ACTIN7 Is Required for Perinuclear Clustering of Chloroplasts during Arabidopsis Protoplast Culture
Abstract
1. Introduction
2. Results
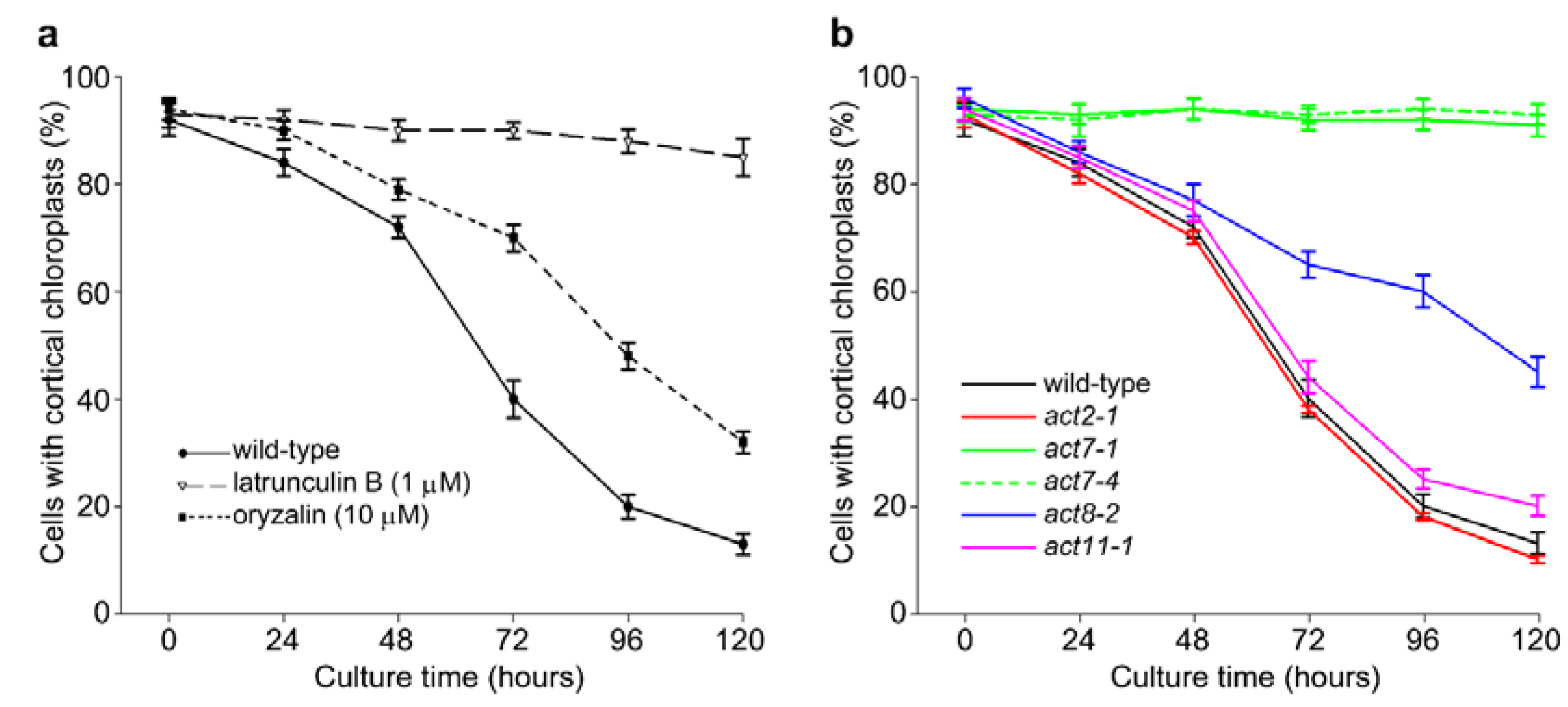
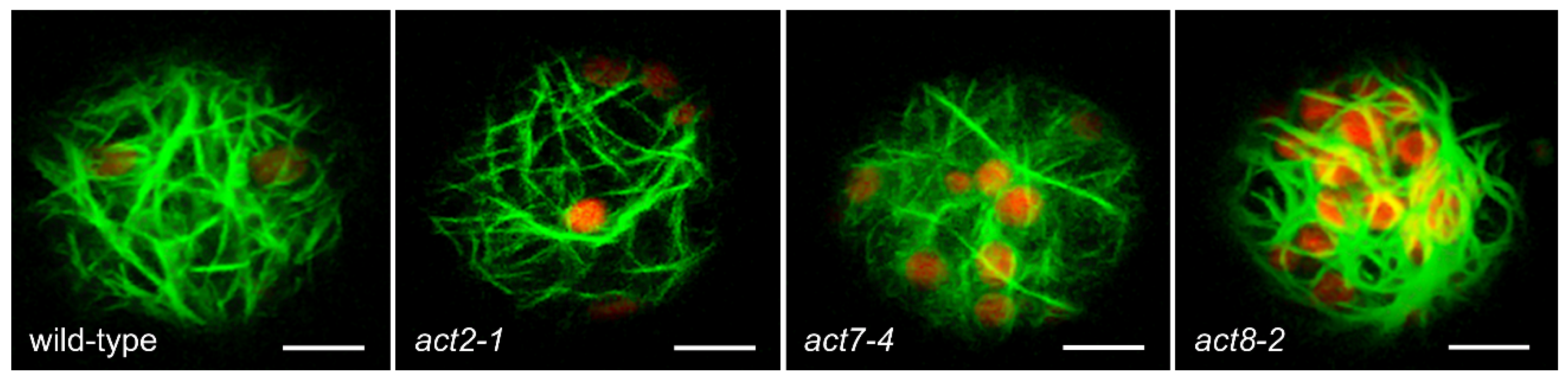
2.1. Actin-Dependent Clustering of Chloroplasts during Protoplast Culture
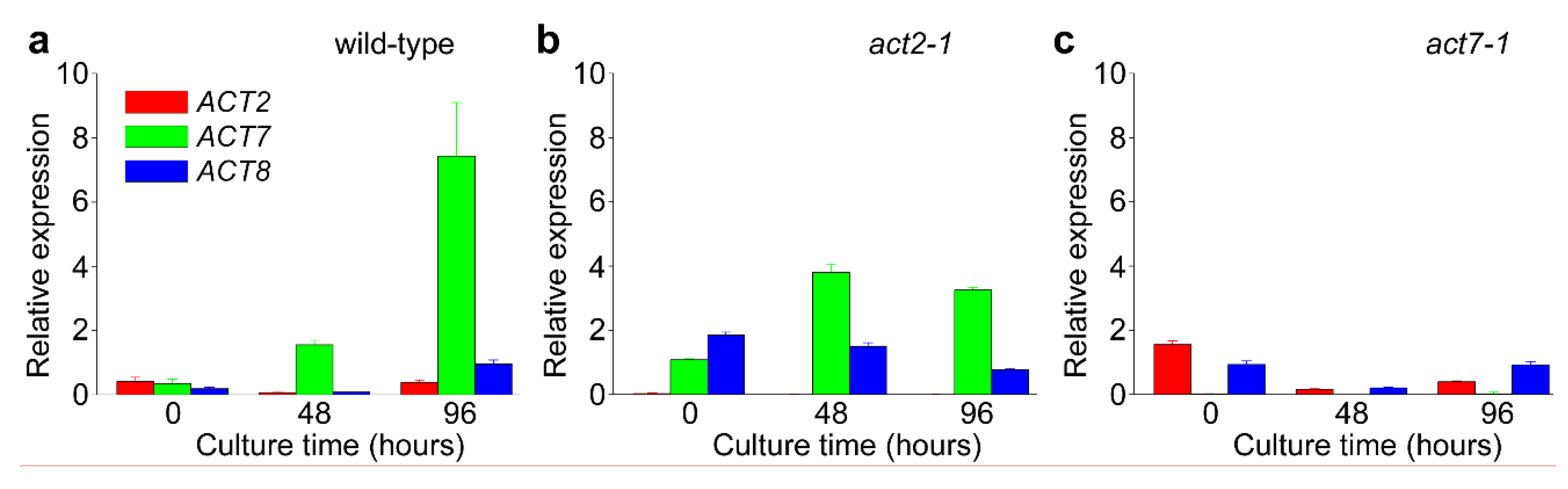
2.2. ACT7 Is Up-Regulated before Re-Initiation of Cell Division
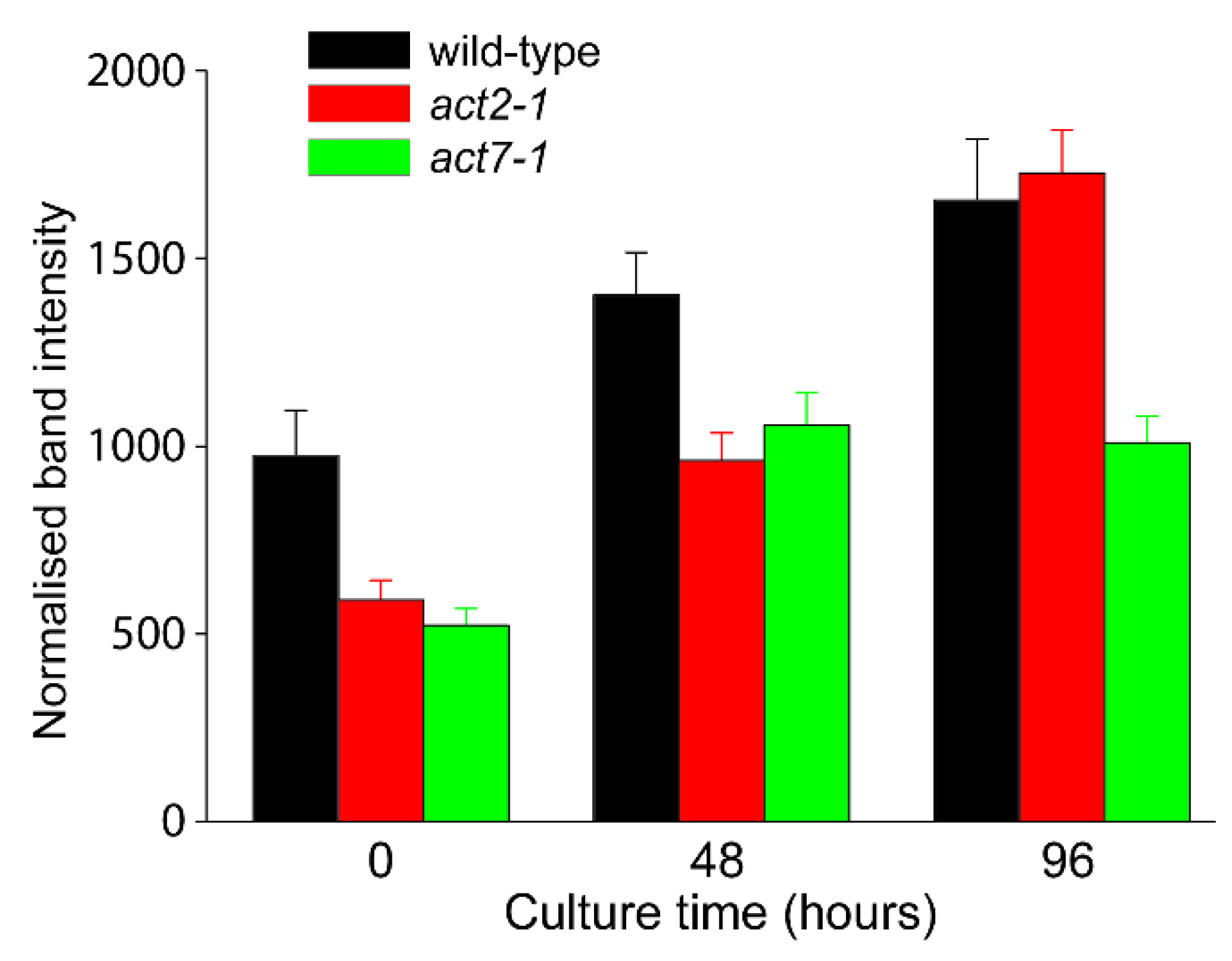
2.3. Analysis of Total Actin
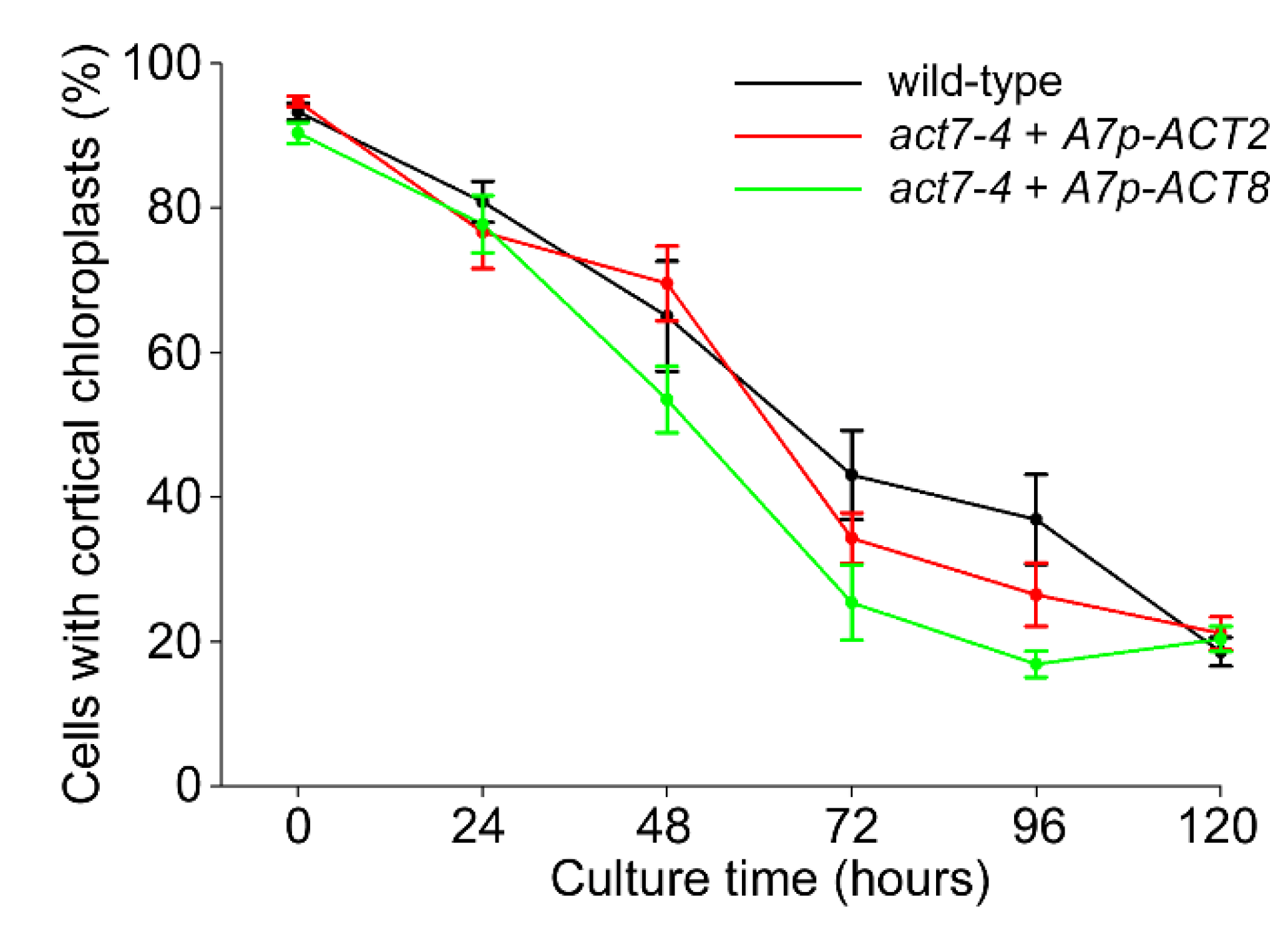
2.4. Over-Expression of Vegetative Actins Suppresses the Act7-4 Phenotype
3. Discussion
4. Materials and Methods
4.1. Plant Growth, Protoplast Isolation and Culture and Reagents
4.2. Microscopy
4.3. Reverse Transcription-Quantitative PCR (RT-qPCR)
4.4. Immunoblotting
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ng, R.; Abelson, J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1980, 77, 3912–3926. [Google Scholar] [CrossRef] [PubMed]
- McDowell, J.M.; Huang, S.; McKinney, E.C.; An, Y.-Q.; Meagher, R.B. Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 1996, 142, 587–602. [Google Scholar] [PubMed]
- Meagher, R.B.; McKinney, E.C.; Kandasamy, M.K. Isovariant dynamics expand and buffer the responses of complex systems: The diverse plant actin gene family. Plant Cell 1999, 11, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.K.; McKinney, E.C.; Roy, E.; Meagher, R.B. Plant vegetative and animal cytoplasmic actins share functional competence for spatial development with protists. Plant Cell 2012, 24, 2041–2057. [Google Scholar] [CrossRef][Green Version]
