Molecular and Morphological Divergence of Australian Wild Rice
Abstract
1. Introduction
2. Results
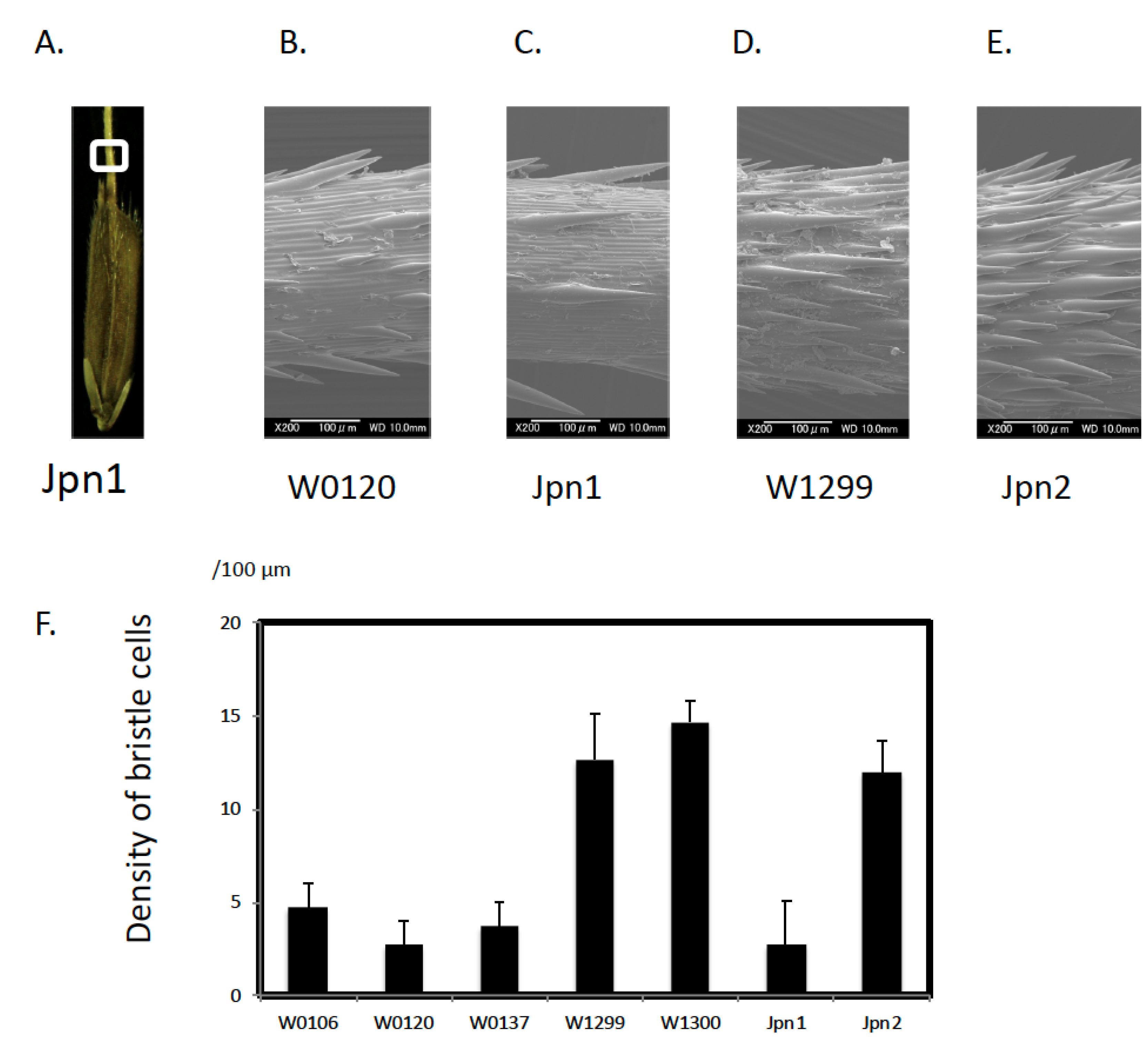
2.1. Morphological Features
2.2. Maternal Lineages
2.3. Reproductive Isolation
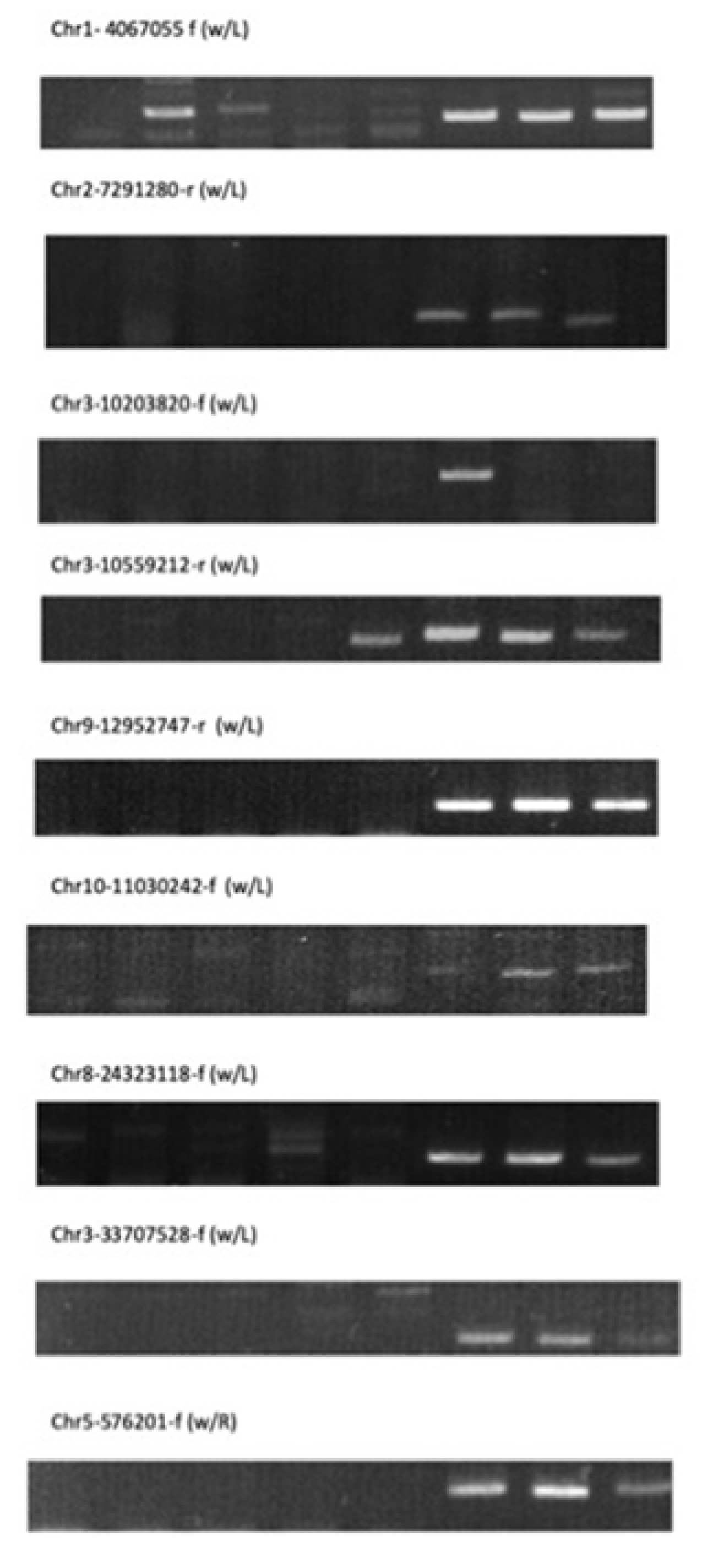
2.4. Unique Insertion of Retrotransposable Element in Jpn2
3. Discussion
3.1. Unique Morphological Traits in Australian Wild Rice
3.2. Maternal Variation
3.3. Reproductive Barriers Among Australian Wild Rice
3.4. Retrotransposable Elements
4. Materials and Methods
4.1. Plant Materials
4.2. Crossing and Fertility Test
4.3. Data Mining from Whole Genome Sequences
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Cp | chloroplast |
| INDEL | insertion/deletion |
| SSR | simple sequence repeat |
References
- Brar, D.S.; Khush, G.D. Wild relative of rice: A valuable genetic resources for genomics and breeding research. In The Wild Oryza Genomes; Mondal, T.K., Henry, R.J., Eds.; Springer International Publishing AG: Basel, Switzerland, 2018; pp. 1–25. [Google Scholar]
- Morishima, H. Phenetic similarity and phylogenetic relationships among strains of Oryza perennis, estimated by methods of numerical taxonomy. Evolution 1969, 23, 429–443. [Google Scholar] [CrossRef]
- Oka, H.-I.; Morishima, H. Variations in the breeding systems of a wild rice, Oryza perennis. Evolution 1967, 21, 249–258. [Google Scholar] [CrossRef]
- Morishima, H.; Oka, H.I.; Chang, W.T. Directions of differentiation in populations of wild rice, O. perennis and O. sativa f. spontanea. Evolution 1961, 15, 326–339. [Google Scholar] [CrossRef]
- Ng, N.Q.; Hawkes, J.G.; William, J.T.; Chang, T.T. The recognition of a new species of rice (Oryza) from Australia. Bot. J. Linn Soc. 1981, 82, 327–330. [Google Scholar] [CrossRef]
- Henry, R.J.; Rice, N.; Waters, D.L.E.; Kasem, S.; Ishikawa, R.; Hao, Y.; Dillon, S.; Crayn, C.; Wing, R.; Vaughan, D. Australian Oryza: Utility and Conservation. Rice 2010, 3, 235–241. [Google Scholar] [CrossRef]
- Cheng, C.; Tsuchimoto, S.; Ohtsubo, H.; Ohtsubo, E. Evolutionary relationships among rice species with AA genome based on SINE insertion analysis. Genes Genet. Syt. 2002, 77, 323–334. [Google Scholar] [CrossRef]
