Using Rapid Chlorophyll Fluorescence Transients to Classify Vitis Genotypes
Abstract
1. Introduction
2. Results
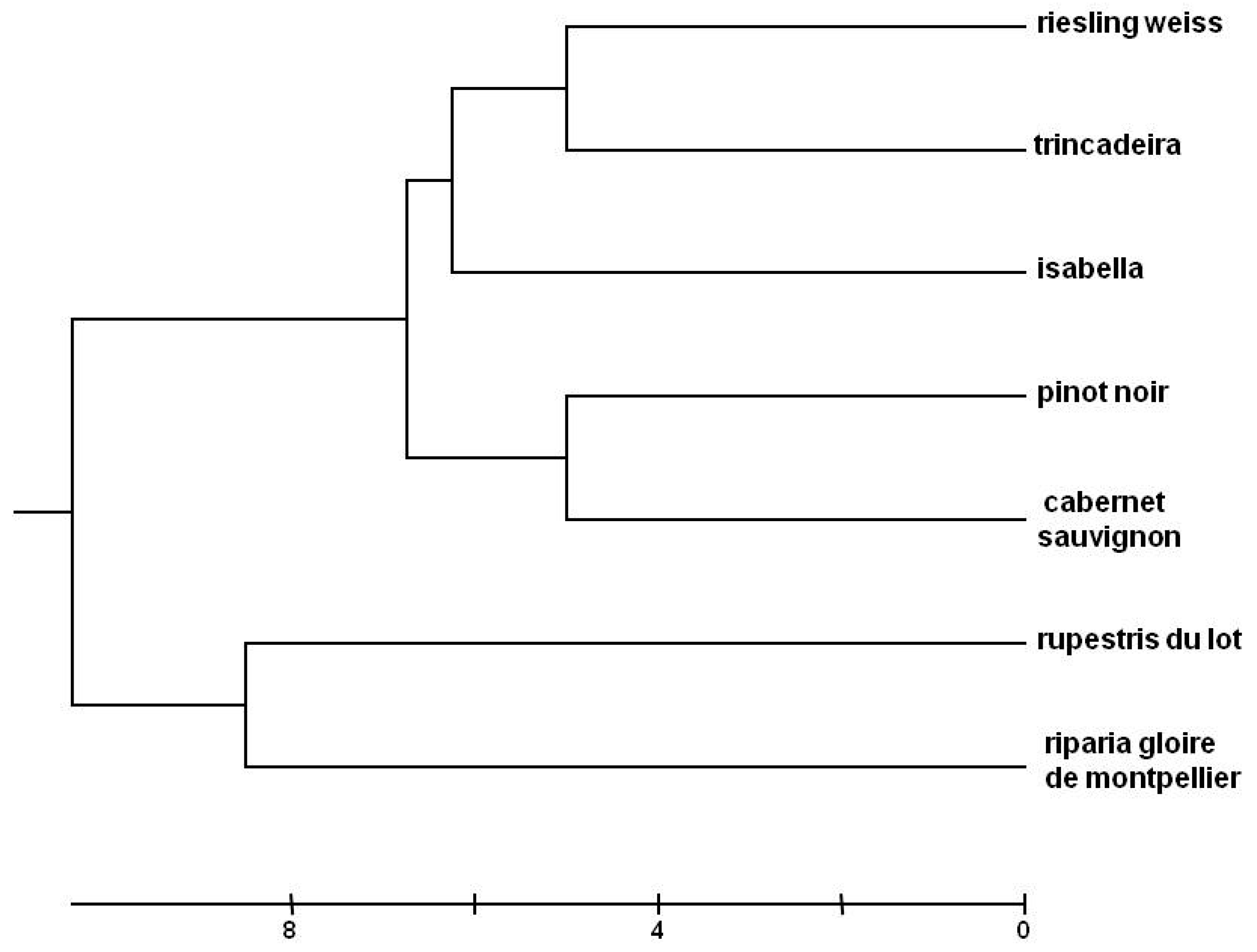
2.1. Genetic Analysis
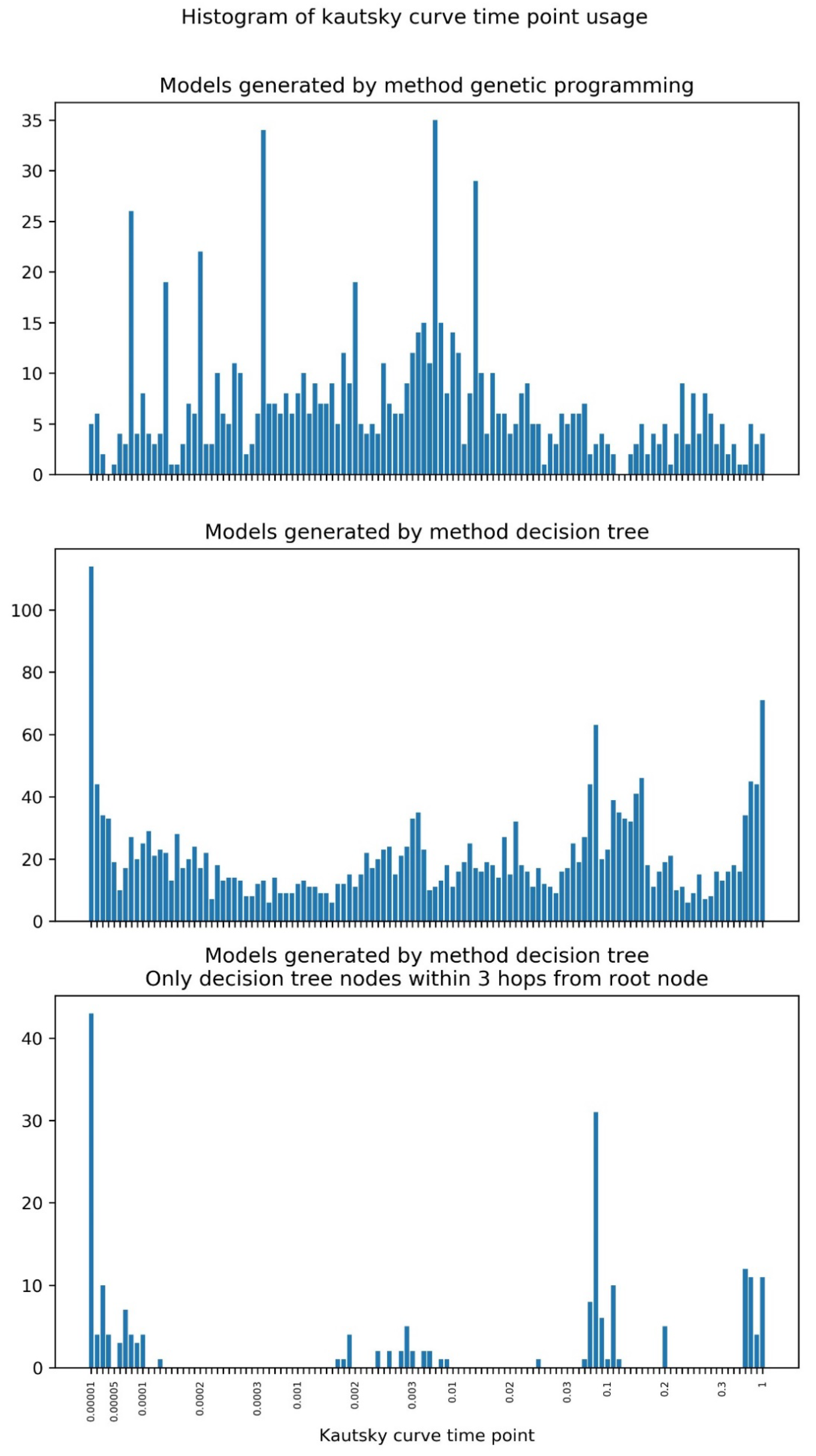
2.2. Machine Learning
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Genetic Analysis
4.3. Fluorescence Measurements
4.4. Machine Learning
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kautsky, H.; Hirsch, A. Neue Versuche zur Kohlensäureassimilation (New experiments on carbonic acid assimilation). Naturwissenschaften 1931, 19, 48. [Google Scholar] [CrossRef]
- Marques da Silva, J. Monitoring photosynthesis by in vivo chlorophyll fluorescence: Application to high-throughput plant phenotyping. In Applied Photosynthesis—New Progress; Najafpour, M.M., Ed.; InTech: Rijeka, Croatia, 2016; pp. 3–22. [Google Scholar]
- Strasser, R.J. Mono-bi-tri- and polypartite models in photosynthesis. Photosynth. Res. 1986, 10, 255–276. [Google Scholar] [CrossRef] [PubMed]
- Strasser, B.J. Donor side capacity of photosystem II probed by chlorophyll a fluorescence transients. Photosynth. Res. 1997, 52, 147–155. [Google Scholar] [CrossRef]
- Strasser, B.J.; Strasser, R.J. Measuring fast fluorescence transients to address environmental questions: The JIP-test. In Photosynthesis: From Light to Biosphere; Mathis, P., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1995; pp. 977–980. [Google Scholar]
- Jee, G. Sixty-three years since Kautsky: Chlorophyll a fluorescence. Aust. J. Plant Physiol. 1995, 22, 131–160. [Google Scholar]