- Šlajcherová, K.; Fišerová, J.; Fischer, L.; Schwarzerová, K. Multiple actin isotypes in plants: Diverse genes for diverse roles? Front. Plant Sci. 2012, 3, 226. [Google Scholar] [CrossRef] [PubMed]
- Perrin, P.J.; Ervasti, J.M. The actin gene family: Function follows isoform. Cytoskeleton 2010, 67, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Röper, K.; Mao, Y.; Brown, N.H. Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. J. Cell Sci. 2005, 118, 3937–3948. [Google Scholar] [CrossRef] [PubMed]
- Brault, V.; Reedy, M.C.; Sauder, U.; Kammerer, R.A.; Aebi, U.; Schoenenberger, C.-A. Substitution of flight muscle-specific actin by human β-cytoplasmic actin in the indirect flight muscle of Drosophila. J. Cell Sci. 1999, 112, 3627–3639. [Google Scholar]
- Kandasamy, M.K.; Gilliland, L.U.; McKinney, E.C.; Meagher, R.B. One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 2001, 13, 1541–1554. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, L.U.; Pawloski, L.C.; Kandasamy, M.K.; Meagher, R.B. Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 2003, 33, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Claes, A.R.; Grebe, T.; Hermkes, R.; Viotti, C.; Ikeda, Y.; Grebe, M. Auxin and ROP GTPase signalling of polar nuclear migration in root epidermal hair cells. Plant Physiol. 2018, 176, 378–391. [Google Scholar] [CrossRef]
