Phytic Acid and Transporters: What Can We Learn from low phytic acid Mutants?
Abstract
1. Introduction
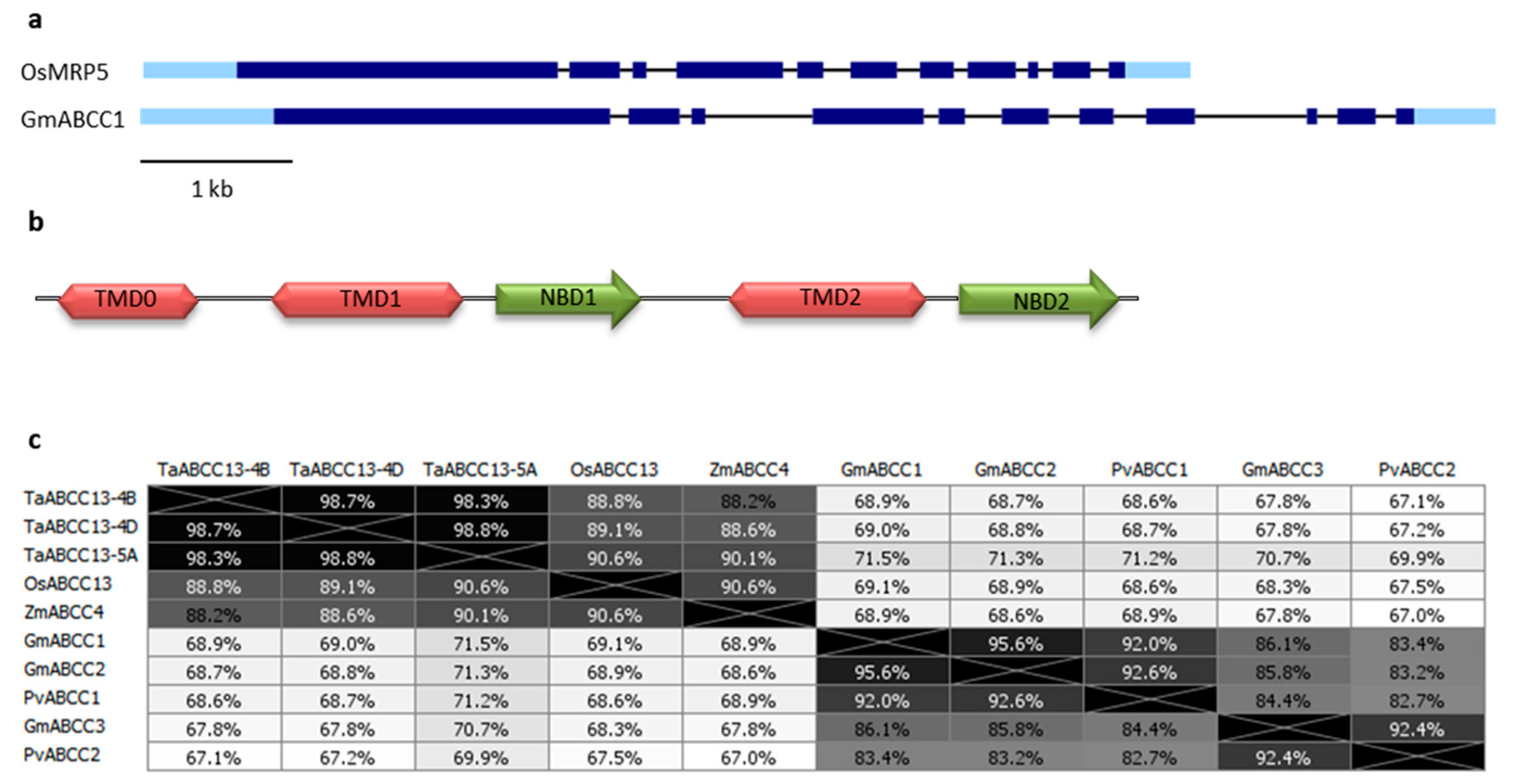
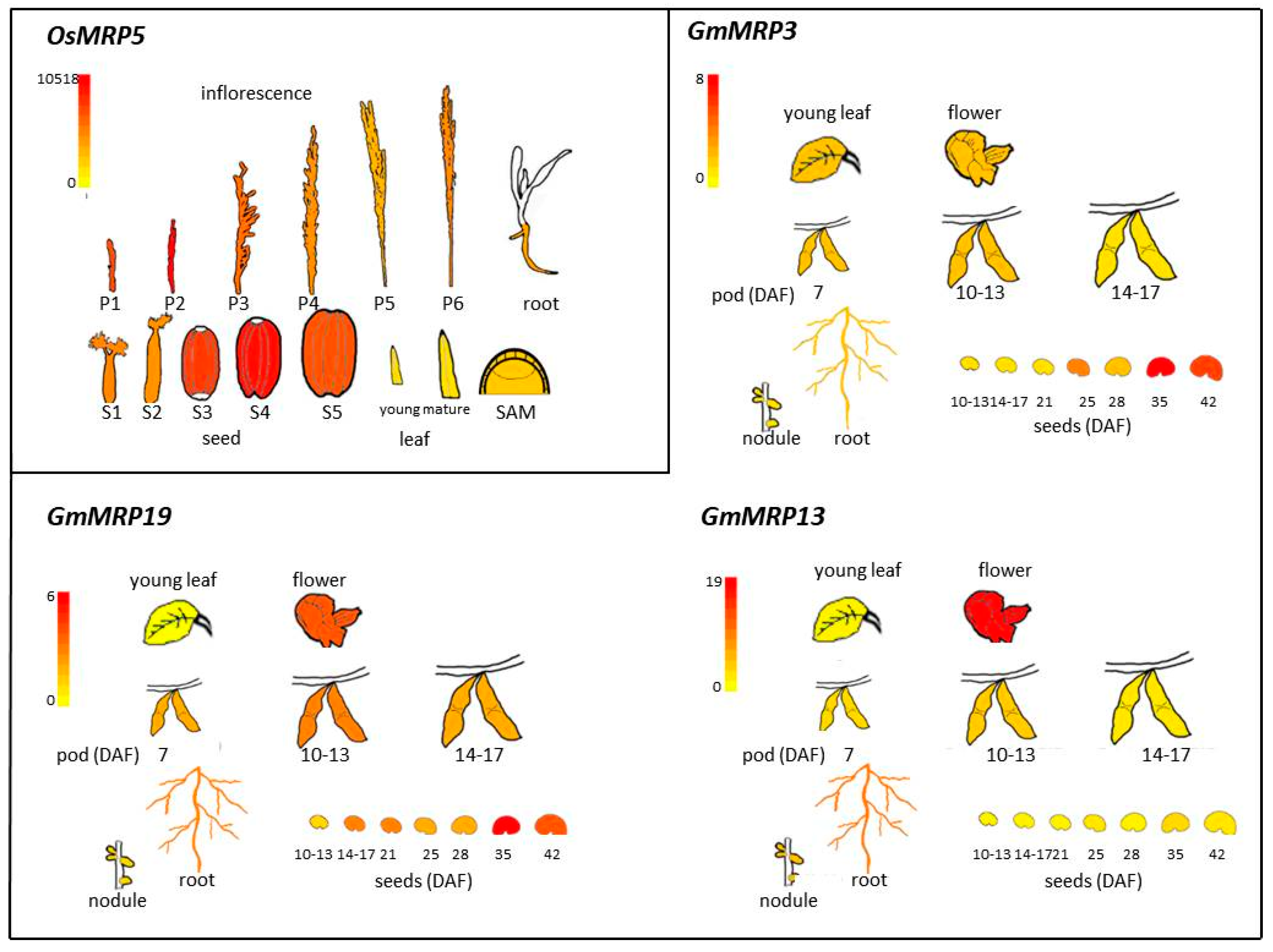
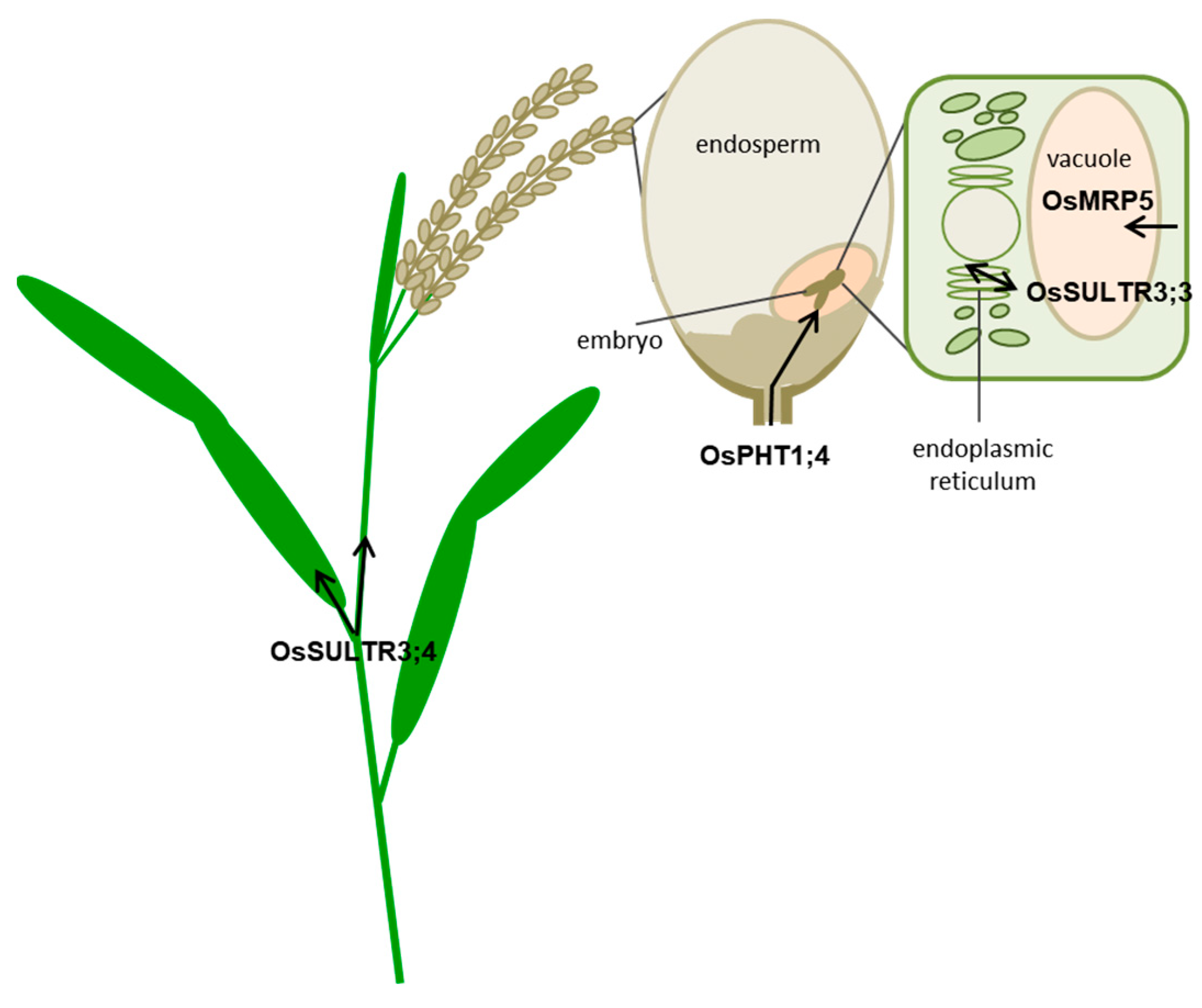
2. PA-MRP Transporters
lpa Mutants in PA-MRP Transporters
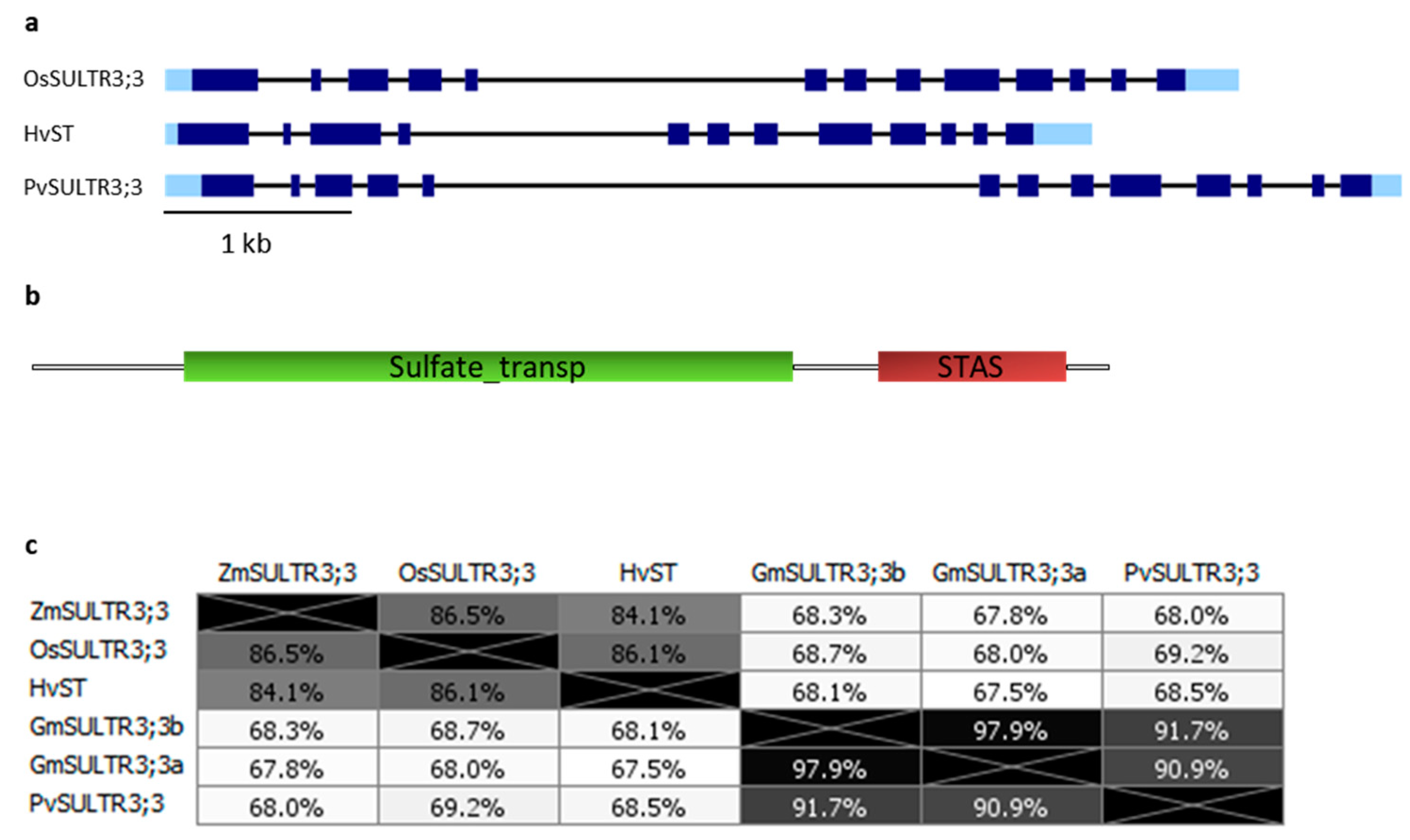
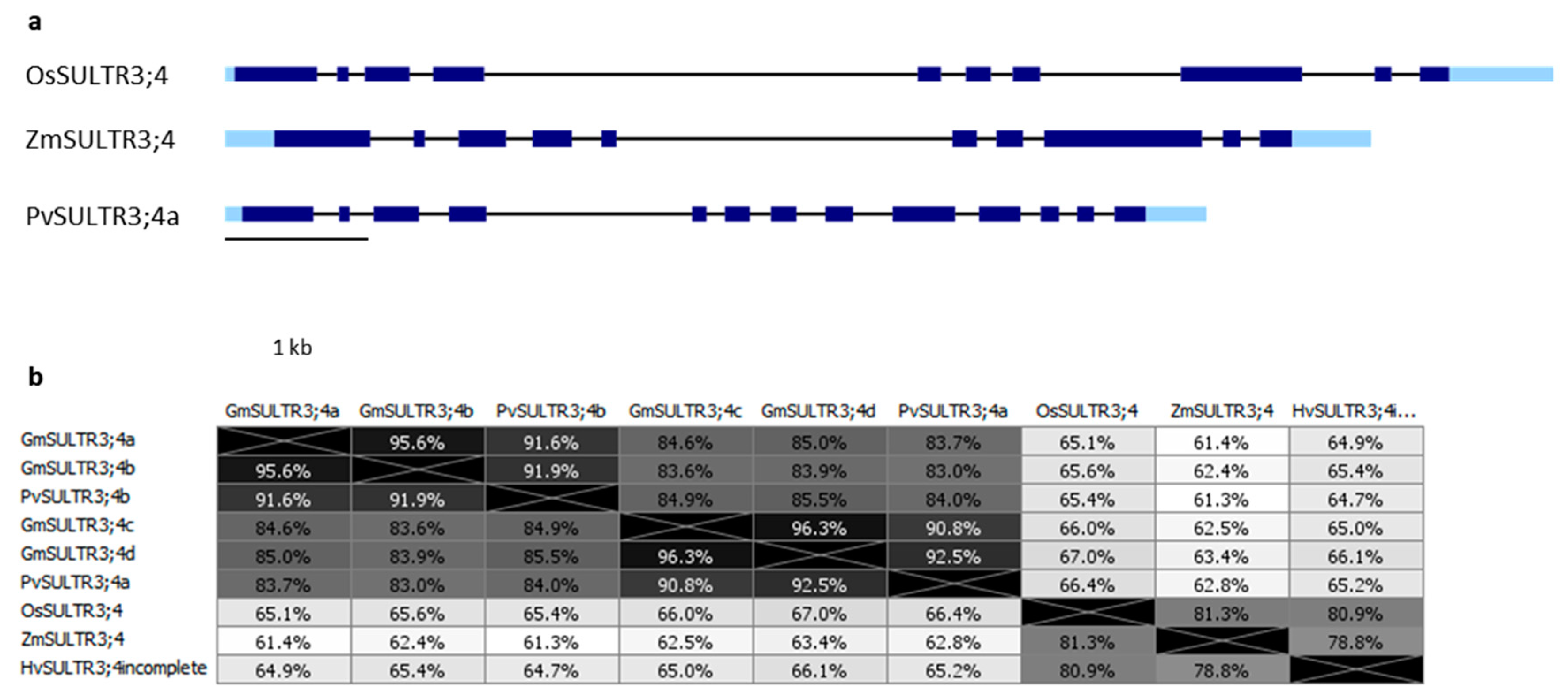
3. SULTR3.3 and SULTR3.4 Transporters Involved in PA Metabolism
3.1. SULTR3;3
3.2. SULTR3;4
3.3. lpa Mutants in SULTR Transporters
3.3.1. Mutants Affected in the SULTR3;3 Genes
3.3.2. The spdt Mutants
4. OsPht1;4 Phosphate Transporter
Ospt4 Mutants, OsPT4 RNAi and Overexpression Lines
5. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Sparvoli, F.; Cominelli, E. Seed biofortification and phytic acid reduction: A conflict of interest for the plant? Plants 2015, 4, 728–755. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.; Tsai, S.; Vitorello, V. Dynamics of inositol phosphate pools (tris-, tetrakis- and pentakisphosphate) in relation to the rate of phytate synthesis during seed development in common bean (Phaseolus vulgaris). J. Plant Physiol. 2005, 162, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hatzack, F.; Johansen, K.; Rasmussen, S. Nutritionally relevant parameters in low-phytate barley (Hordeum vulgare L.) grain mutants. J. Agric. Food Chem. 2000, 48, 6074–6080. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Ockenden, I.; Lott, J. The concentrations and distribution of phytic acid-phosphorus and other mineral nutrients in wild-type and low phytic acid1-1 (lpa1-1) corn (Zea mays L.) grains and grain parts. Can. J. Bot. 2005, 83, 131–141. [Google Scholar] [CrossRef]
- Ockenden, I.; Dorsch, J.; Reid, M.; Lin, L.; Grant, L.; Raboy, V.; Lott, J. Characterization of the storage of phosphorus, inositol phosphate and cations in grain tissues of four barley (Hordeum vulgare L.) low phytic acid genotypes. Plant Sci. 2004, 167, 1131–1142. [Google Scholar] [CrossRef]
- Regvar, M.; Eichert, D.; Kaulich, B.; Gianoncelli, A.; Pongrac, P.; Vogel-Mikus, K.; Kreft, I. New insights into globoids of protein storage vacuoles in wheat aleurone using synchrotron soft X-ray microscopy. J. Exp. Bot. 2011, 62, 3929–3939. [Google Scholar] [CrossRef]
