Soybean Resistance to Soybean Mosaic Virus
Abstract
1. Introduction
2. Biological Properties and Transmission of SMV
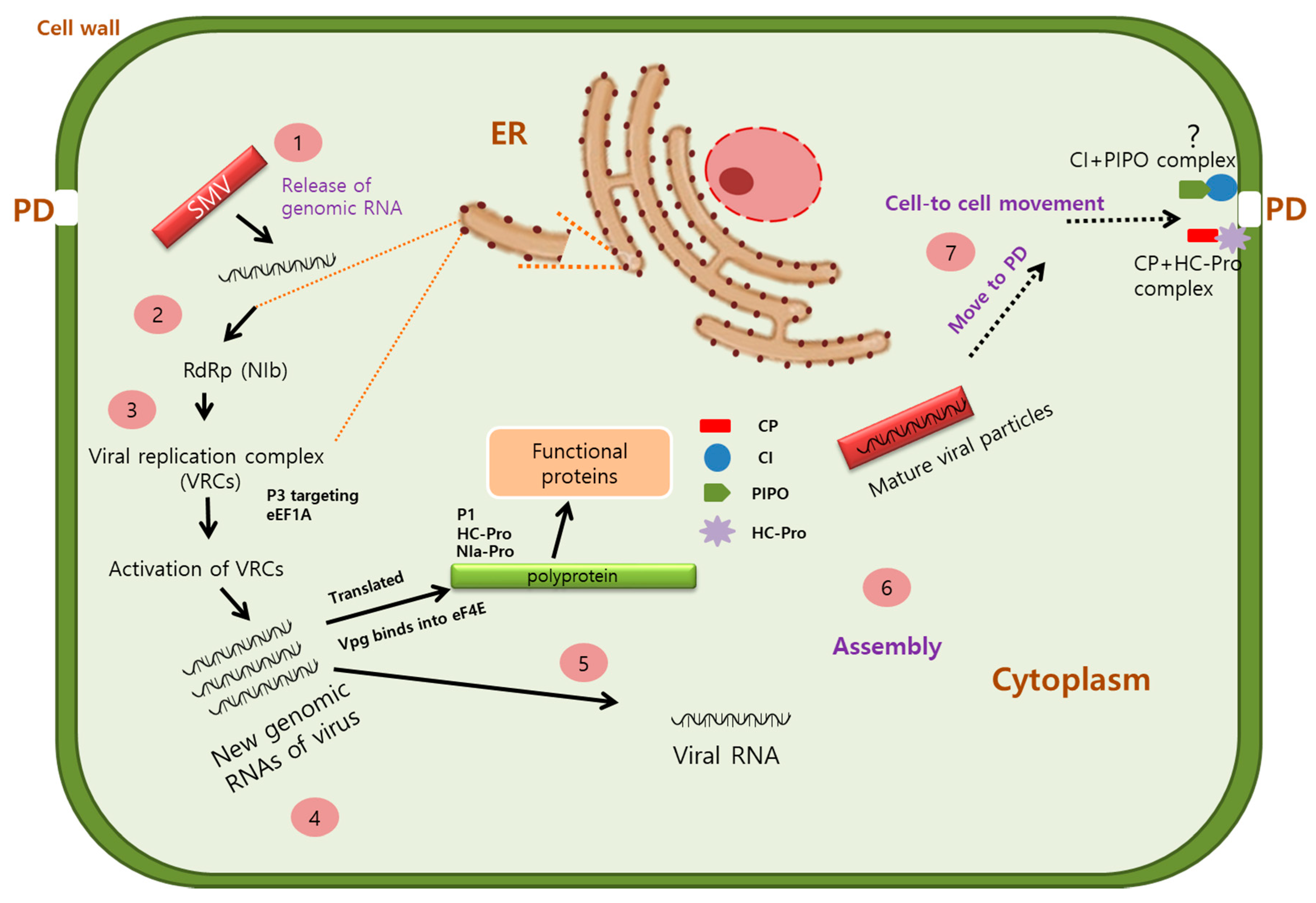
2.1. SMV Genome and Gene Function
2.2. Biological and Molecular Properties of SMV Infection and Transmission
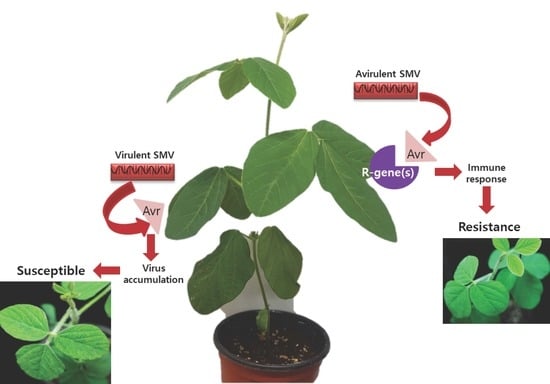
3. Resistance Genes (R-Genes): Soybean Response to SMV Infection
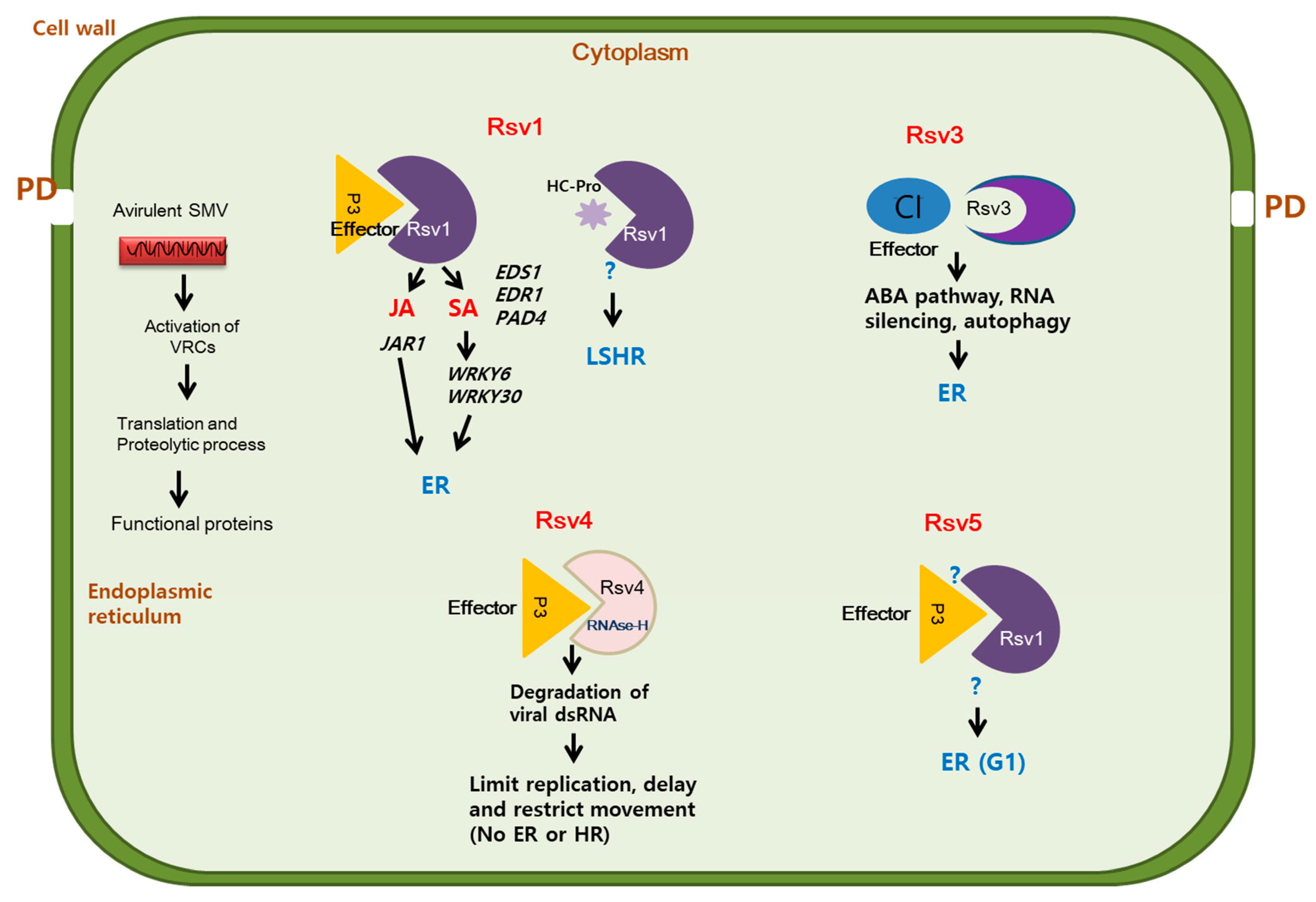
3.1. NLR Gene Family-Mediated Resistance to SMV
3.2. Rsv Genes
3.3. Rsc Genes
4. Independent Host Factors Involved in Soybean-SMV Interaction
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Hartman, G.L.; West, E.D.; Herman, T.K. Crops that feed the World 2. Soybean—Worldwide production, use, and constraints caused by pathogens and pests. J. Food Secur. 2011, 3, 5–17. [Google Scholar] [CrossRef]
- Pagano, M.C.; Miransari, M. The importance of Soybean Production Worldwide; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–26. [Google Scholar]
- Hill, J.H.; Whitham, S.A. Control of virus diseases in soybeans. Adv. Virus Res. 2014, 90, 355–390. [Google Scholar] [PubMed]
- Malapi-Nelson, M.; Wen, R.-H.; Ownley, B.; Hajimorad, M. Co-infection of soybean with Soybean mosaic virus and Alfalfa mosaic virus results in disease synergism and alteration in accumulation level of both viruses. Plant Dis. 2009, 93, 1259–1264. [Google Scholar] [CrossRef] [PubMed]
- Hartman, G.L.; Hill, C.B. 13 Diseases of Soybean and Their Management; CABI: USA, 2010; p. 276. [Google Scholar]
- Hajimorad, M.; Domier, L.L.; Tolin, S.; Whitham, S.; Saghai Maroof, M. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef] [PubMed]
- Galvez, L.C.; Banerjee, J.; Pinar, H.; Mitra, A. Engineered plant virus resistance. Plant Sci. 2014, 228, 11–25. [Google Scholar] [CrossRef]
- Choi, B.K.; Koo, J.M.; Ahn, H.J.; Yum, H.J.; Choi, C.W.; Ryu, K.H.; Chen, P.; Tolin, S. Emergence of Rsv resistance breaking Soybean mosaic virus isolates from Korean soybean cultivars. Virus Res. 2005, 112, 42–51. [Google Scholar] [CrossRef]
- Koo, J.; Choi, B.; Ahn, H.; Yum, H.; Choi, C. First report of an Rsv resistance-breaking isolate of Soybean mosaic virus in Korea. Plant Pathol. 2005, 54, 573. [Google Scholar] [CrossRef]
- Kang, B.-C.; Yeam, I.; Jahn, M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005, 43, 581–621. [Google Scholar] [CrossRef]
- Calvo, M.; Martínez-Turiño, S.; García, J.A. Resistance to Plum pox virus Strain C in Arabidopsis thaliana and Chenopodium foetidum involves genome-linked viral protein and other viral determinants and might depend on compatibility with host translation initiation factors. Mol. Plant-Microbe Interact. 2014, 27, 1291–1301. [Google Scholar] [CrossRef]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front. Microbiol. 2016, 7, 1695. [Google Scholar] [CrossRef]
- Baebler, S.; Witek, K.; Petek, M.; Stare, K.; Tusek-Znidaric, M.; Pompe-Novak, M.; Renaut, J.; Szajko, K.; Strzelczyk-Zyta, D.; Marczewski, W.; et al. Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Y infection in potato. J. Exp. Bot. 2014, 65, 1095–1109. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Eggenberger, A.L.; Hill, J.H. Loss and gain of elicitor function of soybean mosaic virus G7 provoking Rsv1-mediated lethal systemic hypersensitive response maps to P3. J. Virol. 2005, 79, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Hajimorad, M.; Hill, J. Rsv1-mediated resistance against Soybean mosaic virus-N is hypersensitive response-independent at inoculation site, but has the potential to initiate a hypersensitive response-like mechanism. Mol. Plant Microbe Interact. 2001, 14, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; Lin, N.S. Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Grosic, S.; Whitham, S.A.; Hill, J.H. The requirement of multiple defense genes in soybean Rsv1–mediated extreme resistance to Soybean mosaic virus. Mol. Plant Microbe Interact. 2012, 25, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; Tseng, K.-C.; Chang, W.-C.; Seo, J.-K.; Kim, K.-H. Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus. Viruses 2018, 10, 581. [Google Scholar] [CrossRef]
- Alazem, M.; Widyasari, K.; Kim, K.H. An Avirulent Strain of Soybean Mosaic Virus Reverses the Defensive Effect of Abscisic Acid in a Susceptible Soybean Cultivar. Viruses 2019, 11, 879. [Google Scholar] [CrossRef]
- Alazem, M.; He, M.H.; Moffett, P.; Lin, N.S. Abscisic Acid Induces Resistance against Bamboo Mosaic Virus through Argonaute2 and 3. Plant Physiol. 2017, 174, 339–355. [Google Scholar] [CrossRef]
