, M.P., Monasterio, J.I.O., McNab, A., Eds.; CIMMYT: Mexico City, Mexico, 2001; pp. 101–110. [Google Scholar]
- Netondo, G.W.; Onyango, J.C.; Beck, E. Sorghum and salinity: II. Gas exchange and chlorophyll fluorescence of sorghum under salt stress. Crop Sci. 2004, 44, 806–811. [Google Scholar] [CrossRef]
- Ashraf, M. Some important physiological selection criteria for salt tolerance in plants. Flora 2004, 199, 361–376. [Google Scholar] [CrossRef]
- Golldack, D.; Lüking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef]
- Rehman, S.; Abbas, G.; Shahid, M.; Saqib, M.; Farooq, A.B.U.; Hussain, M.; Murtaza, B.; Amjad, M.; Naeem, M.A.; Farooq, A. Effect of salinity on cadmium tolerance, ionic homeostasis and oxidative stress responses in conocarpus exposed to cadmium stress: Implications for phytoremediation. Ecotoxicol. Environ. Saf. 2019, 171, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.B.; Kim, Y.H.; Lee, H.S.; Kim, K.Y.; Deng, X.P.; Kwak, S.S. Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses. Plant Physiol. Biochem. 2009, 47, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Tsugane, K.; Kobayashi, K.; Niwa, Y.; Ohba, Y.; Wada, K.; Kobayashi, H. A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 1999, 11, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Lakkineni, K.; Zhang, Z.; Verma, D.P.S. Removal of feedback inhibition of D1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 2000, 122, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Assaha, D.V.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef] [PubMed]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Malhotra, CH.; Kapoor, R.; Ganjewala, D. Alleviation of abiotic and biotic stresses in plants by silicon supplementation. Sci. Agric. 2016, 13, 59–73. [Google Scholar]
- Ibrahim, M.; Merwad, A.; Elnaka, E.; Burras, C.; Follett, L. Application of silicon ameliorated salinity stress and improved wheat yield. J. Soil Sci. Environ. Manag. 2016, 7, 81. [Google Scholar]
- Manchanda, G.; Garg, N. Salinity and its effects on the functional biology of legumes. Acta Physiol. Plant. 2008, 30, 595–618. [Google Scholar] [CrossRef]
- Sommer, M.; Kaczorek, D.; Kuzyakov, Y.; Breuer, T. Silicon pools and fluxes in soils and landscapes: A review. J. Plant Nutr. Soil Sci. 2006, 169, 310–329. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Gong, H.J. Beneficial effects of silicon on salt and drought tolerance in plants. Agron. Sustain. Dev. 2014, 34, 455–472. [Google Scholar] [CrossRef]
- Jones, L.; Handreck, K. Silica in soils, plants, and animals. In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 1967; pp. 107–149. [Google Scholar]
- Ma, J.F.; Takahashi, E. Soil, Fertilizer, and Plant Silicon Research in Japan; Elsevier: Amsterdam, The Netherlands, 2002. [Google Scholar]
- Epstein, E.; Bloom, A.J. Mineral Nutrition of Plants: Principles and Persperctives, 2nd ed.; Sinauer Associates Inc.: Sunderland, UK, 2005. [Google Scholar]
- Ma, J.F.; Yamaji, N. Functions and transport of silicon in plants. Cell. Mol. Life Sci. 2008, 65, 3049–3057. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.; Suvarchala, V.; Rajesh, Y.B.R.D.; Srinivasa Prasad, M.; Padmakumari, A.P.; Voleti, S.R. Effects of silicon sources on its deposition, chlorophyll content, and disease and pest resistance in rice. Biol. Plant. 2006, 50, 713–716. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.C.; Flowers, T.J.; Gong, H.J. Silicon decreases chloride transport in rice (Oryza sativa L.) in saline conditions. J. Plant Physiol. 2013, 170, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Dragisic Maksimovic, J.; Mojovic, M.; Maksimovic, V.; Romheld, V.; Nikolic, M. Silicon ameliorates manganese toxicity in cucumber by decreasing hydroxyl radical accumulation in the leaf apoplast. J. Exp. Bot. 2012, 63, 2411–2420. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Chen, Q.; Liu, Q.; Zhang, W.; Ding, R. Exogenous silicon (Si) increases antioxidant enzyme activity and reduces lipid peroxidation in roots of salt-stressed barley (Hordeum vulgare L.). J. Plant Physiol. 2003, 160, 1157–1164. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Yin, C.C.; Ma, B.; Collinge, D.P.; Pogson, B.J.; He, S.J.; Xiong, Q. Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase mediated abscisic acid pathway. Plant Cell. 2015, 27, 1061–1081. [Google Scholar] [CrossRef]
- Ahmad, M.; Zahir, Z.A.; Nazli, F.; Akram, F.; Arshad, M.; Khalid, M. Effectiveness of halo-tolerant, auxin producing pseudomonas and Rhizobium strains to improve osmotic stress tolerance in mung bean (Vigna radiata L.). Braz. J. Microbiol. 2013, 44, 1341–1348. [Google Scholar] [CrossRef]
- Hashiguchi, A.; Ahsan, N.; Komatsu, S. Proteomics application of crops in the context of climatic changes. Food Res. Int. 2010, 43, 1803–1813. [Google Scholar] [CrossRef]
- Nam, M.H.; Huh, S.M.; Kim, K.M.; Park, W.J.; Seo, J.B.; Cho, K. Comparative proteomic analysis of early salt stress-responsive proteins in roots of SnRK2 transgenic rice. Proteome Sci. 2012, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Perez-Clemente, R.M.; Vives, V.; Zandalinas, S.I.; Lopez-Climent, M.F.; Munoz, V.; Cadenas, A. Biotechnological approaches to study plant responses to stress. Biomed Res. Int. 2013, 2013, 654120. [Google Scholar] [CrossRef] [PubMed]