- Krishnan, H. Preparative procedures markedly influence the appearance and structural integrity of protein storage vacuoles in soybean seeds. J. Agric. Food Chem. 2008, 56, 2907–2912. [Google Scholar] [CrossRef]
- O’Dell, B.L.; de Boland, A.R.; Koirtyohann, S.T. Distribution of phytate and nutritionally important elements among the morphological components of cereal grains. J. Agric. Food Chem. 1972, 20, 718–721. [Google Scholar] [CrossRef]
- Ariza-Nieto, M.; Blair, M.; Welch, R.; Glahn, R. Screening of iron bioavailability patterns in eight bean (Phaseolus vulgaris L.) genotypes using the caco-2 cell in vitro model. J. Agric. Food Chem. 2007, 55, 7950–7956. [Google Scholar] [CrossRef]
- Raboy, V. myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 2003, 64, 1033–1043. [Google Scholar] [CrossRef]
- Raboy, V. Seeds for a better future: ‘low phytate’, grains help to overcome malnutrition and reduce pollution. Trends Plant Sci. 2001, 6, 458–462. [Google Scholar] [CrossRef]
- Schlemmer, U.; Frølich, W.; Prieto, R.M.; Grases, F. Phytate in foods and significance for humans: Food sources, intake, processing, bioavailability, protective role and analysis. Mol. Nutr. Food Res. 2009, 53 (Suppl. 2), S330–S375. [Google Scholar] [CrossRef] [PubMed]
- Leytem, A.B.; Maguire, R.O. Environmental implications of inositol phosphates in animal manures. In Inositol Phosphates: Linking Agriculture and the Environment; Turner, B.L., Richardson, A.E., Mullaney, E.J., Eds.; CAB International: Wallingford, CT, USA; Oxfordshire, UK, 2007; pp. 150–168. [Google Scholar]
- Raboy, V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009, 177, 281–296. [Google Scholar] [CrossRef]
- Martinoia, E. Vacuolar transporters—Companions on a longtime journey. Plant Physiol. 2018, 176, 1384–1407. [Google Scholar] [CrossRef]
- Nagy, R.; Grob, H.; Weder, B.; Green, P.; Klein, M.; Frelet-Barrand, A.; Schjoerring, J.; Brearley, C.; Martinoia, E. The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 2009, 284, 33614–33622. [Google Scholar] [CrossRef]