- Alazem, M.; Lin, N.S. Antiviral Roles of Abscisic Acid in Plants. Front. Plant Sci. 2017, 8, 1760. [Google Scholar] [CrossRef]
- Xie, K.; Li, L.; Zhang, H.; Wang, R.; Tan, X.; He, Y.; Hong, G.; Li, J.; Ming, F.; Yao, X.; et al. Abscisic acid negatively modulates plant defence against rice black-streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ. 2018, 41, 2504–2514. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, L.; Yu, K.; Wang, A. Pathogenesis of Soybean mosaic virus in soybean carrying Rsv1 gene is associated with miRNA and siRNA pathways, and breakdown of AGO1 homeostasis. Virology 2015, 476, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Niu, H.; Luo, J.; Zhi, H. Soybean Cytochrome b5 Is a Restriction Factor for Soybean Mosaic Virus. Viruses 2019, 11, 546. [Google Scholar] [CrossRef] [PubMed]
- Xun, H.; Yang, X.; He, H.; Wang, M.; Guo, P.; Wang, Y.; Pang, J.; Dong, Y.; Feng, X.; Wang, S.; et al. Over-expression of GmKR3, a TIR–NBS–LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol. Biol. 2019, 99, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Maroof, S.; Jeong, S.C.; Gunduz, I.; Tucker, D.; Buss, G.; Tolin, S. Pyramiding of soybean mosaic virus resistance genes by marker-assisted selection. Crop Sci. 2008, 48, 517–526. [Google Scholar] [CrossRef]
- Shakiba, E.; Chen, P.; Shi, A.; Li, D.; Dong, D.; Brye, K. Two novel alleles at the Rsv 3 locus for resistance to Soybean mosaic virus in PI 399091 and PI 61947 soybeans. Crop Sci. 2012, 52, 2587–2594. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Jiang, H.; Ren, R.; Li, C.; Zhi, H.; Chen, S.; Gai, J. Inheritance, fine-mapping, and candidate gene analyses of resistance to soybean mosaic virus strain SC5 in soybean. Mol. Genet. Genomics 2017, 292, 811–822. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Li, C.; Yin, J.; Li, N.; Yang, Y.; Song, Y.; Ren, R.; Zhi, H.; Gai, J.; et al. Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus strain SC20 in soybean. Theor. Appl. Genet. 2018, 131, 461–476. [Google Scholar] [CrossRef]
- Rui, R.; Liu, S.; Karthikeyan, A.; Wang, T.; Niu, H.; Yin, J.; Yang, Y.; Wang, L.; Yang, Q.; Zhi, H.; et al. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus. Theor. Appl. Genet. 2017, 130, 2395–2410. [Google Scholar] [CrossRef]
- Wang, D.; Ma, Y.; Liu, N.; Yang, Z.; Zheng, G.; Zhi, H. Fine mapping and identification of the soybean RSC4 resistance candidate gene to soybean mosaic virus. Plant Breed. 2011, 130, 653–659. [Google Scholar] [CrossRef]
- Wang, D.; Ma, Y.; Yang, Y.; Liu, N.; Li, C.; Song, Y.; Zhi, H. Fine mapping and analyses of R SC8 resistance candidate genes to soybean mosaic virus in soybean. Theor. Appl. Genet. 2011, 122, 555–565. [Google Scholar] [CrossRef]
- Wang, D.-G.; Lin, Z.; Kai, L.; Ying, M.; Wang, L.-Q.; Yang, Y.-Q.; Yang, Y.-H.; Zhi, H.-J. Marker-assisted pyramiding of soybean resistance genes RSC4, RSC8, and RSC14Q to soybean mosaic virus. J. Integr. Agric. 2017, 16, 2413–2420. [Google Scholar] [CrossRef]
- Cho, E.-K.; Goodman, R.M. Strains of soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology 1979, 69, 467–470. [Google Scholar] [CrossRef]
- Li, K.; Yang, Q.; Zhi, H.; Gai, J. Identification and distribution of soybean mosaic virus strains in southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef]
- Wang, D.; Tian, Z.; Li, K.; Li, H.; Huang, Z.; Hu, G.; Zhang, L.; Zhi, H. Identification and variation analysis of soybean mosaic virus strains in Shandong, Henan and Anhui provinces of China. Soybean Science 2013, 32, 806–809. [Google Scholar]
- Riechmann, J.L.; Lain, S.; García, J.A. Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 1992, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Olspert, A.; Chung, B.Y.W.; Atkins, J.F.; Carr, J.P.; Firth, A.E. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 2015, 16, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Revers, F.; García, J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar]
- Chung, B.Y.-W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef]
- Verchot, J.; Carrington, J.C. Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J. Virol. 1995, 69, 3668–3674. [Google Scholar] [CrossRef]
- Kapust, R.B.; Tözsér, J.; Copeland, T.D.; Waugh, D.S. The P1′ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 2002, 294, 949–955. [Google Scholar] [CrossRef]