- Xu, J.-H.; Osawa, I.; Tsuchimoto, S.; Ohtsubo, H.; Ohtsubo, E. Two new SINE elements, p-SINE2 and p-SINE3, from rice. Genes Genet. Syst. 2005, 80, 161–171. [Google Scholar] [CrossRef][Green Version]
- Juliano, A.B.; Elizabeth, M.; Naredo, B.; Lu, B.-R.; Jackson, M.T. Genetic differentiation in Oryza meridionalis Ng based on Molecular and Crossability Analyses. Genet. Resour. Crop Evol. 2005, 52, 435–445. [Google Scholar] [CrossRef]
- Elizabeth, M.; Naredo, B.; Juliano, A.B.; Lu, B.-R.; Jackson, M.T. Hybridization of AA genome rice species from Asia and Australia, I. Crosses and development of hybrids. Genet. Resour. Crop Evol. 1997, 44, 17–23. [Google Scholar]
- Lu, B.-R.; Elizabeth, M.; Naredo, B.; Juliano, A.B.; Jackson, M.T. Hybridization of AA genome rice species from Asia and Australia II. Meiotic analysis of Oryza meridionalis and its hybrids. Genet. Resour. Crop Evol. 1997, 44, 25–31. [Google Scholar] [CrossRef]
- Sano, Y.; Morishima, H. Variation in resources allocation and adaptive strategy of a wild rice, Oryza perennis Moench. Bot. Gaz. 1982, 143, 518–523. [Google Scholar] [CrossRef]
- Vaughan, D.A. The Wild Relatives of Rice-A Genetic Resources Handbook; IRRI: Manila, Philippines, 1994; p. 64. [Google Scholar]
- Sotowa, M.; Ootsuka, K.; Kobayashi, Y.; Hao, Y.; Tanaka, K.; Ichitani, K.; Flowers, J.M.; Purugganan, M.D.; Nakamura, I.; Sato, Y.-I.; et al. Molecular relationships between Australian annual wild rice, Oryza meridionalis, and two related perennial forms. Rice 2013, 6, 26. [Google Scholar] [CrossRef]
- Waters, D.; Nock, C.J.; Ishikawa, R.; Rice, N.; Henry, R.J. Chloroplast genome sequence confirms distinctiness of Australian and Asian wild rice. Ecol. Evol. 2012, 2, 211–217. [Google Scholar] [CrossRef]
- Brozynska, M.; Omar, E.S.; Furtado, A.; Crayn, D.; Simon, B.; Ishikawa, R.; Henry, R.J. Chloroplast Genome of Novel Rice Germplasm Identified in Northern Australia. Trop. Plant Biol. 2014, 7, 111–120. [Google Scholar] [CrossRef]
- Brozynska, M.; Copetti, D.; Furtado, A.; Wing, R.; Crayn, D.; Fox, G.; Ishikawa, R.; Henry, R.J. Sequencing of Australian wild rice genomes reveals ancestral relationships with domesticated rice. Plant Biot. J. 2016, 15, 765–774. [Google Scholar]