- Sparvoli, F.; Cominelli, E. Phytate Transport by MRPs. In Plant ABC Transporters; Geisler, M., Ed.; Springer: Cham, Switzerland, 2014; pp. 19–38. [Google Scholar]
- Takahashi, H.; Buchner, P.; Yoshimoto, N.; Hawkesford, M.J.; Shiu, S.H. Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2011, 2, 119. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Zhang, X.; Broughton, S.; Westcott, S.; Wu, D.; Lance, R.; Li, C. A nonsense mutation in a putative sulphate transporter gene results in low phytic acid in barley. Funct. Integr. Genom. 2011, 11, 103–110. [Google Scholar] [CrossRef]
- Yamaji, N.; Takemoto, Y.; Miyaji, T.; Mitani-Ueno, N.; Yoshida, K.T.; Ma, J.F. Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 2017, 541, 92–95. [Google Scholar] [CrossRef]
- Zhang, F.; Sun, Y.; Pei, W.; Jain, A.; Sun, R.; Cao, Y.; Wu, X.; Jiang, T.; Zhang, L.; Fan, X.; et al. Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 2015, 82, 556–569. [Google Scholar] [CrossRef]
- Ye, Y.; Yuan, J.; Chang, X.; Yang, M.; Zhang, L.; Lu, K.; Lian, X. The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLoS ONE 2015, 10, e0126186. [Google Scholar] [CrossRef]
- Hwang, J.U.; Song, W.Y.; Hong, D.; Ko, D.; Yamaoka, Y.; Jang, S.; Yim, S.; Lee, E.; Khare, D.; Kim, K.; et al. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol. Plant 2016, 9, 338–355. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, H.; Schellin, K.; Li, B.; Faller, M.; Stoop, J.; Meeley, R.; Ertl, D.; Ranch, J.; Glassman, K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 2007, 25, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Gaedeke, N.; Klein, M.; Kolukisaoglu, U.; Forestier, C.; Muller, A.; Ansorge, M.; Becker, D.; Mamnun, Y.; Kuchler, K.; Schulz, B.; et al. The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J. 2001, 20, 1875–1887. [Google Scholar] [CrossRef]
- Klein, M.; Perfus-Barbeoch, L.; Frelet, A.; Gaedeke, N.; Reinhardt, D.; Mueller-Roeber, B.; Martinoia, E.; Forestier, C. The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signalling and water use. Plant J. 2003, 33, 119–129. [Google Scholar] [CrossRef]