- Kalaji, H.M.; Schansker, G.; Ladle, R.J.; Goltsev, V.; Bosa, K.; Allakhverdiev, S.I.; Brestic, M.; Bussotti, F.; Calatayud, A.; Dąbrowski, P.; et al. Frequently asked questions about in vivo chlorophyll fluorescence: Practical issues. Photosynth. Res. 2014, 122, 121–158. [Google Scholar] [CrossRef]
- Srivastava, A.; Guissé, B.; Greppin, H.; Strasser, R.J. Regulation of antenna structure and electron transport in photosystem II of Pisum sativum under elevated temperature probed by the fast polyphasic chlorophyll a fluorescence transient: OKJIP. Biochim. Biophys. Acta 1997, 1320, 95–106. [Google Scholar] [CrossRef]
- Tsimilli-Michael, M.; Eggenberg, P.; Biro, B.; Köves Pechy, K.; Vörös, I.; Strasser, R.J. Synergistic and antagonistic effects of arbuscular mycorrhizal fungi and Azospirillum and Rizhobium nitrogen-fixers on the photosynthetic activity of alfalfa probed by the polyphasic chlorophyll a fluorescence transient OJIP. Appl. Soil Ecol. 2000, 15, 169–182. [Google Scholar] [CrossRef]
- Demetriou, G.; Neonaki, C.; Navakoudis, E.; Kotzabasis, K. Salt stress impact on the molecular structure and function of the photosynthetic apparatus—The protective role of polyamines. Biochim. Biophys. Acta 2007, 1767, 272–280. [Google Scholar] [CrossRef]
- Zivcák, M.; Brestic, M.; Olsovská, K.; Slamka, P. Performance index as a sensitive indicator of water stress in Triticum aestivum L. Plant Soil Environ. 2008, 54, 133–139. [Google Scholar] [CrossRef]