- Ringli, C.; Baumberger, N.; Diet, A.; Frey, B.; Keller, B. ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol. 2002, 129, 1464–1472. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 2009, 21, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Kijima, S.T.; Hirose, K.; Kong, S.G.; Wada, M.; Uyeda, T.Q.P. Distinct biochemical properties of Arabidopsis thaliana actin isoforms. Plant Cell Physiol. 2016, 57, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Kijima, S.T.; Staiger, C.J.; Katoh, K.; Nagasaki, A.; Ito, K.; Uyeda, T.Q.P. Arabidopsis vegetative actin isoforms, AtACT2 and AtACT7, generate distinct filament arrays in living plant cells. Sci. Rep. 2018, 8, 4381. [Google Scholar] [CrossRef]
- Thitamadee, S.; Tuchihara, K.; Hashimoto, T. Microtubule basis for left-handed helical growth in Arabidopsis. Nature 2002, 417, 193–196. [Google Scholar] [CrossRef]
- Ishida, T.; Kaneko, Y.; Iwano, M.; Hashimoto, T. Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 8544–8549. [Google Scholar] [CrossRef]
- Sunohara, H.; Kawai, T.; Shimizu-Sato, S.; Sato, Y.; Sato, K.; Kitano, H. A dominant mutation of TWISTED DWARF1 encoding an α-tubulin protein causes severe dwarfism and right helical growth in rice. Genes Gene Syst. 2009, 84, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Jöst, M.; Esfeld, K.; Burian, A.; Cannarozzi, G.; Chanyalew, S.; Kuhlemeier, C.; Assefa, K.; Tadele, Z. Semi-dwarfism