- Suh, S.J.; Wang, Y.F.; Frelet, A.; Leonhardt, N.; Klein, M.; Forestier, C.; Mueller-Roeber, B.; Cho, M.H.; Martinoia, E.; Schroeder, J.I. The ATP binding cassette transporter AtMRP5 modulates anion and calcium channel activities in Arabidopsis guard cells. J. Biol. Chem. 2007, 282, 1916–1924. [Google Scholar] [CrossRef]
- Liu, Q.; Xu, X.; Ren, X.; Fu, H.; Wu, D.; Shu, Q. Generation and characterization of low phytic acid germplasm in rice (Oryza sativa L.). Theor. Appl. Genet. 2007, 114, 803–814. [Google Scholar] [CrossRef]
- Maroof, M.; Glover, N.; Biyashev, R.; Buss, G.; Grabau, E. Genetic basis of the low-phytate trait in the soybean line CX1834. Crop Sci. 2009, 49, 69–76. [Google Scholar] [CrossRef]
- Gillman, J.; Pantalone, V.; Bilyeu, K. The low phytic acid phenotype in soybean line CX1834 is due to mutations in two homologs of the maize low phytic acid gene. Plant Genome 2009, 2, 179–190. [Google Scholar] [CrossRef]
- Gillman, J.; Baxter, I.; Bilyeu, K. Phosphorus partitioning of soybean lines containing different mutant alleles of two soybean seed-specific adenosine triphosphate-binding cassette phytic acid transporter paralogs. Plant Genome 2013, 6. [Google Scholar] [CrossRef]
- Panzeri, D.; Cassani, E.; Doria, E.; Tagliabue, G.; Forti, L.; Campion, B.; Bollini, R.; Brearley, C.A.; Pilu, R.; Nielsen, E.; et al. A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol. 2011, 191, 70–83. [Google Scholar] [CrossRef]
- Bhati, K.K.; Alok, A.; Kumar, A.; Kaur, J.; Tiwari, S.; Pandey, A.K. Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 2016, 67, 4379–4389. [Google Scholar] [CrossRef] [PubMed]
- Boncompagni, E.; Orozco-Arroyo, G.; Cominelli, E.; Gangashetty, P.I.; Grando, S.; Kwaku Zu, T.T.; Daminati, M.G.; Nielsen, E.; Sparvoli, F. Antinutritional factors in pearl millet grains: Phytate and goitrogens content variability and molecular characterization of genes involved in their pathways. PLoS ONE 2018, 13, e0198394. [Google Scholar] [CrossRef] [PubMed]