- Mathur, S.; Mehta, P.; Jajoo, A. Effects of dual stress (high salt and high temperature) on the photochemical efficiency of wheat leaves (Triticum aestivum). Physiol. Mol. Biol. Plants 2013, 19, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Silvestre, S.; Araújo, S.S.; Vaz Patto, M.C.; Marques da Silva, J. Performance index: An expeditious tool to screen for improved drought resistance in the Lathyrus genus. J. Integr. Plant Biol. 2014, 56, 610–621. [Google Scholar] [CrossRef]
- Costa, J.M.; Marques da Silva, J.; Pinheiro, C.; Barón, M.; Mylona, P.; Centritto, M.; Haworth, M.; Loreto, F.; Uzilday, B.; Turkan, I.; et al. Opportunities and limitations of crop phenotyping in Southern European countries. Front. Plant Sci. 2019, 10, 1125. [Google Scholar] [CrossRef]
- USDA/NSF. Phenomics: Genotype to Phenotype; NIFA-NSF Phenomics Workshop Report; USDA/NSF: St. Louis, MO, USA, 2011. [Google Scholar]
- Tyystjarvi, E.; Koski, A.; Keranen, M.; Nevalainen, O. The Kautsky curve is a built-in bar code. Biophys. J. 1999, 77, 1159–1167. [Google Scholar] [CrossRef]
- OIV—Organisation Internationale de la Vigne et du Vin. State of the Vitiviniculture World Market. 2018. Available online: http://www.oiv.int/public/medias/5958/oiv-state-of-the-vitiviniculture-world-market-april-2018.pdf (accessed on 15 January 2020).
- MAMAOT Portaria n° 380/2012, de 22 de novembro, do Ministério da Agricultura, do Mar, do Ambiente e do Ordenamento do Território (MAMAOT). In Diário da República; 1.ª Série—N.° 226; Imprensa Nacional—Casa da Moeda: Lisbon, Portugal, 2012.