- Barkla, B.J.; Vera-Estrella, R.; Raymond, C. Single-cell-type quantitative proteomic and ionomic analysis of epidermal bladder cells from the halophyte model plant Mesembryanthemum crystallinum to identify salt-responsive proteins. BMC Plant Biol. 2016, 16, 110. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.C. Effects of silicon on enzyme activity, and sodium, potassium and calcium concentration in barley under salt stress. Plant Soil 1999, 209, 217–224. [Google Scholar] [CrossRef]
- Yeo, A.R.; Flowers, S.A.; Rao, G.; Welfare, K.; Senanayke, N.; Flowers, J.F. Silicon reduces sodium uptake in rice (Oryza sativa L.) in saline conditions and this is accounted for by a reduction in the transpirational bypass flow. Plant Cell Environ. 1999, 22, 559–565. [Google Scholar] [CrossRef]
- Romero-Aranda, M.R.; Jurado, O.; Cuartero, J. Silicon alleviates the deleterious salt effect on tomato plant growth by improving plant water status. J. Plant Physiol. 2006, 163, 847–855. [Google Scholar] [CrossRef]
- Zhu, Z.; Wei, G.; Li, J.; Qian, Q.; Yu, J. Silicon alleviates salt stress and increases antioxidant enzymes activity in leaves of salt-stressed cucumber (Cucumis sativus L.). Plant Sci. 2004, 167, 527–533. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, M.; Li, F.; Lv, H.; Li, C.; Xia, G.A. A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat. Mol. Cell. Proteom. 2009, 8, 2676–2686. [Google Scholar] [CrossRef]
- Bandehagh, A.; Salekdeh, G.H.; Toorchi, M.; Mohammadi, A.; Komatsu, S. Comparative proteomic analysis of canola leaves under salinity stress. Proteomics 2011, 11, 1965–1975. [Google Scholar] [CrossRef]
- Wakeel, A.; Asif, A.R.; Pitann, B.; Schubert, S. Proteome analysis of sugar beet (Beta vulgaris L.) elucidates constitutive adaptation during the first phase of salt stress. J. Plant Physiol. 2011, 168, 519–526. [Google Scholar] [CrossRef]
- Sobhanian, H.; Razavizadeh, R.; Nanjo, Y.; Ehsanpour, A.A.; RastgarJazii, F.; Motamed, N.; Komatsu, S. Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Sci. 2010, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Srivastava, S.; Sarin, N.B.; Kav, N.N.V. Proteomics reveals elevated levels of PR10 proteins in saline-tolerant peanut (Arachis hypogaea) calli. Plant Physiol. Biochem. 2006, 44, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Swami, A.K.; Alam, S.I.; Sengupta, N.; Sarin, R. Differential proteomic analysis of salt response in Sorghum bicolor leaves. Environ. Exp. Bot. 2011, 71, 321–328. [Google Scholar] [CrossRef]
- Chen, S.; Gollop, N.; Heuer, B. Proteomic analysis of salt-stressed tomato (Solanumly copersicum) seedlings: Effect of genotype and exogenous application of glycine betaine. J. Exp. Bot. 2009, 60, 2005–2019. [Google Scholar] [CrossRef]
- Aghaei, K.; Ehsanpour, A.A.; Komatsu, S. Proteome analysis of potato under salt stress. J. Proteome Res. 2008, 7, 4858–4868. [Google Scholar] [CrossRef]
- Du, C.X.; Fan, H.F.; Guo, S.R.; Tezuka, T.; Li, J. Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 2010, 71, 1450–1459. [Google Scholar] [CrossRef]
- Dat, J.; Vandenabeele, S.; Vranová, E.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell Mol. Life Sci. 2000, 57, 779–795. [Google Scholar] [CrossRef]
- Fryer, M.J.; Oxborough, K.; Mullineaux, P.M.; Baker, N.R. Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 2002, 53, 1249–1254. [Google Scholar]
- Hideg, E.; Barta, C.; Kalai, T.; Vass, I.; Hideg, K.; Asada, K. Detection of singlet oxygen and superoxide with fluorescent sensors in leaves under stress by photo inhibition or UV radiation. Plant Cell Physiol. 2002, 43, 1154–1164. [Google Scholar] [CrossRef]
- Lechno, S.; Zamski, E.; Telor, E. Salt Stress-induced responses in cucumber plants. J. Plant Physiol. 1997, 1, 206–211. [Google Scholar] [CrossRef]
- Boveris, A.; Chance, B. Mitochondrial production of superoxide radical and hydrogenperoxide. In Tissue Hypoxia and Ischemia; Reivich, M., Coburn, R., Lahiri, S., Chance, B., Eds.; Plenum: New York, NY, USA, 1977; pp. 67–82. [Google Scholar]
- Takeshiga, K.; Minakami, S. NADH and NADPH dependent formation of superoxide anions by bovine heart sub mitochondrial particles and NADH-ubiquinone reductase preparation. Biochem. J. 1979, 180, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Beyer, R. An analysis of the role of coenzyme Q in free radical generation and as an antioxidant. Biochem. Cell Biol. 1992, 70, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Grene, R. Oxidative stress and acclimation mechanisms in plants. In The Arabidopsis Book; Somerville, C.R., Myerowitz, E.M., Eds.; American Society of Plant Biologists: Rockville, MD, USA, 2002. [Google Scholar]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- López-Huertas, E.; Corpas, F.J.; Sandalio, L.M.; del Río, L.A. Characterization of membrane polypeptides from pea leaf peroxisomes involved in superoxide radical generation. Biochem. J. 1999, 337, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.; Graham, I. Plant Peroxisomes: Biochemistry, Cell Biology and Biotechnological Applications; Kluwer: Dordrecht, The Netherlands, 2002. [Google Scholar]
- Hernández, J.A.; Ferrer, M.A.; Jiménez, A.; Barcelo, A.R.; Sevilla, F. Antioxidant systems andO2−/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 2001, 127, 817–831. [Google Scholar] [CrossRef]
- Bolwell, G.P.; Wojtaszek, P. Mechanisms for the generation of reactive oxygen species in plant defense—Broad perspective. Physiol. Mol. Plant Pathol. 1997, 51, 347–366. [Google Scholar] [CrossRef]
- Hu, X.; Bidney, D.L.; Yalpani, N.; Duvick, J.P.; Crasta, O.; Folkerts, O.; Lu, G.H. Overexpression of a gene encoding hydrogen peroxide-generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol. 2003, 133, 170–181. [Google Scholar] [CrossRef]