- Cominelli, E.; Confalonieri, M.; Carlessi, M.; Cortinovis, G.; Daminati, M.G.; Porch, T.G.; Losa, A.; Sparvoli, F. Phytic acid transport in Phaseolus vulgaris: A new low phytic acid mutant in the PvMRP1 gene and study of the PvMRPs promoters in two different plant systems. Plant Sci. 2018, 270, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ofori, P.A.; Mizuno, A.; Suzuki, M.; Martinoia, E.; Reuscher, S.; Aoki, K.; Shibata, D.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854. [Google Scholar] [CrossRef]
- Raboy, V.; Gerbasi, P.F.; Young, K.A.; Stoneberg, S.D.; Pickett, S.G.; Bauman, A.T.; Murthy, P.P.; Sheridan, W.F.; Ertl, D.S. Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 2000, 124, 355–368. [Google Scholar] [CrossRef]
- Pilu, R.; Panzeri, D.; Gavazzi, G.; Rasmussen, S.K.; Consonni, G.; Nielsen, E. Phenotypic, genetic and molecular characterization of a maize low phytic acid mutant (lpa241). Theor. Appl. Genet. 2003, 107, 980–987. [Google Scholar] [CrossRef]
- Cerino Badone, F.; Amelotti, M.; Cassani, E.; Pilu, R. Study of Low Phytic Acid1-7 (lpa1-7), a New ZmMRP4 Mutation in Maize. J. Hered. 2012, 103, 598–605. [Google Scholar] [CrossRef]
- Liu, K.; Peterson, K.; Raboy, V. Comparison of the phosphorus and mineral concentrations in bran and abraded kernel fractions of a normal barley (Hordeum vulgare) cultivar versus four low phytic acid isolines. J. Agric. Food Chem. 2007, 55, 4453–4460. [Google Scholar] [CrossRef]
- Xu, X.; Zhao, H.; Liu, Q.; Frank, T.; Engel, K.; An, G.; Shu, Q. Mutations of the multi-drug resistance-associated protein ABC transporter gene 5 result in reduction of phytic acid in rice seeds. Theor. Appl. Genet. 2009, 119, 75–83. [Google Scholar] [CrossRef]
- Li, W.; Zhao, H.; Pang, W.; Cui, H.; Poirier, Y.; Shu, Q. Seed-specific silencing of OsMRP5 reduces seed phytic acid and weight in rice. Transgenic Res. 2014, 23, 585–599. [Google Scholar] [CrossRef]
- Wilcox, J.; Premachandra, G.; Young, K.; Raboy, V. Isolation of high seed inorganic P, low-phytate soybean mutants. Crop Sci. 2000, 40, 1601–1605. [Google Scholar] [CrossRef]