- Maliogka, V.I.; Salvador, B.; Carbonell, A.; Saenz, P.; León, D.S.; Oliveros, J.C.; Delgadillo, M.O.; García, J.A.; Simón-Mateo, C. Virus variants with differences in the P1 protein coexist in a Plum pox virus population and display particular host-dependent pathogenicity features. Mol. Plant Pathol. 2012, 13, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Salvador, B.; Saenz, P.; Yangüez, E.; Quiot, J.B.; Quiot, L.; Delgadillo, M.O.; Garcia, J.A.; Simón-Mateo, C. Host-specific effect of P1 exchange between two potyviruses. Mol. Plant Pathol. 2008, 9, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Carrington, J.C. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 2001, 285, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Kadoury, D.; Gal-On, A.; Huet, H.; Wang, Y.; Raccah, B. Mutations in the HC-Pro gene of zucchini yellow mosaic potyvirus: Effects on aphid transmission and binding to purified virions. J. Gen. Virol. 1998, 79, 897–904. [Google Scholar] [CrossRef]
- Seo, J.K.; Kang, S.H.; Seo, B.Y.; Jung, J.K.; Kim, K.H. Mutational analysis of interaction between coat protein and helper component-proteinase of Soybean mosaic virus involved in aphid transmission. Mol. Plant Pathol. 2010, 11, 265–276. [Google Scholar] [CrossRef]
- Kasschau, K.D.; Carrington, J.C. A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 1998, 95, 461–470. [Google Scholar] [CrossRef]
- Wen, R.-H.; Khatabi, B.; Ashfield, T.; Maroof, M.S.; Hajimorad, M. The HC-Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol. Plant Microbe Interact. 2013, 26, 203–215. [Google Scholar] [CrossRef]
- Eggenberger, A.; Hajimorad, M.; Hill, J. Gain of virulence on Rsv1-genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC-Pro. Mol. Plant Microbe Interact. 2008, 21, 931–936. [Google Scholar] [CrossRef]
- Luan, H.; Shine, M.; Cui, X.; Chen, X.; Ma, N.; Kachroo, P.; Zhi, H.; Kachroo, A. The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 2016, 172, 221–234. [Google Scholar] [CrossRef]
- Hajimorad, M.; Eggenberger, A.; Hill, J. Strain-specific P3 of Soybean mosaic virus elicits Rsv1-mediated extreme resistance, but absence of P3 elicitor function alone is insufficient for virulence on Rsv1-genotype soybean. Virology 2006, 345, 156–166. [Google Scholar] [CrossRef][Green Version]
- Chowda-Reddy, R.; Sun, H.; Chen, H.; Poysa, V.; Ling, H.; Gijzen, M.; Wang, A. Mutations in the P3 protein of Soybean mosaic virus G2 isolates determine virulence on Rsv4-genotype soybean. Mol. Plant Microbe Interact. 2011, 24, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.-H.; Hajimorad, M. Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 2010, 400, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.-Y.; Chen, J.; Shi, Y.-H.; Chen, J.-P. The ‘6K1’protein of a strain of Soybean mosaic virus localizes to the cell periphery. Arch. Virol. 2007, 152, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Jensen, P.E.; Schaad, M.C. Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J. 1998, 14, 393–400. [Google Scholar] [CrossRef]
- Seo, J.-K.; Lee, S.-H.; Kim, K.-H. Strain-specific cylindrical inclusion protein of Soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Mol. Plant Microbe Interact. 2009, 22, 1151–1159. [Google Scholar] [CrossRef]
- Zhang, C.; Hajimorad, M.; Eggenberger, A.L.; Tsang, S.; Whitham, S.A.; Hill, J.H. Cytoplasmic inclusion cistron of Soybean mosaic virus serves as a virulence determinant on Rsv3-genotype soybean and a symptom determinant. Virology 2009, 391, 240–248. [Google Scholar] [CrossRef]
- Wei, T.; Wang, A. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI-and COPII-dependent manner. J. Virol. 2008, 82, 12252–12264. [Google Scholar] [CrossRef]
- Wei, T.; Huang, T.-S.; McNeil, J.; Laliberté, J.-F.; Hong, J.; Nelson, R.S.; Wang, A. Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 2010, 84, 799–809. [Google Scholar] [CrossRef]
- Michon, T.; Estevez, Y.; Walter, J.; German-Retana, S.; Le Gall, O. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 2006, 273, 1312–1322. [Google Scholar] [CrossRef]
- Lellis, A.D.; Kasschau, K.D.; Whitham, S.A.; Carrington, J.C. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF (iso) 4E during potyvirus infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef]
- Carrington, J.C.; Freed, D.D.; Leinicke, A.J. Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell 1991, 3, 953–962. [Google Scholar] [PubMed]