- Walters, D.R. Polyamines and plant disease. Phytochemistry 2003, 64, 97–107. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F.V. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Miyake, C.; Asada, K. Ferredoxin-dependent photoreduction of monodehydro ascorbate radicals in spinach thylakoids. Plant Cell Physiol. 1994, 35, 539–549. [Google Scholar] [CrossRef]
- Krasnovsky, A.A., Jr. Singlet molecular oxygen in photo biochemical systems: IR phosphorescence studies. Membr. Cell Biol. 1998, 12, 665–690. [Google Scholar]
- Buettner, G.R.; Jurkiewicz, B.A. Chemistry and biochemistry of ascorbic acid. In Handbook of Antioxidants; Cadenas, E., Packer, L., Eds.; Dekker: New York, NY, USA, 1996; pp. 91–115. [Google Scholar]
- Pang, C.H.; Zhang, S.J.; Gong, Z.Z.; Wang, B.S. NaCl treatment markedly enhances H2O2-scavenging system in leaves of halophyte Suaeda salsa. Physiol. Plant. 2005, 125, 490–499. [Google Scholar]
- Zhang, Q.F.; Li, Y.Y.; Pang, C.H.; Lu, C.M.; Wang, B.S. NaCl enhances thylakoid-bound SOD activity in the leaves of C3 halophyte Suaeda salsa L. Plant Sci. 2005, 168, 23–430. [Google Scholar]
- Badawi, G.H.; Kawano, N.; Yamauchi, Y.; Shimada, E.; Sasaki, R.; Kubo, A.; Tanaka, K. Overexpression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol. Plant. 2004, 121, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.Y.; Jeong, Y.J.; Lee, H.S.; Kim, J.S.; Cho, K.Y.; Allen, R.D.; Kwak, S.S. Enhanced tolerances of transgenic tobacco plants expressing both superoxide dismutase and ascorbate peroxidase in chloroplasts against methyl viologen-mediated oxidative stress. Plant Cell Environ. 2002, 25, 873–882. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Monde, R.A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.M. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 561–591. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; Hernández, J.A.; del Río, L.A.; Sevilla, F. Evidence for the presence of the ascorbateglutathionecycle in mitochondria and peroxisomes of pea leaves. Plant Physiol. 1997, 114, 275–284. [Google Scholar] [CrossRef]
- Chew, O.; Whelan, J.; Millar, A.H. Molecular definition of the ascorbate–glutathione cycle inArabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 2003, 278, 46869–46877. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Foyer, C.H. Roles for reactive oxygen species and antioxidants in plantmitochondria. In Plant Mitochondria: From Genome to Function; Advances in Photosynthesis and Respiration; Day, D.A., Millar, A.H., Whelan, J., Eds.; Kluwer: Dordrecht, The Netherlands, 2004; Volume 1, pp. 307–320. [Google Scholar]
- Purvis, A.C.; Shewfelt, R.L. Does the alternative pathway ameliorate chilling injury insensitive plant tissues? Physiol. Plant 1993, 88, 712–718. [Google Scholar] [CrossRef]
- Hourton-Cabassa, C.; Matos, A.R.; Zachowski, A.; Moreau, F. The plant uncoupling protein homologues: A new family of energy-dissipating proteins in plant mitochondria. Plant Physiol. Biochem. 2004, 42, 283–290. [Google Scholar] [CrossRef]
- Mittova, V.; Guy, M.; Tal, M.; Volokita, M. Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. J. Exp. Bot. 2004, 55, 1105–1113. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.A.; Jiménez, A.; Mullineaux, P.; Sevilla, F. Tolerance of pea (Pisum sativum L.) to long term salt stress is associated with induction of antioxidant defenses. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- Bowler, C.; Slooten, L.; Vandenbranden, S.; De Rycke, R.; Botterman, J. Manganese superoxidedismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO J. 1991, 10, 1723–1732. [Google Scholar] [CrossRef] [PubMed]
- del Río, L.A.; Pastori, G.M.; Palma, J.M.; Sandalio, L.M.; Sevilla, F.; Corpas, F.J.; Jiménez, A.; López-Huertas, E.; Hernández, J.A. The activated oxygen role of peroxisomes in senescence. Plant Physiol. 1998, 116, 1195–1200. [Google Scholar] [CrossRef]
- del Río, L.A.; Corpas, F.J.; Sandalio, L.M.; Palma, J.M.; Gómez, M.; Barroso, J.B. Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J. Exp. Bot. 2002, 53, 1255–1272. [Google Scholar] [CrossRef]
- Kużniak, E.; Skłodowska, M. Fungal pathogen-induced changes in the antioxidant systems of leaf peroxisomes from infected tomato plants. Planta 2005, 222, 192–200. [Google Scholar] [CrossRef]
- Corpas, F.J.; Pedrajas, J.R.; Sandalio, L.M.; León, A.M.; Carreras, A.; Palma, J.M.; Valderrama, R.; del Río, L.A.; Barroso, J.B. Localization of peroxiredoxin in peroxisomes from pea leaves. Free Radic. Res. 2003, 37, 19–20. [Google Scholar]
- Platten, D.J.; Cotsaftis, O.; Berthomieu, P.; Bohnert, H.; Davenport, R.J.; Fairbairn, D.J.; Horie, T.; Leigh, R.A.; Lin, H.X.; Luan, S.; et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 2006, 11, 372–374. [Google Scholar] [CrossRef]
- Mahajan, S.; Sopoy, S.K.; Tuteja, N. CBL-CIPK paradigm: Role in calcium and stress signaling in plants. Proc. Indian Natl. Sci. Acad. 2006, 72, 63–78. [Google Scholar]
- Zhang, F.; Liang, Y.C.; He, W.L.; Zhao, X.; Zhang, L.X. Effects of salinity on growth and compatible solutes of callus induced from Populus euphratica. In Vitro Cell. Dev. Biol. 2004, 40, 491–494. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Mechanisms of high salinity tolerance in plants. Methods Enzym. 2007, 428, 419–438. [Google Scholar]