- Almadanim, M.C.; Baleiras-Couto, M.M.; Pereira, H.S.; Carneiro, L.C.; Fevereiro, P.; Eiras-Dias, J.E.; Morais-Cecilio, L.; Viegas, W.; Veloso, M.M. Genetic diversity of the grapevine (Vitis vinifera L.) cultivars most utilized for wine production in Portugal. Vitis 2007, 46, 116–119. [Google Scholar]
- Veloso, M.M.; Almandanim, M.C.; Baleiras-Couto, M.; Pereira, H.S.; Carneiro, L.C.; Fevereiro, P.; Eiras-Dias, J. Microsatellite database of grapevine (Vitis vinifera L.) cultivars used for wine production in Portugal. Ciência Téc. Vitiv. 2010, 25, 53–61. [Google Scholar]
- Eiras-Dias, J.E.; Faustino, R.; Clímaco, P.; Fernandes, P.; Cruz, A.; Cunha, J.; Veloso, M.; Castro, R. Catálogo das Castas Para Vinho Cultivadas Em Portugal. Volume 1. Instituto da Vinha e do Vinho I.P.; Chaves Ferreira—Publicações: Lisboa, Portugal, 2011. [Google Scholar]
- Eiras-Dias, J.E.; Faustino, R.; Clímaco, P.; Fernandes, P.; Cruz, A.; Cunha, J.; Veloso, M.; Castro, R. Catálogo das Castas Para Vinho Cultivadas em Portugal. Volume 2. Instituto da Vinha e do Vinho I.P.; Chaves Ferreira—Publicações: Lisboa, Portugal, 2011. [Google Scholar]
- Cunha, J.; Ibáñez, J.; Teixeira-Santos, M.; Brazão, J.; Fevereiro, P.; Martínez-Zapater, J.M.; Eiras-Dias, J.E. Characterisation of the Portuguese grapevine germplasm with 48 single nucleotide polymorphisms. Aust. J. Grape Wine Res. 2016, 22, 504–516. [Google Scholar] [CrossRef]
- Tomic, L.; Stajner, N.; Javornik, B. Characterization of grapevines by the use of genetic markers. In The Mediterranean Genetic Code—Grapevine and Olive; Sladonja, B., Ed.; InTech: Rijeka, Croatia, 2013. [Google Scholar]
- Sefc, K.M.; Lopes, M.S.; Lefort, F.; Botta, R.; Roubelakis-Angelakis, K.A.; Ibáñez, J.; Pejić, I.; Wagner, H.W.; Glössl, J.; Steinkellner, H. Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Appl. Genet. 2000, 100, 498–505. [Google Scholar] [CrossRef]
- Lopes, M.S.; Rodrigues dos Santos, M.; Eiras-Dias, J.E.; Mendonça, D.; Câmara Machado, A. Discrimination of Portuguese grapevines based on microsatellite markers. J. Biotechnol. 2006, 127, 34–44. [Google Scholar] [CrossRef]
- Gameiro, C.; Pereira, S.; Figueiredo, A.; Bernardes da Silva, A.; Matos, A.R.; Pires, M.C.; Teubig, P.; Burnay, N.; Moniz, L.; Mariano, P.; et al. Preliminary results on the use of chlorophyll fluorescence and artificial intelligence techniques to automatically characterize plant water status. In Proceedings of the Actas del XIII Simposio Hispano-Portugués de Relaciones Hídricas en las Plantas—Aprendiendo a Optimizar el uso del Agua en las Plantas Para Hacer de Nuestro Entorno un Ambiente Más Soastenible, Pamplona, Espanha, 18–20 October 2016; pp. 15–18. Available online: https://www.unav.edu/documents/10990541/0/resumenes_simposio.pdf/be5b4c16-ff51-4cf9-a10f-aefc3f474fa4 (accessed on 19 November 2019).
- Harris, E.H.; Boynton, J.E.; Gillham, N.W. Chloroplast ribosomes and protein synthesis. Microbiol. Rev. 1994, 58, 700–754. [Google Scholar] [CrossRef]
- Woodson, J.D.; Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008, 9, 383–395. [Google Scholar] [CrossRef]
- Mitchell, T.M. Machine Learning; McGraw-Hill: New York, NY, USA, 1997. [Google Scholar]
- Sipper, M.; Fu, W.; Ahuja, K.; Moore, J.H. Investigating the parameter space of evolutionary algorithms. Biodata Min. 2018, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Sipper, M.; Fu, W.; Ahuja, K.; Moore, J.H. Correction to: Investigating the parameter space of evolutionary algorithms. BioData Min. 2019, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Zivcák, M.; Brestic, M.; Kalaji, H.M.; Govindjee. Photosynthetic responses of sun- and shade-grown barley leaves to high light: Is the lower PS II connectivity in shade leaves associated with protection against excess of light? Photosynth. Res. 2014, 119, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Öz, M.T.; Turan, Ö.; Kayihan, C.; Eyidoğan, F.; Ekmekçi, Y.; Yücel, M.; Öktem, H.A. Evaluation of photosynthetic performance of wheat cultivars exposed to boron toxicity by the JIP fluorescence test. Photosynthetica 2014, 52, 555–563. [Google Scholar] [CrossRef]
- Jedmowski, C.; Brüggemann, W. Imaging of fast chlorophyll fluorescence induction curve (OJIP) parameters, applied in a screening study with wild barley (Hordeum spontaneum) genotypes under heat stress. J. Photochem. Photobiol. B Biol. 2015, 151, 153–160. [Google Scholar] [CrossRef]
- Fernandes, A.; Utkin, A.; Eiras-Dias, J.; Silvestre, J.; Cunha, J.; Melo-Pinto, P. Assessment of grapevine variety discrimination using stem hyperspectral data and AdaBoost of random weight neural networks. Appl. Soft Comput. 2018, 72, 140–155. [Google Scholar] [CrossRef]
- Odilbekov, F.; Armoniené, R.; Henriksson, T.; Chawade, A. Proximal phenotyping and machine learning methods to identify Septoria tritici blotch disease symptoms in wheat. Front. Plant Sci. 2018, 9, 685. [Google Scholar] [CrossRef]
- Vitis International Catalogue of Varieties. Available online: www.vivc.de (accessed on 1 March 2018).