- Adams, M.J.; Antoniw, J.F.; Beaudoin, F. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 2005, 6, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Saruta, M.; Shimizu, T.; Shu, M.; Anai, T.; Komatsu, K.; Yamada, N.; Katayose, Y.; Ishikawa, M.; Ishimoto, M.; et al. Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 2019, 10, 4033. [Google Scholar] [CrossRef] [PubMed]
- Charon, J.; Theil, S.; Nicaise, V.; Michon, T. Protein intrinsic disorder within the Potyvirus genus: From proteome-wide analysis to functional annotation. Mol. Biosyst. 2016, 12, 634–652. [Google Scholar] [CrossRef]
- Hong, Y.; Hunt, A.G. RNA polymerase activity catalyzed by a potyvirus-encoded RNA-dependent RNA polymerase. Virology 1996, 226, 146–151. [Google Scholar] [CrossRef]
- Kang, S.-H.; Lim, W.-S.; Hwang, S.-H.; Park, J.-W.; Choi, H.-S.; Kim, K.-H. Importance of the C-terminal domain of soybean mosaic virus coat protein for subunit interactions. J. Gen. Virol. 2006, 87, 225–229. [Google Scholar] [CrossRef]
- Steinlage, T.A.; Hill, J.; Nutter Jr, F. Temporal and spatial spread of Soybean mosaic virus (SMV) in soybeans transformed with the coat protein gene of SMV. Phytopathology 2002, 92, 478–486. [Google Scholar] [CrossRef]
- Helm, M.; Qi, M.; Sarkar, S.; Yu, H.; Whitham, S.A.; Innes, R.W. Engineering a decoy substrate in soybean to enable recognition of the Soybean Mosaic Virus NIa protease. Mol. Plant Microbe Interact. 2019, 32, 760–769. [Google Scholar] [CrossRef]
- Reagan, B.C.; Burch-Smith, T.M. Viruses Reveal the Secrets of Plasmodesmal Cell Biology. Mol. Plant Microbe Interact. 2019, 33, 26–39. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Lockhart, B. Plant cell wall dynamics in compatible and incompatible potato response to infection caused by Potato virus Y (PVYNTN). Int. J. Mol. Sci. 2018, 19, 862. [Google Scholar] [CrossRef]
- Allie, F.; Pierce, E.J.; Okoniewski, M.J.; Rey, C. Transcriptional analysis of South African cassava mosaic virus-infected susceptible and tolerant landraces of cassava highlights differences in resistance, basal defense and cell wall associated genes during infection. BMC Genomics 2014, 15, 1006. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Zhang, C.; Hong, J.; Xiong, R.; Kasschau, K.D.; Zhou, X.; Carrington, J.C.; Wang, A. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 2010, 6, e1000962. [Google Scholar] [CrossRef] [PubMed]
- Vijayapalani, P.; Maeshima, M.; Nagasaki-Takekuchi, N.; Miller, W.A. Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 2012, 8, e1002639. [Google Scholar] [CrossRef] [PubMed]
- Nachappa, P.; Culkin, C.T.; Saya, P.M.; Han, J.; Nalam, V.J. Water stress modulates soybean aphid performance, feeding behavior, and virus transmission in soybean. Front. Plant Sci. 2016, 7, 552. [Google Scholar] [CrossRef] [PubMed]
- Jossey, S.; Hobbs, H.A.; Domier, L.L. Role of Soybean mosaic virus–encoded proteins in seed and aphid transmission in soybean. Phytopathology 2013, 103, 941–948. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Lou, Y. PRRs and NB-LRRs: From signal perception to activation of plant innate immunity. Int. J. Mol. Sci. 2019, 20, 1882. [Google Scholar] [CrossRef]
- Wu, C.-H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118. [Google Scholar] [CrossRef]
- Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018, 219, 17–24. [Google Scholar] [CrossRef]
- Adachi, H.; Derevnina, L.; Kamoun, S. NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 2019, 50, 121–131. [Google Scholar] [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef]
- Deslandes, L.; Olivier, J.; Theulières, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Morgante, M.; Michelmore, R.W. TIR-X and TIR-NBS proteins: Two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2002, 32, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-Z.; Fang, Y.; Pang, H. The current status of the soybean-soybean mosaic virus (SMV) pathosystem. Front. Microbiol. 2016, 7, 1906. [Google Scholar] [CrossRef] [PubMed]
- Khatabi, B.; Fajolu, O.; Wen, R.H.; Hajimorad, M. Evaluation of N orth A merican isolates of S oybean mosaic virus for gain of virulence on Rsv-genotype soybeans with special emphasis on resistance-breaking determinants on Rsv4. Mol. Plant Pathol. 2012, 13, 1077–1088. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Chen, S.; Shu, L.; Palmer, R.G.; Xing, G.; Li, Y.; Yang, S.; Yu, D.; Zhao, T. Exploration of presence/absence variation and corresponding polymorphic markers in soybean genome. J. Integr. Plant Biol. 2014, 56, 1009–1019. [Google Scholar] [CrossRef]