- Moner, A.M.; Furtado, A.; Chivers, I.; Fox, G.; Crayn, D.; Henry, R.J. Diversity and evolution of rice progenitors in Australia. Ecol. Evol. 2018, 8, 4360–4366. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Brozynska, M.; Furtado, A.; Waters, D.; Henry, R.J. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci. Rep. 2015, 5, 13957. [Google Scholar] [CrossRef]
- Tang, L.; Zou, X.H.; Achoundong, G.; Potgieter, C.; Second, G.; Zhang, D.Y.; Ge, S. Phylogeny and biogeography of the rice tribe (Oryzeae): Evidence from combined analysis of 20 chloroplast fragments. Mol. Phylogenetics Evol. 2010, 54, 266–277. [Google Scholar] [CrossRef]
- Kim, K.; Lee, S.C.; Lee, J.; Yu, Y.; Yang, K.; Choi, B.S.; Koh, H.-J.; Waminal, N.E.; Choi, H.-I.; Kim, N.-H.; et al. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci. Rep. 2015, 5, 15655. [Google Scholar] [CrossRef]
- Zhang, Q.-J.; Zhua, T.; Xiaa, E.-H.; Shia, C. Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc. Natl. Acad. Sci. USA 2014, 46, E4954–E4962. [Google Scholar] [CrossRef]
- Xu, J.-H.; Kurta, N.; Akimoto, M.; Ohtsubo, H.; Ohtsubo, E. Identification and characterization of Australian wild strains of Oryza meridionalis and Oryza rufipogon by SINE insertion polymorphism. Genes Genet. Syst. 2005, 80, 129–134. [Google Scholar] [CrossRef][Green Version]
- Nonomura, K.I.; Morishima, H.; Miyabayashi, T.; Yamaki, S.; Eiguchi, M.; Kubo, T.; Kurata, N. The wild Oryza collection in National BioResource Project (NBRP) of Japan: History, biodiversity and utility. Breed. Sci. 2010, 60, 502–508. [Google Scholar] [CrossRef]
- Kumar, S.; Dudley, J.; Nei, M.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
| No. of | GPS Data | ||||
|---|---|---|---|---|---|
| Sites | Populations | Plants | S | E | Year |
| P26 | P26a | 8 | S16.5332 | E145.2138 | 2011 |
| P26b | 8 | S16.5529 | E145.2136 | 2011 | |
| P26c | 8 | S16.5541 | E145.2130 | 2011 | |
| P26d | 8 | S16.5539 | E145.2128 | 2011 | |
| P26e | 8 | S16.5536 | E145.2125 | 2011 | |
| P26f | 8 | S16.5536 | E145.2124 | 2011 | |