- Thomas, M.R.; Matsumoto, S.; Cain, P.; Scott, N.S. Repetitive DNA of grapevine: Classes present and sequences suitable for cultivar identification. Appl. Genet. 1993, 86, 173–180. [Google Scholar] [CrossRef]
- OIV—Organisation Internationale de la Vigne et du Vin. Descriptor List for Grapevine Cultivars and Vitis Species, 2nd ed.; Organisation Internationale de la Vigne et du Vin: Paris, France, 2009. [Google Scholar]
- Alifragkis, A.; Cunha, J.; Pereira, J.; Fevereiro, P.; Eiras Dias, J.E. Identity, Synonymies and Homonynies of Minor Grapevine Cultivars Maintained in the Portuguese Ampelographic Collection. Ciência Téc. Vitiv. 2015, 30, 43–52. [Google Scholar] [CrossRef]
- Thomas, M.R.; Scott, N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Appl. Genet. 1993, 86, 985–990. [Google Scholar] [CrossRef]
- Bowers, J.E.; Meredith, C.P. The parentage of classic wine grape: Cabernet Sauvignon. Nat. Genet. 1996, 16, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Dang, L.; Gerald, S.; Meredith, C.P. Development and characterization of additional microsatellite DNA markers for grape. Am. J. Enol. Vitic. 1999, 50, 243–246. [Google Scholar]
- Sefc, K.M.; Regner, F.; Turetschek, E.; Glössl, J.; Steinkellner, H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 1999, 42, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Doligez, A.; Adam-Blondon, A.F.; Cipriani, G.; Di Gaspero, G.; Laucou, V.; Merdinoglu, D.; Meredith, C.P.; Riaz, S.; Roux, C.; This, P. An integrated SSR map of grapevine based on five mapping populations. Appl. Genet. 