and lodging tolerance in tef (Eragrostis tef) is linked to a mutation in the α-Tubulin 1 gene. J. Exp. Bot. 2015, 66, 933–944. [Google Scholar] [CrossRef]
- Vaškebová, L.; Šamaj, J.; Ovečka, M. Single-point ACT2 gene mutation in the Arabidopsis root hair mutant der1-3 affects overall actin organization, root growth and plant development. Ann. Bot. 2018, 122, 889–901. [Google Scholar] [CrossRef]
- Kato, T.; Morita, M.T.; Tasaka, M. Defects in dynamics and functions of actin filament in Arabidopsis caused by the dominant-negative actin fiz1-induced fragmentation of actin filament. Plant Cell Physiol. 2010, 51, 333–338. [Google Scholar] [CrossRef]
- Sheahan, M.B.; Rose, R.J.; McCurdy, D.W. Organelle inheritance in plant cell division: The actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 2004, 37, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, M.B.; McCurdy, D.W.; Rose, R.J. Mechanisms of organelle inheritance in dividing plant cells. In Molecular Cell Biology of the Growth and Differentiation of Plant Cells; Rose, R.J., Ed.; CRC Press: Boca Raton, FL, USA, 2016; pp. 66–88. [Google Scholar]
- Kandasamy, M.K.; Meagher, R.B. Actin-organelle interaction: Association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil. Cytoskel. 1999, 44, 110–118. [Google Scholar] [CrossRef]