| P26g | 8 | S16.5535 | E145.2123 | 2011 | |
| P26h | 8 | S16.5534 | E145.2122 | 2011 | |
| P26i | 8 | S16.5533 | E145.2122 | 2011 | |
| Jpn1 | Jpn1 | 8 | S16.3809 | E145.1936 | 2011 |
| Jpn2 | Jpn2 | 8 | S15.2622 | E144.1239 | 2011 |
| Jpn3 | Jpn3 | 8 | S15.0431 | E143.4321 | 2011 |
| P6 | P6 | 8 | S15 41519 | E145 02473 | 2011 |
| P7 | P7E | 8 | S15 42003 | E145 04219 | 2011 |
| P7L | 8 | S15 42003 | E145 04219 | 2011 | |
| P8 | P8W | 4 | S15 41302 | E145 07237 | 2011 |
| P8R | 4 | S15 41302 | E145 07237 | 2011 | |
| P10 | P10H | 8 | S15.41416 | E145.09040 | 2011 |
| P10L | 8 | S15.41416 | E145.09040 | 2011 | |
| P12 | P12 | 8 | S15.31486 | E144.22564 | 2011 |
| P17 | P17 | 8 | S15.09256 | E143.49481 | 2011 |
| P21 | P21 | 8 | S15.23047 | E144.07228 | 2011 |
| P22 | P22 | 8 | S15.78099 | E144.14333 | 2011 |
| P23 | P23 | 8 | S15.31198 | E144.22079 | 2011 |
| P26j | P26j | 8 | S16.55118 | E145.2136 | 2011 |
| P26PL | P26PL | 8 | S16.15579 | E145.2060 | 2011 |
| P27 | P27 | ||||
| P5 | P5O | 8 | S15 45329 | E144 59419 | 2010 |
| P5N | 8 | S15 45329 | E144 59419 | 2010 | |
| P5W | 8 | S15 45329 | E144 59419 | 2010 | |
| Core collection of NBRP | |||||
| Oryza meridinoalis | 19* | ||||
| Oryza rufipogon | 32 | ||||
| Anther Length (mm) | Spikelet Length (mm) | Spikelet Width (mm) | Density of Bristle Cells (per 200 μm2) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Accession | Life History | n= | mean | ± | SD | mean | ± | SD | mean | ± | SD | mean | ± | SD | ||||
| O.rufipogon | |||||||||||||||||||
| W0106 | Annual | 5 | 1.50 | ± | 0.07 | ** | 7.01 | ± | 0.63 | ** | 2.42 | ± | 0.20 | 4.67 | ± | 1.92 | ** | ||
| W0120 | Perennial | 5 | 1.88 | ± | 0.10 | ** | 7.06 | ± | 0.16 | ** | 2.34 | ± | 0.11 | 2.67 | ± | 1.92 | ** | ||
| W0137 | Perennial | 5 | 2.31 | ± | 0.05 | ** | 6.95 | ± | 0.25 | ** | 2.60 | ± | 0.11 | 3.67 | ± | 1.92 | ** | ||
| O.meridionalis | |||||||||||||||||||
| W1299 | Annual | 5 | 1.42 | ± | 0.05 | ** | 6.47 | ± | 0.27 | ** | 2.32 | ± | 0.12 | 12.67 | ± | 3.42 | |||
| W1300 | Annual | 5 | ND | 6.91 | ± | 0.19 | ** | 2.13 | ± | 0.05 | ** | 14.67 | ± | 1.60 | |||||
| Australian wild rice | |||||||||||||||||||