- Campion, B.; Sparvoli, F.; Doria, E.; Tagliabue, G.; Galasso, I.; Fileppi, M.; Bollini, R.; Nielsen, E. Isolation and characterisation of an lpa (low phytic acid) mutant in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2009, 118, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, Q.; Ren, X.; Wu, D.; Shu, Q. Gene identification and allele-specific marker development for two allelic low phytic acid mutations in rice (Oryza sativa L.). Mol. Breed. 2008, 22, 603–612. [Google Scholar] [CrossRef]
- Gene Structure Display Server. Available online: http://gsds.cbi.pku.edu.cn/ (accessed on 20 November 2019).
- Patel, R.; Nahal, H.; Breit, R.; Provart, N. BAR expressolog identification: Expression profile similarity ranking of homologous genes in plant species. Plant J. 2012, 71, 1038–1050. [Google Scholar] [CrossRef]
- Bhati, K.; Aggarwal, S.; Sharma, S.; Mantri, S.; Singh, S.; Bhalla, S.; Kaur, J.; Tiwari, S.; Roy, J.; Tuli, R.; et al. Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Sci. 2014, 224, 74–85. [Google Scholar] [CrossRef]
- Latrasse, D.; Jegu, T.; Meng, P.; Mazubert, C.; Hudik, E.; Delarue, M.; Charon, C.; Crespi, M.; Hirt, H.; Raynaud, C.; et al. Dual function of MIPS1 as a metabolic enzyme and transcriptional regulator. Nucleic Acids Res. 2013, 41, 2907–2917. [Google Scholar] [CrossRef]
- Donahue, J.; Alford, S.; Torabinejad, J.; Kerwin, R.; Nourbakhsh, A.; Ray, W.; Hernick, M.; Huang, X.; Lyons, B.; Hein, P.; et al. The Arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 2010, 22, 888–903. [Google Scholar] [CrossRef]
- Chen, H.; Xiong, L. myo-inositol-1-phosphate synthase is required for polar auxin transport and organ development. J. Biol. Chem. 2010, 285, 24238–24247. [Google Scholar] [CrossRef]
- Sato, Y.; Yazawa, K.; Yoshida, S.; Tamaoki, M.; Nakajima, N.; Iwai, H.; Ishii, T.; Satoh, S. Expression and functions of myo-inositol monophosphatase family genes in seed development of Arabidopsis. J. Plant Res. 2011, 124, 385–394. [Google Scholar] [CrossRef]
- Nourbakhsh, A.; Collakova, E.; Gillaspy, G.E. Characterization of the inositol monophosphatase gene family in Arabidopsis. Front. Plant Sci. 2014, 5, 725. [Google Scholar] [CrossRef]