- Wu, Y.; Lei, D.; Zhu, J.K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 1996, 8, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Park, HJ.; Kim, WY.; Yun, DJ. A new insight of salt stress signaling in plant. Mol. Cells. 2016, 39, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Schumaker, K.; Zhu, J.K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J. Exp. Bot. 2004, 55, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought and salt stress. Plant Cell 2002, 14, 165–183. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997, 115, 327–334. [Google Scholar] [CrossRef]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176. [Google Scholar] [CrossRef]
- Jayakannan, M.; Bose, J.; Babourina, O.; Rengel, Z.; Shabala, S. Salicylic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K+ loss via a GORK channel. J. Exp. Bot. 2013, 64, 2255–2268. [Google Scholar] [CrossRef]
- El-Mashad, A.A.A.; Mohamed, H.I. Brassinolide alleviates salt stress and increases antioxidant activity of cowpea plants (Vigna sinensis). Protoplasma 2012, 249, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotech. 2005, 16, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Harinasut, P.; Tsutsui, K.; Takabe, T.; Nomura, M.; Kishitani, S. Exogenous glycine beatin accumulation and increased salt tolerance in rice seedlings. Biosci. Biotech. Biochem. 1996, 60, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lu, C. Photosynthesis is improved by exogenous glycine-betain in salt-stressed maize plants. Physiol. Plant. 2005, 124, 343–352. [Google Scholar] [CrossRef]
- Delauney, A.J.; Verma, D.P.S. Proline biosynthesis and osmoregulation in plants. Plant J. 1993, 4, 215–223. [Google Scholar] [CrossRef]
- Kishor, P.B.K.; Hong, Z.; Miao, G.H.; Hu, C.A.A.; Verma, D.P.S. Overexpression of [delta]-pyrroline-5-carboxylate synthetase increases proline production and confers osmo-tolerance in transgenic plants. Plant Physiol. 1995, 108, 1387–1394. [Google Scholar] [CrossRef]
- Yan, H.; Gong, L.Z.; Zhao, C.Y.; Guo, W.Y. Effects of exogenous proline on the physiology of soybean plantlets regenerated from embryos in vitro and on the ultrastructure of their mitochondria under NaCl stress. Soybean Sci. 2000, 19, 314–319. [Google Scholar]
- Kurdali, F.; Al-Chammaa, M.; Al-Ain, F. Growth and N2fixation in Saline and/or Water Stressed Sesbania aculeata Plants in Response to Silicon Application. Silicon 2019, 11, 781–788. [Google Scholar] [CrossRef]
- Alexandre, A.; Meunier, J.D.; Colin, F.; Koud, J.M. Plant impact on the biogeochemical cycle of silicon and related weathering processes. Geochim. Cosmochim. Acta 1997, 61, 677–682. [Google Scholar] [CrossRef]
- Guntzer, F.; Keller, C.; Poulton, P.R.; McGrath, S.P.; Meunier, J.D. Long term removal of wheat straw decreases soil amorphous silica at Broadbalk, Rothamsted. Plant Soil 2012, 352, 173–184. [Google Scholar] [CrossRef]
- Rizwan, M.; Meunier, J.D.; Miche, H.; Keller, C. Effect of silicon on reducing cadmium toxicity in durum wheat (Triticum turgidum L. cv. Claudio W.) grown in a soil with aged contamination. J. Hazard. Mater. 2012, 30, 209–210, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Sauer, D.; Saccone, L.; Conley, D.J.; Herrmann, L.; Sommer, M. Review of methodologies for extracting plant-available and amorphous Si from soils and aquatic sediments. Biogeochemistry 2006, 80, 89–108. [Google Scholar] [CrossRef]
- Liang, Y.; Nikolic, M.; Bélanger, R.; Gong, H.; Song, A. Silicon in Agriculture: From Theory to Practice; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Raven, J.A. Silicon transport at the cell and tissue level. In Silicon in Agriculture; Datnoff, L.E., Snyder, G.H., Korndorfer, G.H., Eds.; Elsevier: Amsterdam, The Netherlands, 2001; pp. 41–55. [Google Scholar]
- Knight, C.T.G.; Kinrade, S.D. A primer on the aqueous chemistry of silicon. In Silicon in Agriculture, Studies in Plant Science; Datnoff, L.E., Snyder, G.H., Korndörfer, G.H., Eds.; Elsevier: Amsterdam, The Netherlands, 2001; Volume 8, pp. 57–84. [Google Scholar]
- Epstein, E. Silicon. Annu. Rev. Plant Biol. 1999, 50, 641–664. [Google Scholar] [CrossRef] [PubMed]
- Broadley, M.; Brown, P.; Cakmak, I.; Ma, J.F.; Rengel, Z.; Zhao, F. Beneficial elements. In Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Marschner, P., Ed.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 257–261. [Google Scholar]
- Ma, J.F.; Tamai, K.; Yamaji, N.; Mitani, N.; Konishi, S.; Katsuhara, M.; Ishiguro Murata, Y.; Yano, M. A silicon transporter in rice. Nature 2006, 440, 688–691. [Google Scholar] [CrossRef]
- Ma, J.F.; Miyake, Y.; Takahashi, E. Silicon as a beneficial element for crop plants. In Silicon in Agriculture; Datnoff, L.E., Snyder, G.H., Korndorfer, G.H., Eds.; Elsevier: Amsterdam, The Netherlands, 2001; pp. 17–39. [Google Scholar]
- Hodson, M.; White, P.; Mead, A.; Broadley, M. Phylogenetic variation in the silicon composition of plants. Ann. Bot. 2005, 96, 1027–1046. [Google Scholar] [CrossRef]
- Takahashi, E.; Ma, J.F.; Miyake, Y. The possibility of silicon as an essential element for higher plants. Comments Agric. Food Chem. 1990, 2, 99–122. [Google Scholar]
- Cornelis, J.T.; Delvauz, B.; Georg, R.B.; Lucas, Y.; Ranger, J.; Opfergelt, S. Tracing the origin of dissolved silicon transferred from various soil-plant systems towards rivers: A review. Biogeosciences 2011, 8, 89–112. [Google Scholar] [CrossRef]
- Ma, J.F.; Yamaji, N. Silicon uptake and accumulation in higher plants. Trends Plant Sci. 2006, 11, 392–397. [Google Scholar] [CrossRef]