| Greenhouse | |||||||||||||||||||
| Jpn1 | Perennial | 5 | 3.48 | ± | 0.20 | 7.13 | ± | 0.15 | ** | 2.02 | ± | 0.16 | ** | 2.67 | ± | 3.42 | ** | ||
| Jpn2 | Perennial | 5 | 1.64 | ± | 0.07 | ** | 8.28 | ± | 0.14 | * | 2.62 | ± | 0.11 | 12.00 | ± | 2.34 | |||
| Field | |||||||||||||||||||
| Jpn1 | Perennial | 5 | 3.79 | ± | 0.20 | 7.62 | ± | 0.17 | ** | 2.00 | ± | 0.07 | ** | ND | |||||
| Jpn2 | Perennial | 5 | 1.74 | ± | 0.19 | ** | 8.60 | ± | 0.23 | 2.40 | ± | 0.23 | ND | ||||||
| Genotype (Based on Relative Migration Distance) | INDEL Start Position in Nipponbare cp Genome | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker Type | Nipponbare | W0106 | W0120 | W0137 | W1299 | W1300 | Jpn1 | Jpn2 | INDEL | Forward | Reverse | |
| INDEL1 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | -C | 1605 | CTATTCCGAAGAGGAAGTCTAC | TCTCCGTATCAATGATCTGGTG |
| SSR1 | 1 | 1 | 1 | 1 | 2 | 2 | 3 | 2 | +AA | 3535 | CTTTTGACTTTGGGATACAGTC | GATTAGTGCCTGATGTAGGG |
| INDEL2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | -CAATC | 5852 | GGAATTTCCATCCTCAACAGA | GTTTTGTTACGGAAAAATGGTATG |
| SSR2 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +A | 6098 | TTCTCGTATTTCTTCGACTCG | GATAAGAACTGCTCGTTAGATAG |
| INDEL3 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +AGAAA | 8192 | GCCGCTTTAGTCCACTCAGCCATC | TCAATGCCTTTTTTCAATGGTCTC |
| SSR3 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +A | 11441 | CTGGCTCGGTTATTCTATC | GAAAACCGGTATAGTTCTAGG |
| INDEL4 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | -AGGG | 12669 | GCAACAGGGTTCCCTAAACCG | GCCAAATTGAGCAGGTTGCG |
| INDEL5 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | -T | 13566 | GCTTCGCGACTCTGTACTCA | TACTTAAGGCGTCCTTAAGG |
| INDEL6 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | -AC | 14011 | GAAATCTGGGCCATAGAGAA | CTAAGCAGAGACATTCAGAATC |
| INDEL7 | NA | -TATTTCTAAGA | 14527 | - | - | |||||||
| SSR4 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | -A | 17099 | GAAAAAATCCATGGAGGGAGAG | CCCAACATATCGCACATTTTCC |
| SSR5 | 2 | 1 | 1 | 1 | 3 | 3 | 3 | 3 | -TTTCTA | 17336 | GGTCGCTTCTAGTAGCGATTATG | TGCCGAACTTTATTCTTTCTCTC |
| SSR6 | including SSR5 to INDEL10 | -TTTCTA | 17358 | |||||||||