- Zhan, Y.; Zhi, H.; Yu, D.; Gai, J. Identification and distribution of SMV strains in huang-huai valleys. Agric. Sci. China 2006, 39, 2009–2015. [Google Scholar]
- Hayes, A.; Jeong, S.; Gore, M.; Yu, Y.; Buss, G.; Tolin, S.; Maroof, M.S. Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 2004, 166, 493–503. [Google Scholar] [CrossRef]
- Shi, A.; Chen, P.; Li, D.; Zheng, C.; Zhang, B.; Hou, A. Pyramiding multiple genes for resistance to soybean mosaic virus in soybean using molecular markers. Mol. Breed. 2009, 23, 113. [Google Scholar] [CrossRef]
- Wang, J.; Shine, M.; Gao, Q.-M.; Navarre, D.; Jiang, W.; Liu, C.; Chen, Q.; Hu, G.; Kachroo, A. Enhanced disease susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiol. 2014, 165, 1269–1284. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef]
- Pandey, S.P.; Roccaro, M.; Schön, M.; Logemann, E.; Somssich, I.E. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 2010, 64, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Hwang, S.H.; Fang, I.R.; Kwon, S.I.; Park, S.R.; Ahn, I.; Kim, J.B.; Hwang, D.J. Molecular characterization of Oryza sativa WRKY 6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 2015, 208, 846–859. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.J.; Bowman, B.C.; Jeong, N.; Yang, K.; Kastl, C.; Tolin, S.A.; Maroof, M.; Jeong, S.-C. The Rsv3 locus conferring resistance to soybean mosaic virus is associated with a cluster of coiled-coil nucleotide-binding leucine-rich repeat genes. Plant Genome 2011, 4, 55–64. [Google Scholar] [CrossRef]
- Tran, P.-T.; Widyasari, K.; Seo, J.-K.; Kim, K.-H. Isolation and validation of a candidate Rsv3 gene from a soybean genotype that confers strain-specific resistance to soybean mosaic virus. Virology 2018, 513, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Gunduz, I. Genetic Analysis of Soybean Mosaic Virus Resistance in Soybean; Virginia Tech: Blacksburg, VA, USA, 2000. [Google Scholar]
- Jeong, S.; Kristipati, S.; Hayes, A.; Maughan, P.; Noffsinger, S.; Gunduz, I.; Buss, G.; Maroof, M. Genetic and sequence analysis of markers tightly linked to the soybean mosaic virus resistance gene, Rsv 3. Crop Sci. 2002, 42, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-K.; Kwon, S.-J.; Cho, W.K.; Choi, H.-S.; Kim, K.-H. Type 2C protein phosphatase is a key regulator of antiviral extreme resistance limiting virus spread. Sci. Rep. 2014, 4, 5905. [Google Scholar] [CrossRef]
- Hayes, A.J.; Ma, G.; Buss, G.R.; Maroof, M. Molecular marker mapping of Rsv 4, a gene conferring resistance to all known strains of soybean mosaic virus. Crop Sci. 2000, 40, 1434–1437. [Google Scholar] [CrossRef]
- Maroof, M.; Tucker, D.M.; Skoneczka, J.A.; Bowman, B.C.; Tripathy, S.; Tolin, S.A. Fine mapping and candidate gene discovery of the soybean mosaic virus resistance gene, Rsv4. Plant Genome 2010, 3, 14–22. [Google Scholar] [CrossRef]
- Gunduz, I.; Buss, G.R.; Chen, P.; Tolin, S.A. Genetic and phenotypic analysis of Soybean mosaic virus resistance in PI 88788 soybean. Phytopathology 2004, 94, 687–692. [Google Scholar] [CrossRef]
- Wang, Y.; Khatabi, B.; Hajimorad, M. Amino acid substitution in P 3 of S oybean mosaic virus to convert avirulence to virulence on R sv4-genotype soybean is influenced by the genetic composition of P 3. Mol. Plant Pathol. 2015, 16, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Klepadlo, M.; Chen, P.; Shi, A.; Mason, R.E.; Korth, K.L.; Srivastava, V.; Wu, C. Two tightly linked genes for Soybean mosaic virus resistance in soybean. Crop Sci. 2017, 57, 1844–1853. [Google Scholar] [CrossRef]
- Zheng, C.; Chen, P.; Gergerich, R. Characterization of resistance to Soybean mosaic virus in diverse soybean germplasm. Crop Sci. 2005, 45, 2503–2509. [Google Scholar] [CrossRef]
- Shi, A.; Chen, P.; Zheng, C.; Hou, A.; Zhang, B. A PCR-based marker for the Rsv1 locus conferring resistance to soybean mosaic virus. Crop Sci. 2008, 48, 262–268. [Google Scholar] [CrossRef]
- Zheng, C.; Chen, P.; Li, D.; Gergerich, R. New sources of resistance to Soybean mosaic virus in soybean. Can. J. Plant Pathol. 2008, 30, 595–603. [Google Scholar] [CrossRef]
- Buss, G.; Ma, G.; Chen, P.; Tolin, S. Registration of V94-5152 soybean germplasm resistant to soybean mosaic potyvirus. Crop Sci. 1997, 37, 1987–1988. [Google Scholar] [CrossRef]
- Ma, G.; Chen, P.; Buss, G.; Tolin, S. Genetic characteristics of two genes for resistance to soybean mosaic virus in PI486355 soybean. Theor. Appl. Genet. 1995, 91, 907–914. [Google Scholar] [CrossRef]