- Chiba, Y.; Mitani, N.; Yamaji, N.; Ma, J.F. HvLsi1 is a silicon in flux transporter in barley. Plant J. 2009, 57, 810–818. [Google Scholar] [CrossRef]
- Mitani, N.; Chiba, Y.; Yamaji, N.; Ma, J.F. Identification and characterization of maize and barley Lsi-2-like silicon efflux transporters reveals a distinct silicon uptake system from that in rice. Plant Cell 2009, 21, 2133–2142. [Google Scholar] [CrossRef]
- Mitani, N.; Yamaji, N.; Ma, J.F. Identification of maize silicon influx transporters. Plant Cell Physiol. 2009, 50, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.K.; Vivancos, J.; Guérin, V.; Sonah, H.; Labbé, C.; Belzile, F.; Bélanger, R.R. Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol. Biol. 2013, 83, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, C.; Rémus-Borel, W.; Vivancos, J.; Labbé, C.; Belzile, F.; Bélanger, R.R. Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense. Plant J. 2012, 72, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Vivancos, J.; Deshmukh, R.; Grégoire, C.; Rémus-Borel, W.; Belzile, F.; Bélanger, R.R. Identification and characterization of silicon efflux transporters in horsetail (Equisetum arvense). J. Plant Physiol. 2016, 200, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Yamaji, N.; Mitani, M.; Tamai, K.; Konishi, S.; Fujiwara, T.; Katsuhara, M.; Yano, M. An efflux transporter of silicon in rice. Nature 2007, 448, 209–212. [Google Scholar] [CrossRef]
- Yamaji, N.; Mitatni, N.; Ma, J.F. A transporter regulating silicon distribution in rice shoots. Plant Cell 2008, 20, 1381–1389. [Google Scholar] [CrossRef]
- Ma, J.F.; Yamaji, N.; Mitani-Ueno, N. Transport of silicon from roots to panicles in plants. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 377–385. [Google Scholar] [CrossRef]
- Gao, X.; Zou, C.; Wang, L.; Zhang, F. Silicon improves water use deficiency in maize plants. J. Plant Nutr. 2005, 27, 1457–1470. [Google Scholar] [CrossRef]
- Savvas, D.; Ntatsi, G.G. Bio stimulant activity of silicon in horticulture. Sci. Hort. 2015, 196, 66–81. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Abbas, T.; Balal, R.M.; Shahid, M.A.; Pervez, M.A.; Ayyub, C.M.; Aqueel, M.A.; Javaid, M.M. Silicon-induced alleviation of NaCl toxicity in okra (Abelmoschus esculentus) is associated with enhanced photosynthesis, osmoprotectant and antioxidant metabolism. Acta Physiol. Plant. 2015, 37, 1–15. [Google Scholar] [CrossRef]
- Wang, X.; Wei, Z.; Liu, D.; Zhao, G. Effects of NaCl and silicon on activities of antioxidative enzymes in roots, shoots and leaves of alfalfa. Afr. J. Biotech. 2011, 10, 545–549. [Google Scholar]
- Muneer, S.; Jeong, B.R. Proteomic analysis of salt-stress responsive proteins in roots of tomato (Lycopersicon esculentum L.) plants towards silicon efficiency. Plant Growth Regul. 2015, 77, 1–14. [Google Scholar] [CrossRef]
- Li, H.; Zhu, Y.; Hu, Y.; Han, W.; Gong, H. Beneficial effects of silicon in alleviating salinity stress of tomato seedlings grown under sand culture. Acta Physiol. Plant. 2015, 37, 1–9. [Google Scholar] [CrossRef]
- Kim, Y.H.; Khan, A.L.; Waqas, M.; Shim, J.K.; Kim, D.H.; Lee, K.Y.; Lee, I.J. Silicon application to rice root zone influenced the phytohormonal and antioxidant responses under salinity stress. J. Plant Growth Regul. 2014, 33, 137–149. [Google Scholar] [CrossRef]
- Liu, P.; Yin, L.; Wang, S.; Zhang, M.; Deng, X.; Zhang, S.; Tanaka, K. Enhanced root hydraulic conductance by aquaporin regulation accounts for silicon alleviated salt-induced osmotic stress in Sorghum bicolor L. Environ. Exp. Bot. 2015, 111, 42–51. [Google Scholar] [CrossRef]
- Soylemezoglu, G.; Demir, K.; Inal, A.; Gunes, A. Effect of silicon on antioxidant and stomatal response of two grapevine (Vitis vinifera L.) rootstocks grown in boron toxic, saline and boron toxic-saline soil. Sci. Hort. 2009, 123, 240–246. [Google Scholar] [CrossRef]
- Tuna, A.L.; Kaya, C.; Higgs, D.; Murillo-Amador, B.; Aydemir, S.; Girgin, A.R. Silicon improves salinity tolerance in wheat plants. Environ. Exp. Bot. 2008, 62, 10–16. [Google Scholar] [CrossRef]
- Abdel-Haliem, M.E.F.; Hegazy, H.S.; Hassan, N.S.; Naguib, D.M. Effect of silica ions and nano silica on rice plants under salinity stress. Ecol. Eng. 2017, 99, 282–289. [Google Scholar] [CrossRef]
- Li, Y.T.; Zang, W.J.; Cui, J.J.; Lang, D.Y.; Li, M.; Zhao, Q.P.; Zhang, X.H. Silicon nutrition alleviates the lipid peroxidation and ion imbalance of Glycyrrhiza uralensis seedlings under salt stress. Acta Physiol. Plant. 2016, 38, 96–105. [Google Scholar] [CrossRef]
- Garg, N.; Bhandari, P. Silicon nutrition and mycorrhizal inoculations improve growth, nutrient status, K+/Na+ ratio and yield of Cicer arietinum L. genotypes under salinity stress. Plant Growth Regul. 2016, 78, 371–387. [Google Scholar] [CrossRef]
- Guerrier, G. Fluxes of Na+, K+ and Cl−, and osmotic adjustment in Lycopersicon pimpinellifolium and L. esculentum during short- and long-term exposures to NaCl. Physiol. Plant. 1996, 97, 583–591. [Google Scholar] [CrossRef]
- Khan, M.A.; Ungar, I.A.; Showalter, A.M. Effects of sodium chloride treatments on growth and ion accumulation of the halophyte Haloxylon recurvum. Communication in soil sci. Plant Anal. 2000, 31, 2763–2774. [Google Scholar] [CrossRef]
- Wang, X.S.; Han, J.G. Effects of NaCl and silicon on ion distribution in the roots, shoots and leaves of two alfalfa cultivars with different salt tolerance. Soil Sci. Plant Nutr. 2007, 53, 278–285. [Google Scholar] [CrossRef]
- Abdalla, M.M. Impact of diatomite nutrition on two Trifolium alexandrinum cultivars differing in salinity tolerance. Int. J. Plant Physiol. Biochem. 2011, 3, 233–246. [Google Scholar]