2006, 113, 369–382. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Domingos, P. The Master Algorithm: How the Quest for the Ultimate Learning Machine Will Remake Our World; Basic Books: New York, NY, USA, 2015. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Silva, S. GPLAB—A Genetic Programming Toolbox for MATLAB (Version 4.04). 2018. Available online: http://gplab.sourceforge.net (accessed on 15 January 2020).
- Poli, R.; Langdon, W.B.; McPhee, N.F. A Field Guide to Genetic Programming. 2008. Available online: http://www.gp-field-guide.org.uk (accessed on 18 November 2019).
- Muñoz, L.; Silva, S.; Trujillo, L. M3GP—Multiclass classification with GP. In Proceedings of the European Conference on Genetic Programming, Copenhagen, Denmark, 8–10 April 2015; pp. 78–91. [Google Scholar]
- Silva, S.; Muñoz, L.; Trujillo, L.; Ingalalli, V.; Castelli, M.; Vanneschi, L. Multiclass classification through multidimensional clustering. In Genetic Programming Theory and Practice XIII; Springer: Cham, Switzerland, 2016; pp. 219–239. [Google Scholar]

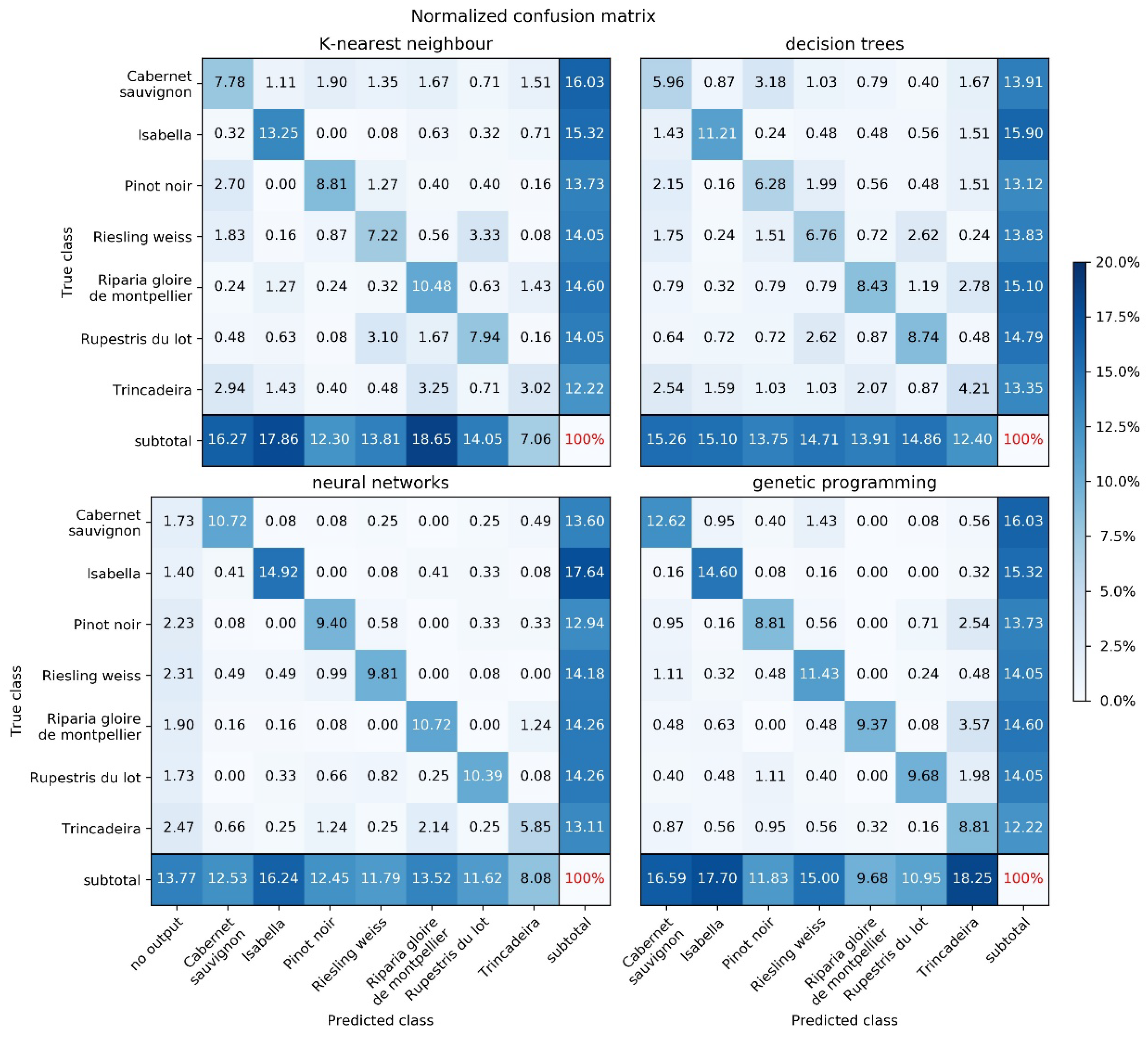
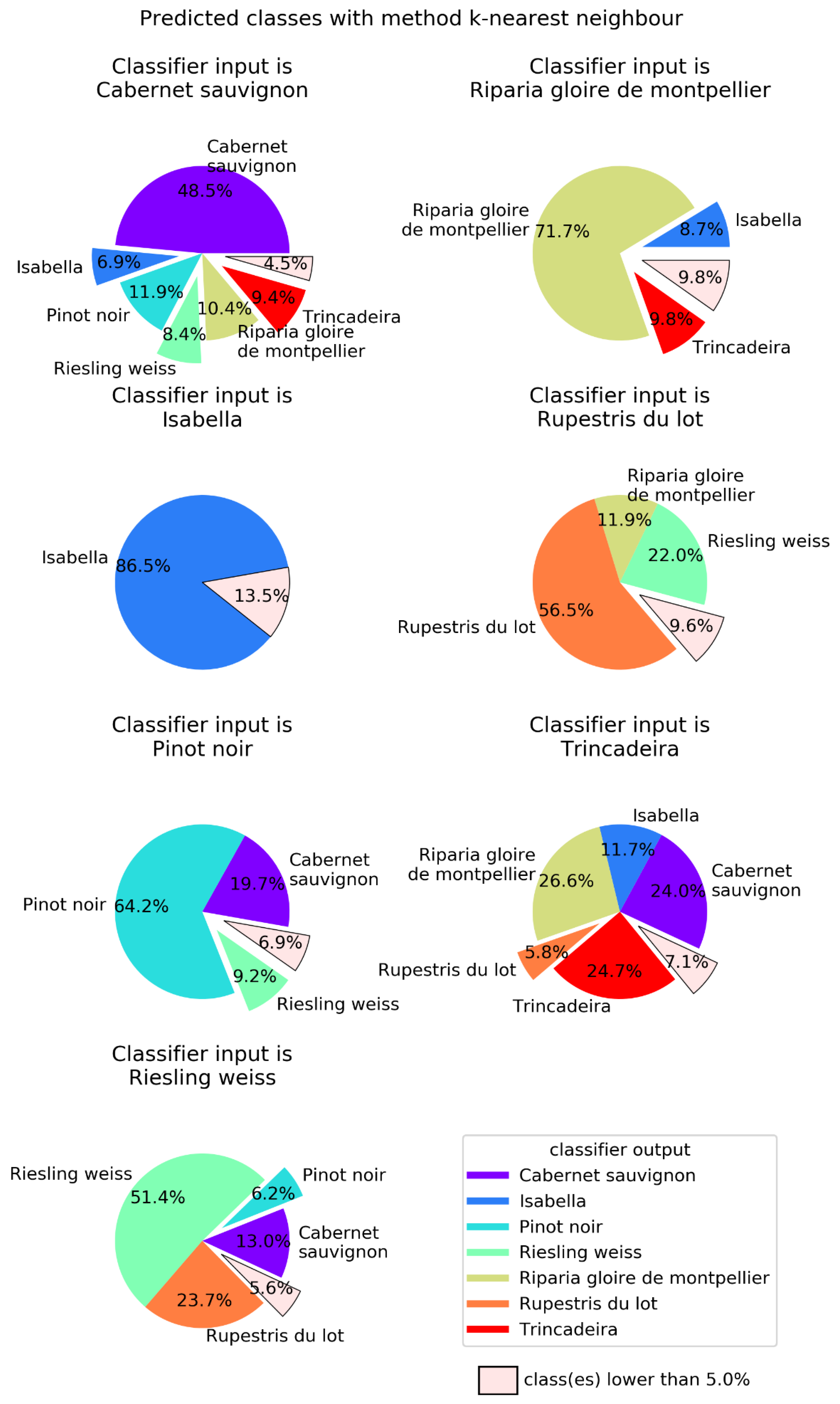
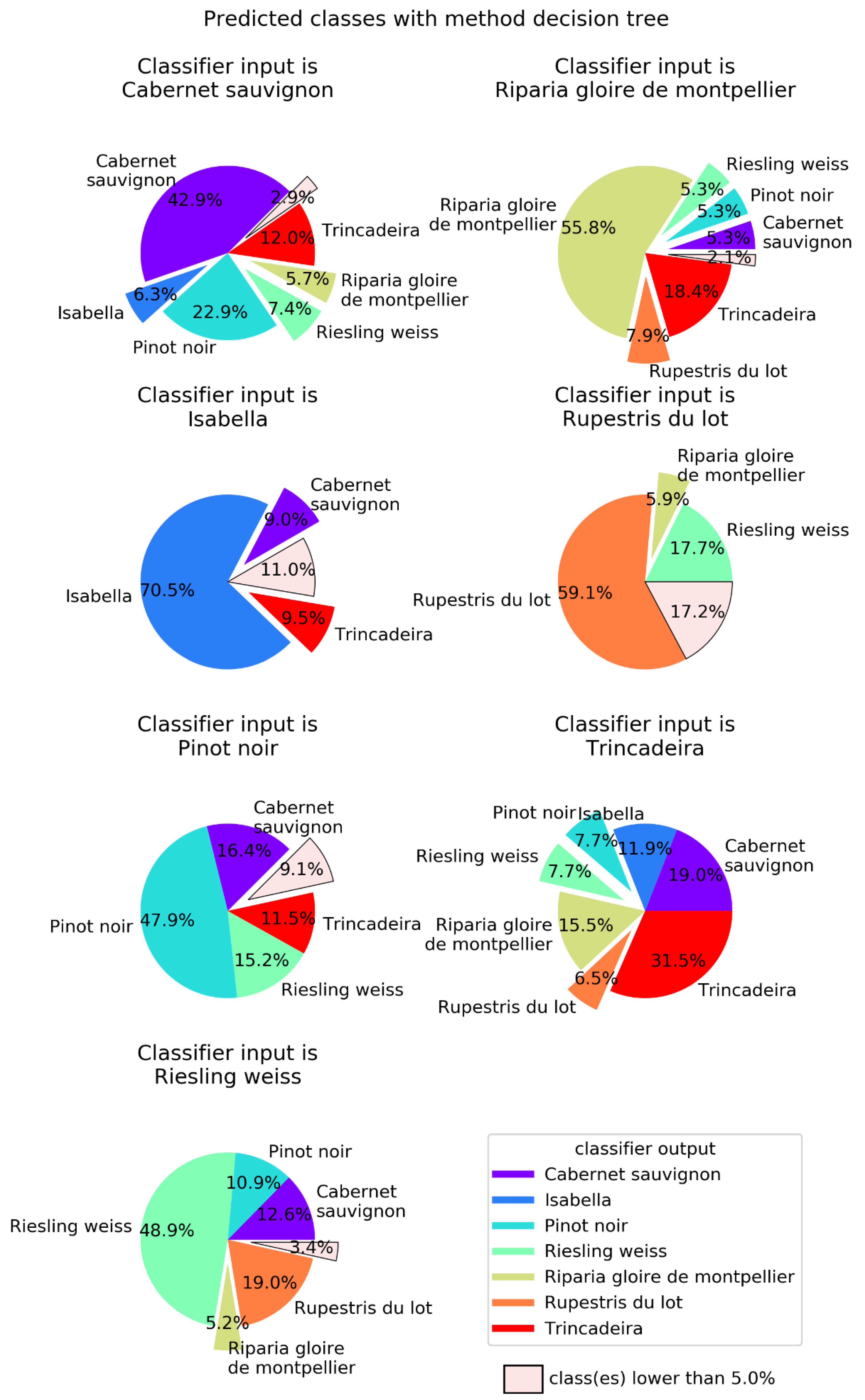


| Method | Success Rate | Main Parameters |
|---|---|---|
| K-nearest neighbors | 58.5% | Number of neighbors: 5 |