- Ilut, D.C.; Lipka, A.E.; Jeong, N.; Bae, D.N.; Kim, D.H.; Kim, J.H.; Redekar, N.; Yang, K.; Park, W.; Kang, S.-T.; et al. Identification of haplotypes at the Rsv4 genomic region in soybean associated with durable resistance to soybean mosaic virus. Theor. Appl. Genet. 2016, 129, 453–468. [Google Scholar] [CrossRef]
- Fu, S.; Zhan, Y.; Zhi, H.; Gai, J.; Yu, D. Mapping of SMV resistance gene Rsc-7 by SSR markers in soybean. Genetica 2006, 128, 63–69. [Google Scholar] [CrossRef]
- Yan, H.; Wang, H.; Cheng, H.; Hu, Z.; Chu, S.; Zhang, G.; Yu, D. Detection and fine-mapping of SC7 resistance genes via linkage and association analysis in soybean. J. Integr. Plant Biol. 2015, 57, 722–729. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, D.; Zhang, H.; Shen, Y.; Yang, Y.; Li, K.; Wang, L.; Yang, Y.; Zhi, H. Fine mapping of the RSC8 locus and expression analysis of candidate SMV resistance genes in soybean. Plant Breed. 2016, 135, 701–706. [Google Scholar] [CrossRef]
- Ma, Y.; Li, H.; Wang, D.; Liu, N.; Zhi, H. Molecular mapping and marker assisted selection of soybean mosaic virus resistance gene RSC12 in soybean. Legume Genomics Genet. 2010, 1. [Google Scholar] [CrossRef]
- Li, C.; Adhimoolam, K.; Yuan, Y.; Yin, J.; Ren, R.; Yang, Y.; Zhi, H. Identification of candidate genes for resistance to Soybean mosaic virus strain SC3 by using fine mapping and transcriptome analyses. Crop Pasture Sci. 2017, 68, 156–166. [Google Scholar] [CrossRef]
- Zheng, G.-J.; Yang, Y.-Q.; Ying, M.; Yang, X.-F.; Chen, S.-Y.; Rui, R.; Wang, D.-G.; Yang, Z.-L.; Zhi, H.-J. Fine mapping and candidate gene analysis of resistance gene RSC3Q to soybean mosaic virus in Qihuang 1. J. Integr. Agric. 2014, 13, 2608–2615. [Google Scholar] [CrossRef]
- Li, H.C.; Zhi, H.J.; Gai, J.Y.; Guo, D.Q.; Wang, Y.W.; Li, K.; Bai, L.; Yang, H. Inheritance and gene mapping of resistance to soybean mosaic virus strain SC14 in soybean. J. Integr. Plant Biol. 2006, 48, 1466–1472. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, D.-G.; Li, H.-C.; Zheng, G.-J.; Yang, Y.-Q.; Li, H.-W.; Zhi, H.-J. Fine mapping of the R SC14Q locus for resistance to soybean mosaic virus in soybean. Euphytica 2011, 181, 127–135. [Google Scholar] [CrossRef]
- Li, K.; Ren, R.; Adhimoolam, K.; Gao, L.; Yuan, Y.; Liu, Z.; Zhong, Y.; Zhi, H. Genetic analysis and identification of two soybean mosaic virus resistance genes in soybean [Glycine max (L.) Merr]. Plant Breed. 2015, 134, 684–695. [Google Scholar] [CrossRef]
- Li, N.; Yin, J.L.; Li, C.; Wang, D.G.; Yang, Y.Q.; Karthikeyan, A.; Luan, H.X.; Zhi, H.J. NB-LRR gene family required for Rsc4-mediated resistance to Soybean mosaic virus. Crop Pasture Sci. 2016, 67, 541–552. [Google Scholar] [CrossRef]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; Wilson, C.; et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Liu, J.-Z.; Horstman, H.D.; Braun, E.; Graham, M.A.; Zhang, C.; Navarre, D.; Qiu, W.-L.; Lee, Y.; Nettleton, D.; Hill, J.H.; et al. Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 2011, 157, 1363–1378. [Google Scholar] [CrossRef]
- Liu, J.-Z.; Braun, E.; Qiu, W.-L.; Shi, Y.-F.; Marcelino-Guimarães, F.C.; Navarre, D.; Hill, J.H.; Whitham, S.A. Positive and negative roles for soybean MPK6 in regulating defense responses. Mol. Plant-Microbe Interact. 2014, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Kontra, L.; Csorba, T.; Tavazza, M.; Lucioli, A.; Tavazza, R.; Moxon, S.; Tisza, V.; Medzihradszky, A.; Turina, M.; Burgyán, J.; et al. Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathog. 2016, 12, e1005935. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Sahu, P.P.; Puranik, S.; Prasad, M. Recent advances in plant–virus interaction with emphasis on small interfering RNAs (siRNAs). Mol. Biotechnol. 2013, 55, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Li, M.L.; Weng, K.F.; Shih, S.R.; Brewer, G. The evolving world of small RNAs from RNA viruses. Wiley Interdiscip. Rev. RNA 2016, 7, 575–588. [Google Scholar] [CrossRef]
- Alazem, M.; Kim, K.H.; Lin, N.S. Effects of Abscisic Acid and Salicylic Acid on Gene Expression in the Antiviral RNA Silencing Pathway in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2538. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, C. The role of plant small RNAs in NB-LRR regulation. Brief. Funct. Genomics 2015, 14, 268–274. [Google Scholar] [CrossRef]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef]