- Ali, A.; Basra, S.M.; Ahmad, R.; Wahid, A. Optimizing silicon application to improve salinity tolerance in wheat. Soil Environ. 2009, 2, 136–144. [Google Scholar]
- Ali, A.; Basra, S.M.; Iqbal, J.; Hussain, S.; Subhani, M.N.; Sarwar, M.; Haji, A. Silicon mediated biochemical changes in wheat under salinized and non-salinized solution cultures. Afr. J. Biotech. 2012, 11, 606–615. [Google Scholar]
- Gurmani, A.R.; Bano, A.; Najeeb, U.; Zhang, J.; Khan, S.U.; Flowers, T.J. Exogenously applied silicate and abscisic acid ameliorates the growth of salinity stressed wheat (Triticum aestivum L.) seedlings through Na+ exclusion. Aust. J. Crop Sci. 2013, 7, 1123–1130. [Google Scholar]
- Liang, Y.C.; Ding, R.X. Influence of silicon on micro distribution of mineral ions in roots of salt-stressed barley as associated with salt tolerance in plants. Sci. China Earth Sci. 2002, 45, 298–308. [Google Scholar]
- Gong, H.J.; Randall, D.P.; Flowers, T.J. Silicon deposition in root reduces sodium uptake in rice (Oryza sativa L.) seedlings by reducing bypass flow. Plant Cell Environ. 2006, 29, 1970–1979. [Google Scholar] [CrossRef]
- Shahzad, M.; Zörb, C.; Geilfus, C.M.; Mühling, K.H. Apoplastic Na+ in Vicia faba leaves rises after short-term salt stress and is remedied by silicon. J. Agron. Crop Sci. 2013, 199, 161–170. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Matloob, A.; Khan, F.A.; Khaliq, A.; Saud, S.; Huang, J. Phytohormones and plant responses to salinity stress: A review. Plant Growth Regul. 2015, 75, 391–404. [Google Scholar] [CrossRef]
- Tahir, M.A.; Aziz, T.; Farooq, M.; Sarwar, G. Silicon-induced changes in growth, ionic composition, water relations, chlorophyll contents and membrane permeability in two salt-stressed wheat genotypes. Arch. Agron. Soil Sci. 2012, 58, 247–256. [Google Scholar] [CrossRef]
- Rohanipoor, A.; Norouzi, M.; Moezzi, A.; Hassibi, P. Effect of silicon on some physiological properties of maize (Zea mays) under salt stress. J. Biol. Environ. Sci. 2013, 7, 71–79. [Google Scholar]
- Yin, L.; Wang, S.; Li, J.; Tanaka, K.; Oka, M. Application of silicon improves salt tolerance through ameliorating osmotic and ionic stresses in the seedling of Sorghum bicolor. Acta Physiol. Plant. 2013, 35, 3099–3107. [Google Scholar] [CrossRef]
- Esmaeili, S.; Salehi, H.; Eshghi, S. Silicon ameliorates the adverse effects of salinity on turf grass growth and development. J. Plant Nutr. 2015, 12, 1885–1901. [Google Scholar] [CrossRef]
- Chen, D.; Yin, L.; Deng, X.; Wang, S. Silicon increases salt tolerance by influencing the two-phase growth response to salinity in wheat (Triticum aestivum L.). Acta Physiol. Plant. 2014, 36, 2531–2535. [Google Scholar] [CrossRef]
- Yue, Y.; Zhang, M.; Zhang, J.C.; Duan, L.S.; Li, Z.H. SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J. Plant Physiol. 2012, 169, 255–261. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ exchanger. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef]
- Liang, Y.C.; Zhang, W.H.; Chen, Q.; Ding, R.X. Effects of silicon on H+-ATPase and H+-PPase activity, fatty acid composition and fluidity of tonoplast vesicles from roots of salt-stressed barley (Hordeum vulgare L.). Environ. Exp. Bot. 2005, 53, 29–37. [Google Scholar] [CrossRef]
- Mali, M.; Aery, N.C. Influence of silicon on growth, relative water contents and uptake of silicon, calcium and potassium in wheat grown in nutrient solution. J. Plant Nutr. 2008, 31, 1867–1876. [Google Scholar] [CrossRef]
- Gzik, A. Accumulation of proline and pattern of α-amino acids in sugar beet plants in response to osmotic, water and salt stress. Environ. Exp. Bot. 1997, 36, 29–38. [Google Scholar] [CrossRef]
- Mansour, M.M.F. Protection of plasma membrane of onion epidermal cells by glycine betaine and proline against NaCl stress. Plant Physiol. Biochem. 1998, 36, 767–772. [Google Scholar] [CrossRef]
- Kumar, A.P.; Bandhu, A.D. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar]
- Balibrea, M.E.; Rus-alvarez, A.M.; Bolarfn, M.C.; Pérez-alfocea, F. Fast changes in soluble carbohydrates and proline contents in tomato seedlings in response to ionic and non-ionic iso-osmotic stresses. J. Plant Physiol. 1997, 151, 221–226. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- An, Y.Y.; Liang, Z.S. Drought tolerance of Periploca sepium during seed germination: Antioxidant defense and compatible solutes accumulation. Acta Physiol. Plant. 2013, 35, 959–967. [Google Scholar] [CrossRef]
- Seckin, B.; Sekmen, A.H.; Türkan, İ. An enhancing effect of exogenous mannitol on the antioxidant enzyme activities in roots of wheat under salt stress. J. Plant Growth Regul. 2009, 28, 12–20. [Google Scholar] [CrossRef]
- Flowers, T.J.; Hajibagueri, M.A.; Clipson, N.C.W. Halophytes. Q. Rev. Plant Biol. 1986, 61, 313–337. [Google Scholar] [CrossRef]
- Watanabe, S.; Kojima, K.; Ide, Y.; Sasaki, S. Effects of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro. Plant Cell Tissue Organ 2000, 63, 199–206. [Google Scholar] [CrossRef]
- Lee, S.K.; Sohn, E.Y.; Hamayun, M.; Yoon, J.Y.; Lee, I.J. Effect of silicon on growth and salinity stress of soybean plant grown under hydroponic system. Agrofor. Syst. 2010, 80, 333–340. [Google Scholar] [CrossRef]
- Gunes, A.; Inal, A.; Bagci, E.G.; Coban, S. Silicon-mediated changes on some physiological and enzymatic parameters symptomatic of oxidative stress in barley grown in sodic-B toxic soil. J. Plant Physiol. 2007, 164, 807–811. [Google Scholar] [CrossRef]