| Decision tree | 51.6% | Split criterion: entropy Maximum tree depth: 19 Minimum number of samples in a node: 5 |
| Neural network | 71.8% | Number of neurons: 5000 Activation function: logistic |
| Genetic programming | 75.3% | Number of individuals: 250 Number of generations: 100 |
| Genotype | Variety | Acession PRT051 | VIVC | Photo (VIVC) * | Leaf Colour | Leaf Bright | Country of Origin |
|---|---|---|---|---|---|---|---|
| Vitis rupestris Scheele | Rupestris du Lot | 13,821 | 10,389 |  | Light green | bright | France |
| Vitis riparia Michaux | Riparia Gloire de Montpellier | 13,822 | 4824 |  | Dark green | dull | France |
| Vitis interspecific crossing | Isabella | 13,619 | 5560 |  | Dark green | dull | United States of America |
| Vitis vinifera Linné subsp. vinifera | Pinot Noir | 10,918 | 9279 |  | green | dull | France |
| Vitis vinifera Linné subsp. vinifera | Cabernet Sauvignon | 10,714 | 1929 |  | Light green | Slightly bright | France |
| Vitis vinifera Linné subsp. vinifera | Riesling Weiss | 13,413 | 10,077 |  | Dark green | Slightly bright | Germany |
| Vitis vinifera Linné subsp. vinifera | Trincadeira | 11,402 | 15,685 |  | Dark green | Very bright | Portugal |
| SSR Name | Linkage Group | Microsatellite Repeat Motif | Reference |
|---|---|---|---|
| VVS2 | 11 | (GA)n | Thomas and Scott [42] |
| VVMD5 | 16 | (CT)nAT(CT)nATAG(AT)n | Bowers and Meredith [43] |