| INDEL8 | -ATAGAA | 17379 | ||||||||||
| INDEL9 | +AGAATTAT | 17385 | ||||||||||
| INDEL10 | +GAATTATATAGAAC | 17392 | ||||||||||
| INDEL11 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | +TGG | 19001 | GAATATCATAAACTGTAAGTGGCAG | CACATGAAATTCTCGGGAACTCC |
| SSR7 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +T | 41464 | GAGGCAAGTGTTCGGATCTATTATG | CTATATTATGCTCAAGGAAAGTAGA |
| SSR8 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | -TATAT | 46086 | CTCTAATTCGCAAATCTATTTTTC | CAAGAAATTCGCATGTTCTC |
| SSR9 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | -T | 46174 | GAGAACATGCGAATTTCTTG | CATACTATAACGCTTGATATTC |
| INDEL12 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | +T | 47211 | GTCGTGAGGGTTCAAGT | CGAGTTAATAATCGACATTCCTTGCC |
| INDEL13 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | +AGGAC | 50351 | GCCTGTCCAGTCTATAAACAAG | GGGTCTTTGAAACAGTTCG |
| SSR10 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +T | 53999 | CATAGAATGTACACAGGGTGTACCC | CTCACAACGACAGGGTCTAC |
| INDEL14 | INDEL14 and SSR11 | -CTTTTTTTTTAGAATA | 57017 | GGATAGAAAGGCCGCGAG including | GACTATTGTATTTTTGAGTTTGC | |||||||
| SSR11 | -A | 57061 | ||||||||||
| INDEL15 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | -CTTTTCAAT | 64815 | CCAGATGCTTTGTCATTCCC | TCATGACTCTAAGGTCCAACC |
| INDEL16 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +TTCCTATTTAATA | 65452 | GTCGTTATTGTCGTAAGCATACGA | GATGAATACCCTCGATACATATG |
| SSR12 | NA | +T | 65615 | - | - | |||||||
| INDEL17 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +t | 66896 | CCAATGGCTTTTGCTACTATAACC | GAAAGAAAGGGCTCCGGTG |
| SSR13 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +A | 71377 | GCACCTGTTATCTCTATCAAG | GTCTGGTTGCGAGGTCTGAATAG |
| SSR14 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | -TTTCTA | 75980 | GATATCCGTTTCAGGGTAAA | CTGATTCGTAGGCGTGGAC |
| SSR15 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | -A | 76232 | CAAATTTTACGAACAGAAGCTC | CCGAAGACTCGAAGGATACC |
| SSR16 | NA | -T | 76574 | CATAACTAAACCCTCGAAAGTAA | CCCGCCTATAGCGGTAATC | |||||||