- Sheahan, M.B.; Staiger, C.J.; Rose, R.J.; McCurdy, D.W. A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 2004, 136, 3968–3978. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, K.; Kasahara, M.; Kiyosue, T.; Kagawa, T.; Suetsugu, N.; Takahashi, F.; Kanegae, T.; Niwa, Y.; Kadota, A.; Wada, M. CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell 2003, 15, 2805–2815. [Google Scholar] [CrossRef]
- Gilliland, L.U.; Kandasamy, M.K.; Pawloski, L.C.; Meagher, R.B. Both vegetative and reproductive actin isovarients complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiol. 2002, 130, 2199–2209. [Google Scholar] [CrossRef] [PubMed]
- Lessard, J.L. Two monoclonal antibodies to actin: One muscle selective and one generally reactive. Cell Motil. Cytoskel. 1989, 10, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Kadota, A.; Yamada, N.; Suetsugu, N.; Hirose, M.; Saito, C.; Shoda, K.; Ichikawa, S.; Kagawa, T.; Nakano, A.; Wada, M. Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 13106–13111. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.-G.; Wada, M. New insights into dynamic actin-based chloroplast photorelocation movement. Mol. Plant 2011, 4, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Collings, D.A.; Lill, A.W.; Himmelspach, R.; Wasteneys, G.O. Hypersensitivity to cytoskeletal antagonists demonstrates microtubule–microfilament cross-talk in the control of root elongation in Arabidopsis thaliana. New Phytol. 2006, 170, 275–290. [Google Scholar] [CrossRef]