- Dodd, I.C.; Davies, W.J. Hormones and the regulation of water balance. In Plant Hormones: Biosynthesis, Signal Transduction, Action, 3rd ed.; Davies, P.J., Ed.; Kluwer: Dordrecht, The Netherlands, 2004; pp. 519–548. [Google Scholar]
- Karmoker, J.L.; Von Steveninck, R.F.M. The effect of abscisic acid on the uptake and distribution of ions in intact seedlings of Phaseolus vulgaris cv. Red land Pioneer. Physiol. Plant. 1979, 45, 453–459. [Google Scholar] [CrossRef]
- Chakrabarti, N.; Mukherji, S. Effect of phytohormone pretreatment on nitrogen metabolism in Vigna radiata under salt stress. Biol. Plant. 2003, 46, 63–66. [Google Scholar] [CrossRef]
- Kim, Y.H.; Khan, A.L.; Kim, D.H.; Lee, S.Y.; Kim, K.M.; Waqas, M.; Jung, H.Y.; Shin, J.H.; Kim, J.G.; Lee, I.J. Silicon mitigates heavy metal stress by regulating P-type heavy metal ATPases, Oryza sativa low silicon genes, and endogenous phytohormones. BMC Plant Boil. 2014, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Song, L.; Huang, Z.; Yang, Y.; Wang, S.; Wang, Z.; Tong, J.; Gu, W.; Ma, H.; Xiao, L. Comparative proteomic analysis reveals molecular mechanism of seedling roots of different salt tolerant soybean genotypes in response to salinity stress. EuPA Open Proteom. 2014, 4, 40–57. [Google Scholar] [CrossRef]
- Pandey, A.; Choudhary, M.K.; Bhushan, D.; Chattopadhyay, A.; Chakraborty, S.; Datta, A.; Chakroborty, N. The nuclear proteome of chick pea (Cicer arietinum L.) reveals predicted and un-expected proteins. J. Proteome Res. 2006, 5, 3301–3311. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, D.; Pandey, A.; Choudhary, M.; Datta, A.; Chakraborty, S.; Chakraborty, N. Comparative proteomic analysis of differentially expressed proteins in Chickpea extracellular matrix during dehydration stress. Mol. Cell. Proteom. 2006, 6, 1868–1884. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 85–212. [Google Scholar] [CrossRef]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Gene Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, P.; Manivannan, A.; Ko, C.H.; Jeong, B.R. Silicon enhanced redox homeostasis and protein expression to mitigate the salinity stress in Rosa hybrida ‘Rock Fire’. J. Plant Growth Regul. 2017, 37, 1–19. [Google Scholar] [CrossRef]
- Nwugo, C.C.; Huerta, A.J. The effect of silicon on the leaf proteome of rice (Oryza sativa L.) Plants under Cadmium-Stress. J. Proteome Res. 2011, 10, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, A.; Soundararajan, P.; Muneer, S.; Ko, C.H.; Jeong, B.R. Silicon mitigates salinity stress by regulating the physiology, antioxidant enzyme activities, and Protein expression in Capsicum annuum ‘Bugwang’. BioMed Res. Int. 2016, 14, 3076357. [Google Scholar]
- Luo, S.; Ishida, H.; Makino, A.; Mae, T. Fe2+-catalyzed site-specific cleavage of the large subunit of ribulose 1,5- bisphosphate carboxylase close to the active site. J. Biol. Chem. 2002, 277, 12382–12387. [Google Scholar] [CrossRef]
- Dong, C.H.; Hu, X.; Tang, W. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 2006, 26, 9533–9543. [Google Scholar] [CrossRef]
| Serial No. | Accession Number | Protein Name | Biological Function | Plant Species | Theo./Exp. pI | Sequence Coverage (%) | Author |
|---|---|---|---|---|---|---|---|
| 1. | Q6K3C7 | Os02g0282000 protein | Defense response | Lycopersicon esculentum L. | 7.86/3.6 | 17 | Muneer and Jeong (2015) |
| 2. | B9IFL3 | COPINE 1 family protein | Defense response | Lycopersicon esculentum L. | 5.54/5.3 | 24 | Muneer and Jeong (2015) |
| 3. | G7IZ85 | Zinc finger A20 and AN1 domain containing stress-associated protein | Stress response | Lycopersicon esculentum L. | 6.28/5.9 | 55 | Muneer and Jeong (2015) |
| 4. | A2TDB3 | Caffeoyl-CoA O-methyltransferase | Stress response | Lycopersicon esculentum L. | 7.86/3.6 | 21 | Muneer and Jeong (2015) |
| 5. | B0JEM1 | NBS-LRR disease resistance protein | Defense response | Lycopersicon esculentum L. | 6.5/4.6 | 12 | Muneer and Jeong (2015) |
| 6. | Q5DUH6 | Pathogenesis-related protein 10 | Defense response | Lycopersicon esculentum L. | 5.1/5.8 | 21 | Muneer and Jeong (2015) |
| 7. | M1C4D6 | Transcription elongation factor | Transcriptional regulation | Lycopersicon esculentum L. | 5.65/5.67 | 56 | Muneer and Jeong (2015) |
| 8. | B9HUZ8 | Transcription elongation factor | Transcriptional Regulation | Lycopersicon esculentum L. | 5.66/6.5 | 56 | Muneer and Jeong (2015) |
| 9. | A9PK54 | Transcription elongation factor SPT4 homolog | Transcriptional Regulation | Lycopersicon esculentum L. | 5.66/5.6 | 56 | Muneer and Jeong (2015) |
| 10. | A9PK54 | Transcription elongation factor SPT4 homolog | Transcriptional Regulation | Lycopersicon esculentum L. | 5.66/5.8 | 56 | Muneer and Jeong (2015) |
| 11. | A9PK54 | Transcription elongation factor SPT4 homolog | Transcriptional Regulation | Lycopersicon esculentum L. | 5.66/6.0 | 56 | Muneer and Jeong (2015) |
| 12. | A9PK54 | Transcription elongation factor SPT4 homolog | Transcriptional Regulation | Lycopersicon esculentum L. | 5.66/6.6 | 56 | Muneer and Jeong (2015) |
| 13. | Q75HP9 | Potassium channel AKT2 | ABA response | Lycopersicon esculentum L. | 6.64/7.0 | 21 | Muneer and Jeong (2015) |
| 14. | H9BAN2 | AKT2/3-like potassium channel | ABA response | Lycopersicon esculentum L. | 4.9/4.5 | 13 | Muneer and Jeong (2015) |
| 15. | B2G4V8 | Gibberellin 20-oxidase | GA mediated signaling | Lycopersicon esculentum L. | 5.97/6.5 | 32 | Muneer and Jeong (2015) |