| INDEL18 | 2 | 3 | 2 | 3 | 1 | 1 | 1 | 1 | -T | 77728 | GCTACATTTAAAAGGGTCTGAGG | CTGCCAGCAAAATGCCC |
| SSR17 | NA | -T | 78423 | - | - | |||||||
| INDEL19 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +T | 80090 | GGGTTGTACCAAGTCTGAA | GCTCGAGGACGTAGTTCTCCCATAA |
| SSR18 | NA | -C | 93004 | - | - | |||||||
| SSR19 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +C | 93534 | GTTCGTCCTCAATGGGAAAATG | GGGAAGTCCTATTGATTGCTG |
| INDEL20 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | +AACA | 104530 | GATCATTTTCTGGCGTCAGCG | GAATATTGTACCGAGGAATTCG |
| INDEL21 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | +G | 121618 | AAGGCTCGAATGGTACGATC | CTTCTCGAGAATCCATACATCCC |
| SSR20 | NA | -G | 122142 | - | - | |||||||
| Pollen and Seed Fertilitty (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Self Pollinated | Crossed with Jpn1 | Crossed with Jpn2 | Crossed with W1297 | ||||
| Pollen | Seed | Pollen | Seed | Pollen | Seed | Pollen | Seed | ||
| W0106 | 97.2 | 30.1 | 87.2 | 15.7 | 10.2 | 0.0 | 0.1 | 0.0 | |
| 98.8 | 8.9 | 95.7 | 18.3 | 0.8 | 0.0 | 0.1 | 0.0 | ||
| Mean* | 98.0 | 19.5 | 91.4 | 17.0 | 5.5 | 0.0 | 0.1 | 0.0 | |
| W0120 | 98.1 | 96.8 | 84.0 | 24.5 | 10.2 | 0.0 | 21.3 | 0.0 | |
| 98.7 | 74.5 | 78.3 | 21.6 | 12.7 | 0.0 | 20.2 | 0.0 | ||
| Mean | 98.4 | 85.6 | 81.2 | 23.1 | 11.5 | 0.0 | 20.8 | 0.0 | |
| W1299 | 95.7 | 20.0 | 0.0 | 0.0 | 29.4 | 0.0 | 99.3 | 42.9 | |
| 96.1 | 24.5 | 4.5 | 0.0 | 39.8 | 0.3 | - | - | ||
| Mean | 95.9 | 22.3 | 2.2 | 0.0 | 34.6 | 0.1 | - | - | |
| W1297 | 95.7 | 54.4 | 5.3 | 0.0 | 53.2 | 0.0 | - | - | |
| 96.9 | 75.9 | 6.4 | 0.0 | 33.8 | 0.0 | - | - | ||
| Mean | 96.3 | 65.2 | 2.2 | 0.0 | 34.6 | 0.1 | - | - | |
| Flanking Primers to Confirm | Insertion (+) / no-Insertion (-) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pSINE1/3 Insertions | Genome | ||||||||||
| with Inner Primers toward Outside | Sequence | Position | Nipponbare | W0106 | W0120 | W0137 | Jpn1 | Jpn2 | W1299 | W1300 | Remarks |
| pSINE1 | |||||||||||
| Jpn2-meridionalis class | |||||||||||
| Chr2-7291280-r (w/L) | TCTCTCTACAGATAATGCTC | 7291535 | - | - | - | - | - | + | + | + | Other O. meridionalis |