- Sweetman, D.; Stavridou, I.; Johnson, S.; Green, P.; Caddick, S.; Brearley, C. Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase 4 (AtITPK4) is an outlier to a family of ATP-grasp fold proteins from Arabidopsis. FEBS Lett. 2007, 581, 4165–4171. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Brearley, C.; Elge, S.; Kaplan, B.; Fromm, H.; Mueller-Roeber, B. Arabidopsis inositol polyphosphate 6-/3-kinase is a nuclear protein that complements a yeast mutant lacking a functional ArgR-Mcm1 transcription complex. Plant Cell 2003, 15, 449–463. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Yang, G.; Arana, F.; Chen, Z.; Li, Y.; Xia, H.J. Arabidopsis inositol polyphosphate 6-/3-kinase (AtIpk2beta) is involved in axillary shoot branching via auxin signaling. Plant Physiol. 2007, 144, 942–951. [Google Scholar] [CrossRef] [PubMed]
- Munnik, T.; Vermeer, J. Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ. 2010, 33, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; VanToai, T.; Pratt, R. Expression and nucleotide sequence of an INS (3) P-1 synthase gene associated with low-phytate kernels in maize (Zea mays L.). J. Agric. Food Chem. 2004, 52, 4565–4570. [Google Scholar] [CrossRef] [PubMed]
- Pilu, R.; Landoni, M.; Cassani, E.; Doria, E.; Nielsen, E. The maize lpa241 mutation causes a remarkable variability of expression and some pleiotropic effects. Crop Sci. 2005, 45, 2096–2105. [Google Scholar] [CrossRef]
- Pilu, R.; Panzeri, D.; Cassani, E.; Badone, F.C.; Landoni, M.; Nielsen, E. A paramutation phenomenon is involved in the genetics of maize low phytic acid1-241 (lpa1-241) trait. Heredity 2009, 102, 236–245. [Google Scholar] [CrossRef]
- Doria, E.; Galleschi, L.; Calucci, L.; Pinzino, C.; Pilu, R.; Cassani, E.; Nielsen, E. Phytic acid prevents oxidative stress in seeds: Evidence from a maize (Zea mays L.) low phytic acid mutant. J. Exp. Bot. 2009, 60, 967–978. [Google Scholar] [CrossRef]
- Cerino Badone, F.; Cassani, E.; Landoni, M.; Doria, E.; Panzeri, D.; Lago, C.; Mesiti, F.; Nielsen, E.; Pilu, R. The low phytic acid 1-241 (lpa1-241) maize mutation alters the accumulation of anthocyanin pigment in the kernel. Planta 2010, 231, 1189–1199. [Google Scholar] [CrossRef]