| 16. | P19312 | Ribulose bisphosphate carboxylase small chain SSU5B | Photosynthesis | Rosa hybrida ‘Rock Fire’ | 7.60/6.50 | 28 | Soundararajan et al. (2017) |
| 17. | A7M975 | Photosystem I assembly protein Ycf4 | Photosynthesis | Rosa hybrida ‘Rock Fire’ | 9.59/4.10 | 28 | Soundararajan et al. (2017) |
| 18. | Q7XKV5 | β-glucosidase 11 | Energy metabolism | Rosa hybrida ‘Rock Fire’ | 7.21/5.90 | 19 | Soundararajan et al. (2017) |
| 19. | Q9SCV4 | β -galactosidase 8 | Energy metabolism | Rosa hybrida ‘Rock Fire’ | 8.09/5.10 | 9 | Soundararajan et al. (2017)) |
| 20. | P12300 | Glucose-1-phosphate adenylyltransferase large subunit | Energy metabolism | Rosa hybrida ‘Rock Fire’ | 6.61/6.70 | 16 | Soundararajan et al. (2017) |
| 21. | P85438 | Acetyl-CoA carboxylase | Energy metabolism | Rosa hybrida ‘Rock Fire’ | 9.99/4.10 | 100 | Soundararajan et al. (2017) |
| 22. | P85438 | Acetyl-CoA carboxylase | Energy metabolism | Rosa hybrida ‘Rock Fire’ | 9.99/5.10 | 96 | Soundararajan et al. (2017) |
| 23. | Q8H2J9 | Glycerol-3-phosphate dehydrogenase (NAD+) | Energy metabolism | Rosa hybrida ‘Rock Fire’ | 9.76/6.80 | 22 | Soundararajan et al. (2017) |
| 24 | A2YMU2 | Ribosome-recycling factor | Transcription/translation | Rosa hybrida ‘Rock Fire’ | 9.35/5.10 | 23 | Soundararajan et al. (2017) |
| 25. | Q32RJ9 | tRNA(Ile)-lysidine synthase | Transcription/translation | Rosa hybrida ‘Rock Fire’ | 9.55/5.87 | 13 | Soundararajan et al. (2017) |
| 26. | Q9FZ48 | Ubiquitin-conjugating enzyme E2 8 | Ubiquitination | Rosa hybrida ‘Rock Fire’ | 6.74/4.47 | 59 | Soundararajan et al. (2017) |
| 27. | P35131 | Ubiquitin-conjugating enzyme E2 36 | Ubiquitination | Rosa hybrida ‘Rock Fire’ | 6.74/4.60 | 59 | Soundararajan et al. (2017) |
| 28. | XP 004249273 | Adenylosuccinate synthetase | Purine metabolism | Capsicum annuum ‘Bugwang’ | 7.5/4.2 | 25 | Manivannan et al. (2016) |
| 29. | XP 008793948 | E3 ubiquitin-protein ligase PUB23-like | Photo morphogenesis | Capsicum annuum ‘Bugwang’ | 8.2/4.1 | 20 | Manivannan et al. (2016) |
| 30. | AHL68475 | Ribulose-1,5-bisphosphate carboxylase/oxygenase, partial (chloroplast) | Carbon fixation | Capsicum annuum ‘Bugwang’ | 6.7/5.0 | 41 | Manivannan et al. (2016) |
| 31. | XP 009398204 | Oxygen-evolving enhancer protein 3-1, chloroplastic-like | Photosynthesis | Capsicum annuum ‘Bugwang’ | 9.5/5.1 | 52 | Manivannan et al. (2016) |
| 32. | XP 003058724 | Nucleoporin-like protein | Plant disease and hormone signaling | Capsicum annuum ‘Bugwang’ | 9.1/4.6 | 33 | Manivannan et al. (2016) |
| 33. | XP 004951624 | Mediator of RNA polymerase II transcription subunit 11-like | Transcription/translation | Capsicum annuum ‘Bugwang’ | 5.6/6.2 | 59 | Manivannan et al. (2016) |
| 34. | AFB70663 | Ribosomal protein L16, partial (chloroplastic) | Transcription/translation | Capsicum annuum ‘Bugwang’ | 11.8/4.7 | 63 | Manivannan et al. (2016) |
| 35. | AAR08850 | Resistance protein candidate | Transcription/translation | Capsicum annuum ‘Bugwang’ | 9.4/5.2 | 100 | Manivannan et al. (2016) |
| 36. | XP 010517956 | Molybdopterin synthase catalytic subunit-like | ABA synthesis | Capsicum annuum ‘Bugwang’ | 6.5/4.8 | 80 | Manivannan et al. (2016) |
| 37. | AAL83898 | Beta-keto acyl reductase | Fatty acid synthesis | Capsicum annuum ‘Bugwang’ | 11.6/6.9 | 87 | Manivannan et al. (2016) |
| 38. | BAB40826 | Reverse transcriptase | Transcription/translation | Capsicum annuum ‘Bugwang’ | 7.9/6.3 | 30 | Manivannan et al. (2016) |
| 39. | KHG25806 | Eukaryotic translation initiation factor 3 subunit D | Transcription/translation | Capsicum annuum ‘Bugwang’ | 8.9/4.4 | 26 | Manivannan et al. (2016) |
| 40. | XP 008677250 | MADS-box transcription factor 26 isoform X2 | Transcription/translation | Capsicum annuum ‘Bugwang’ | 8.8/5.4 | 42 | Manivannan et al. (2016) |
| 41. | CAC87838 | Cullin 1D | Ubiquitin-proteasome pathway | Capsicum annuum ‘Bugwang’ | 5.0/4.5 | 25 | Manivannan et al. (2016) |
| 42. | KIY92373 | Phosphoglycerate kinase, partial | Metabolic processes | Capsicum annuum ‘Bugwang’ | 8.7/4.4 | 51 | Manivannan et al. (2016) |
| 43. | AIF71068 | ATP synthase CF1 alpha subunit, partial (chloroplast) | Metabolic processes | Capsicum annuum ‘Bugwang’ | 8.6/5.8 | 49 | Manivannan et al. (2016) |
| 44. | XP 010046336 | Disease resistance protein RPS2-like | Metabolic processes | Capsicum annuum ‘Bugwang’ | 5.3/7.0 | 19 | Manivannan et al. (2016) |
| 45. | XP 012064817 | Double-stranded RNA-binding protein 2 | Metabolic processes | Capsicum annuum ‘Bugwang’ | 8.7/5.7 | 14 | Manivannan et al. (2016) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Murad, M.; Khan, A.L.; Muneer, S. Silicon in Horticultural Crops: Cross-talk, Signaling, and Tolerance Mechanism under Salinity Stress. Plants 2020, 9, 460. https://doi.org/10.3390/plants9040460
Al Murad M, Khan AL, Muneer S. Silicon in Horticultural Crops: Cross-talk, Signaling, and Tolerance Mechanism under Salinity Stress. Plants. 2020; 9(4):460. https://doi.org/10.3390/plants9040460
Chicago/Turabian StyleAl Murad, Musa, Abdul Latif Khan, and Sowbiya Muneer. 2020. "Silicon in Horticultural Crops: Cross-talk, Signaling, and Tolerance Mechanism under Salinity Stress" Plants 9, no. 4: 460. https://doi.org/10.3390/plants9040460
APA StyleAl Murad, M., Khan, A. L., & Muneer, S. (2020). Silicon in Horticultural Crops: Cross-talk, Signaling, and Tolerance Mechanism under Salinity Stress. Plants, 9(4), 460. https://doi.org/10.3390/plants9040460