| Chr9-12952747-r (w/L) | CACACCCATCTACATCGATG | 12953007 | - | - | - | - | - | + | + | + | Other O. meridionalis |
| Chr10-11030242-f (w/L) | GATTGCCGGCTTCTTTACTAG | 11029860 | |||||||||
| Australia class | |||||||||||
| Chr3-10559212-r (w/L) | ACCTATAACAACTGAGAGAC | 10559538 | - | - | - | - | + | + | + | + | Other O. meridionalis |
| Jpn2-meridionalis-W0106 class | |||||||||||
| Chr1-4067055-f (w/ L) | GAAAGAGATCACAGGTAAAC | 4066713 | - | + | - | - | - | + | + | + | Other O. meridionalis |
| Jpn2-W0180,W1921 class | |||||||||||
| Chr3-10203820-f (w/L) | TCCACCGACTTATAAATCAC | 10203450 | - | - | - | - | - | + | - | - | No other O. meridionalis, but two O. rufipogon,W0180, W1921 |
| Inner primer toward outside | |||||||||||
| pSINE1-L | GAAGACCCCTGGGCGTTTCT | ||||||||||
| (paired primer to amplify pSINE1 insertion) | |||||||||||
| pSINE3 | |||||||||||
| Chr3-33707528-f (w/L) | GTGTAAATATGTATTGTACC | 33702014 | - | - | - | - | - | + | + | + | Other O. meridionalis |
| Chr8-24323118-f (w/L) | GCCTATTACTATCAATCACC | 24322813 | - | - | - | - | - | + | + | + | Other O. meridionalis |
| Chr5-576201-f (w/R) | GATAACTAGGGTAAATGAC | 575868 | - | - | - | - | + | + | + | Other O. meridionalis | |
| Inner primer toward outside | |||||||||||
| pSINE3-R | TCCTTCCTAGATTGGTCCC | ||||||||||
| pSINE3-L | TGCTAGCCGGGAAGACC | ||||||||||
| (paired primer to amplify pSINE3 insertion) | |||||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lam, D.T.; Ichitani, K.; Henry, R.J.; Ishikawa, R. Molecular and Morphological Divergence of Australian Wild Rice. Plants 2020, 9, 224. https://doi.org/10.3390/plants9020224
Lam DT, Ichitani K, Henry RJ, Ishikawa R. Molecular and Morphological Divergence of Australian Wild Rice. Plants. 2020; 9(2):224. https://doi.org/10.3390/plants9020224
Chicago/Turabian StyleLam, Dinh Thi, Katsuyuki Ichitani, Robert J. Henry, and Ryuji Ishikawa. 2020. "Molecular and Morphological Divergence of Australian Wild Rice" Plants 9, no. 2: 224. https://doi.org/10.3390/plants9020224
APA StyleLam, D. T., Ichitani, K., Henry, R. J., & Ishikawa, R. (2020). Molecular and Morphological Divergence of Australian Wild Rice. Plants, 9(2), 224. https://doi.org/10.3390/plants9020224