- Mallory, A.C.; Hinze, A.; Tucker, M.R.; Bouché, N.; Gasciolli, V.; Elmayan, T.; Lauressergues, D.; Jauvion, V.; Vaucheret, H.; Laux, T. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 2009, 5, e1000646. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef]
- Sarma, B.K.; Singh, H.B.; Fernando, D.; Silva, R.N.; Gupta, V.K. Enhancing plant disease resistance without R genes. Trends Biotechnol. 2016, 34, 523–525. [Google Scholar] [CrossRef]
- Gao, L.; Luo, J.; Ding, X.; Wang, T.; Hu, T.; Song, P.; Zhai, R.; Zhang, H.; Zhang, K.; Li, K. Soybean RNA interference lines silenced for eIF4E show broad potyvirus resistance. Mol. Plant Pathol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bastet, A.; Robaglia, C.; Gallois, J.-L. eIF4E resistance: Natural variation should guide gene editing. Trends Plant Sci. 2017, 22, 411–419. [Google Scholar] [CrossRef]
- Zhou, L.; He, H.; Liu, R.; Han, Q.; Shou, H.; Liu, B. Overexpression of GmAKT2 potassium channel enhances resistance to soybean mosaic virus. BMC Plant Biol. 2014, 14, 154. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Niu, L.; Zhang, W.; Yang, J.; Xing, G.; He, H.; Guo, D.; Du, Q.; Qian, X.; Yao, Y. RNAi-mediated SMV P3 cistron silencing confers significantly enhanced resistance to multiple Potyvirus strains and isolates in transgenic soybean. Plant Cell Rep. 2018, 37, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Furutani, N.; Hidaka, S.; Kosaka, Y.; Shizukawa, Y.; Kanematsu, S. Coat protein gene-mediated resistance to soybean mosaic virus in transgenic soybean. Breed. Sci. 2006, 56, 119–124. [Google Scholar] [CrossRef][Green Version]
- Kim, H.J.; Kim, M.-J.; Pak, J.H.; Jung, H.W.; Choi, H.K.; Lee, Y.-H.; Baek, I.-Y.; Ko, J.-M.; Jeong, S.-C.; Pack, I.S.; et al. Characterization of SMV resistance of soybean produced by genetic transformation of SMV-CP gene in RNAi. Plant Biotechnol. Rep. 2013, 7, 425–433. [Google Scholar] [CrossRef]
- Gao, L.; Ding, X.; Li, K.; Liao, W.; Zhong, Y.; Ren, R.; Liu, Z.; Adhimoolam, K.; Zhi, H. Characterization of Soybean mosaic virus resistance derived from inverted repeat-SMV-HC-Pro genes in multiple soybean cultivars. Theor. Appl. Genet. 2015, 128, 1489–1505. [Google Scholar] [CrossRef]
- Redekar, N.; Clevinger, E.; Laskar, M.; Biyashev, R.; Ashfield, T.; Jensen, R.V.; Jeong, S.-C.; Tolin, S.; Maroof, S. Candidate gene sequence analyses toward identifying Rsv3-type resistance to Soybean mosaic virus. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, M.-J.; Pak, J.H.; Im, H.H.; Lee, D.H.; Kim, K.-H.; Lee, J.-H.; Kim, D.-H.; Choi, H.K.; Jung, H.W.; et al. RNAi-Mediated Soybean Mosaic Virus (SMV) Resistance of a Korean Soybean Cultivar; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
| Protein | Function for Virus | Function for Plant |
|---|---|---|
| P1 | Protease [41,42], Viral host range [43,44] | |
| HC-Pro | Long-distance movement [45], a ‘bridge’ between virion particles and aphid stylets in aphid transmission [46,47], suppression of host defense (RNA silencing) [48] | Virulence determinant [49,50] |
| P3 | Targets host elongation factors 1A (eEF1A) to facilitate SMV replication [51] | Effector of Rsv1 [50,52], Effector of Rsv4 [53] |
| PIPO | Movement [54] | |
| 6K1 | Cell-to-cell movement [55] | |
| CI | Required for genome replication and movement (cell-to-cell or long-distance movement) [56] | Effector of Rsv3 [57,58] |
| 6K2 | Formation of the virus replication complex [59,60] | |
| VPg | Binds specifically to eIF4E to initiate polyprotein translation [61,62] | |
| NIa-Pro | Proteinase [63,64] | |
| Nib | The catalytic subunit of RdRp [65,66,67] | |
| CP | A ‘bridge’ between virion particles and aphid stylets in aphid transmission [47], cell-to-cell movement, virus assembly [68,69] |
| R Gene | SMV Strain | Cultivar | Location | Effector | Type of R Gene |
|---|---|---|---|---|---|
| Rsv1 | G1–G6 [90] | Kwanggyo Marshall Odgen PI96983 PI507389 Raiden Suweon97 Kosuzu Susumaru PI39887 Jitsuka Clifford Tousan65 Corcisa PI61944 PI61947 [107,108] | Chromosome 13 | P3 [14,52] HC-pro [49] | NB-LRR-type of R-genes [89] |
| Rsv3 | G5,G6,G7 [98,99] | Columbia Hardee Tosan140 PI 339870 PI399091 L29 [90,108] | Chromosome 14 | CI [57,58] | CC-NB-LRR type or R-gene [96] |
| Rsv4 | G1–G7 [103] | PI486355 V94-5152 P188788 Haman Ilpumgeomjeong KAERI-GNT-220-7 PI 398593 PI438307 Rhosa Beeson [86,108,109,110,111] | Chromosome 2 | P3 [53,86,104] | Non-NLR genes (RNase-H family protein) [65] |
| Rsv5 | G1 [106] | York Dorman Arksoy Riple Calhoun Musen [106] | Chromosome 13 | Possibly P3 | unknown |