- Muller, P.Y.; Janovjak, H.; Miserez, A.R.; Dobbie, Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 2002, 32, 1372–1379. [Google Scholar] [PubMed]
- Simon, P. Q-Gene: Processing quantitative real-time RT-PCR data. Bioinformatics 2003, 19, 1439–1440. [Google Scholar] [CrossRef] [PubMed]
- Q-Gene software. Available online: http://download.gene-quantification.info/ (accessed on 5 January 2020).

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheahan, M.B.; Collings, D.A.; Rose, R.J.; McCurdy, D.W. ACTIN7 Is Required for Perinuclear Clustering of Chloroplasts during Arabidopsis Protoplast Culture. Plants 2020, 9, 225. https://doi.org/10.3390/plants9020225
Sheahan MB, Collings DA, Rose RJ, McCurdy DW. ACTIN7 Is Required for Perinuclear Clustering of Chloroplasts during Arabidopsis Protoplast Culture. Plants. 2020; 9(2):225. https://doi.org/10.3390/plants9020225
Chicago/Turabian StyleSheahan, Michael B., David A. Collings, Ray J. Rose, and David W. McCurdy. 2020. "ACTIN7 Is Required for Perinuclear Clustering of Chloroplasts during Arabidopsis Protoplast Culture" Plants 9, no. 2: 225. https://doi.org/10.3390/plants9020225
APA StyleSheahan, M. B., Collings, D. A., Rose, R. J., & McCurdy, D. W. (2020). ACTIN7 Is Required for Perinuclear Clustering of Chloroplasts during Arabidopsis Protoplast Culture. Plants, 9(2), 225. https://doi.org/10.3390/plants9020225