| VVMD7 | 7 | (CT)n | Bowers and Meredith [43] |
| VVMD25 | 11 | (CT)n | Bowers et al. [44] |
| VVMD27 | 5 | (CT)n | Bowers et al. [44] |
| VVMD28 | 3 | (CT)n | Bowers et al. [44] |
| VVMD32 | 4 | (CT)n | Bowers et al. [44] |
| VRZAG62 | 7 | (GA)n | Sefc et al. [45]/Doligez et al. [46] |
| VRZAG79 | 5 | (GA)n | Sefc et al. [45]/Doligez et al. [46] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marques da Silva, J.; Figueiredo, A.; Cunha, J.; Eiras-Dias, J.E.; Silva, S.; Vanneschi, L.; Mariano, P. Using Rapid Chlorophyll Fluorescence Transients to Classify Vitis Genotypes. Plants 2020, 9, 174. https://doi.org/10.3390/plants9020174
Marques da Silva J, Figueiredo A, Cunha J, Eiras-Dias JE, Silva S, Vanneschi L, Mariano P. Using Rapid Chlorophyll Fluorescence Transients to Classify Vitis Genotypes. Plants. 2020; 9(2):174. https://doi.org/10.3390/plants9020174
Chicago/Turabian StyleMarques da Silva, Jorge, Andreia Figueiredo, Jorge Cunha, José Eduardo Eiras-Dias, Sara Silva, Leonardo Vanneschi, and Pedro Mariano. 2020. "Using Rapid Chlorophyll Fluorescence Transients to Classify Vitis Genotypes" Plants 9, no. 2: 174. https://doi.org/10.3390/plants9020174
APA StyleMarques da Silva, J., Figueiredo, A., Cunha, J., Eiras-Dias, J. E., Silva, S., Vanneschi, L., & Mariano, P. (2020). Using Rapid Chlorophyll Fluorescence Transients to Classify Vitis Genotypes. Plants, 9(2), 174. https://doi.org/10.3390/plants9020174