| R gene | SMV Strain | Cultivar | Location | Candidate Genes |
|---|---|---|---|---|
| Rsc7 | SC7 | Kefeng No.1 [112,113] | Chromosome 2 Linked markers (distance): Satt266 (43.7 cM) Satt634 (18.1 cM) Satt558 (26.6 cM) Satt157 (36.4 cM) Satt698 (37.9 cM) [112] Flanking markers: BARCSOYSSR_02_0621 BARCSOYSSR_02_0632 [113] | 15 candidate genes with one NBS-LRR type gene, one HSP40 gene and one serine carboxypeptidase-type gene [113]. |
| Rsc8 | SC8 | Kefeng No.1 [32] | Chromosome 2 Flanking markers: BARCSOYSSR_02_0610 BARCSOYSSR_02_0616 [32] Other markers: ZL-42 and ZL-52 | Glyma02g13310, Glyma02g13320, Glyma02g13400, Glyma02g13460, Glyma02g13470 [32] Glyma02g121500 and Glyma02g121600 (encoding MADS-box proteins) [114] |
| Rsc5 | SC5 | Kefeng No1 [28] | Chromosome 2 Flanking markers: Bin 352 Bin353 [28] | 11 candidate genes with Glyma02g13495 as the most plausible candidate [28] |
| Rsc20 | SC20 | Qihuang-1 [29] | Chromosome 13 Flanking markers: BARCSOYSSR_13_1099 BARCSOYSSR_13_1185 [29] | TIR-NBS-LRR type R genes: Glyma13g194700 and Glyma13g195100 [29]. |
| Rsc12 | SC12 | Qihuang-22 [115] | Chromosome 13 Flanking marker: Satt334 Sct_033 [115] | |
| Rsc3 | SC3 | Qihuang-1 [116] | Chromosome 13 [116] | Glyma13g25920, Glyma13g25950, Glyma13g25970, and Glyma13g26000 [116]. |
| Rsc3Q | SC3 | Qihuang-1 [117] | Chromosome 13 Flanking markers: BARCSOYSSR_13_1114 BARCSOYSSR_13_1136 [117] | Glyma13g25730, Glyma13g25750, Glyma13g25950, Glyma13g25970, and Glyma13g26000 [117]. |
| Rsc14Q | SC14 | Qihuang-1 [118,119] | Chromosome 13 Flanking markers: Sat_234 Sct_033 [118] Other markers: Satt334 MY750 [119] | |
| Rsc18 | SC18 | Kefeng No.1 [120] Qihuang-22 [120] | Chromosome 2 Flanking marker: BARCSOYSSR_02_0667 BARCSOYSSR_02_0670 [120] Chromosome 13 Flanking marker: SOYHSP176 Satt334 [120] | Glyma02g127800, Glyma02g128200 and Glyma02g128300 [120] |
| Rsc4 | SC4 | Dabaima [121] | Chromosome 14 Flanking markers: BARCSOYSSR_14_1413 BARCSOYSSR_14_1416 [31] | NB-LRR genes: Glyma14g38510 and Glyma14g38560 P450 family gene: Glyma14g38580 [31] |
| Host Factors | Roles in SMV Resistance | Reference |
|---|---|---|
| eEF1A | Targeted by P3, promotes SMV replication | [51] |
| GmEDR1, GmEDS1 GmPAD4 | Induce accumulation of SA, mediated resistance against SMV | [17] |
| GmHSP90 | Reduced the replication and movement of SMV-G2 (Rsv1-mediated resistance) | [17] |
| WRKY6 WRKY30 | Rsv1-mediated resistance against SMV-G2 | [17] |
| GmPP2C3a | Induces callose accumulation, restricts SMV movement | [100] |
| GmPEX14 | Induces burst of H2O2, (Rsc15-mediated resistance) | [30] |
| GmMPK4 | Negatively regulates SA accumulation and defense response | [123] |
| GmMPK6 | Repressor and activator in defense response | [124] |
| GmKR3 | Stimulates ABA accumulation | [25] |
| GmCYB5 | Targets the P3 protein to inhibit SMV accumulation | [24] |
| Tolerance Cultivar | Reference | |
|---|---|---|
| Transgenic GmAKT2 | Alter the level of potassium, reduce the spread of SMV | [136] |
| RNAi-mediated silencing of SMV P3 transgenic soybean | Exhibited stable and enhanced resistance to SMV SC3 and other potyviruses. | [137] |
| Transgenic GmKR3 | Enhances resistance against multiple viruses, including SMV-SC3, via ABA signaling | [25] |
| Attenuated SMV-Coat-protein mediated-resistance transgenic soybean | Highly resistant to SMV strain D and A (in Japan) | [138] |
| SMV-CP-RNAi transgenic soybean | Induces a functional gene silencing system and resulted in a viral-resistant phenotype. | [139] |
| Inverted repeat-SMV-HC-pro transgenic soybean | Induced RNA-mediated resistance via RNAi by targeting SMV-HC-pro | [140] |
| Soybean RNA interfere lines, silenced for eIF4E | Interferes viral replication cycles, increases broad-spectrum resistance against SMV-SC3, SC7,SC-15,SC18, and SMV-R | [134] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Widyasari, K.; Alazem, M.; Kim, K.-H. Soybean Resistance to Soybean Mosaic Virus. Plants 2020, 9, 219. https://doi.org/10.3390/plants9020219
Widyasari K, Alazem M, Kim K-H. Soybean Resistance to Soybean Mosaic Virus. Plants. 2020; 9(2):219. https://doi.org/10.3390/plants9020219
Chicago/Turabian StyleWidyasari, Kristin, Mazen Alazem, and Kook-Hyung Kim. 2020. "Soybean Resistance to Soybean Mosaic Virus" Plants 9, no. 2: 219. https://doi.org/10.3390/plants9020219
APA StyleWidyasari, K., Alazem, M., & Kim, K.-H. (2020). Soybean Resistance to Soybean Mosaic Virus. Plants, 9(2), 219. https://doi.org/10.3390/plants9020219