- Landoni, M.; Badone, F.; Haman, N.; Schiraldi, A.; Fessas, D.; Cesari, V.; Toschi, I.; Cremona, R.; Delogu, C.; Villa, D.; et al. Low phytic acid 1 mutation in maize modifies density, starch properties, cations, and fiber contents in the seed. J. Agric. Food Chem. 2013, 61, 4622–4630. [Google Scholar] [CrossRef]
- Meis, S.; Fehr, W.; Schnebly, S. Seed source effect on field emergence of soybean lines with reduced phytate and raffinose saccharides. Crop Sci. 2003, 43, 1336–1339. [Google Scholar] [CrossRef]
- Bregitzer, P.; Raboy, V. Effects of four independent low-phytate mutations in barley (Hordeum vulgare L.) on seed phosphorus characteristics and malting quality. Cereal Chem. 2006, 83, 460–464. [Google Scholar] [CrossRef]
- Guttieri, M.; Peterson, K.; Souza, E. Mineral distributions in milling fractions of low phytic acid wheat. Crop Sci. 2006, 46, 2692–2698. [Google Scholar] [CrossRef]
- Borlini, G.; Rovera, C.; Landoni, M.; Cassani, E.; Pilu, R. lpa1-5525: A new lpa1 mutant isolated in a mutagenized population by a novel non-disrupting screening method. Plants 2019, 8, 209. [Google Scholar] [CrossRef]
- Raboy, V.; Peterson, K.; Jackson, C.; Marshall, J.; Hu, G.; Saneoka, H.; Bregitzer, P. A substantial fraction of barley (Hordeum vulgare L.) low phytic acid mutations have little or no effect on yield across diverse production environments. Plants 2015, 4, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Graf, E.; Empson, K.; Eaton, J. Phytic acid—A natural antioxidant. J. Biol. Chem. 1987, 262, 11647–11650. [Google Scholar] [PubMed]
- Graf, E.; Eaton, J.W. Antioxidant functions of phytic acid. Free Radic. Biol. Med. 1990, 8, 61–69. [Google Scholar] [CrossRef]
- Empson, K.; Labuza, T.; Graf, E. Phytic acid as food antioxidant. J. Food Sci. 1991, 56, 560–563. [Google Scholar] [CrossRef]
- Dorsch, J.; Cook, A.; Young, K.; Anderson, J.; Bauman, A.; Volkmann, C.; Murthy, P.; Raboy, V. Seed phosphorus and inositol phosphate phenotype of barley low phytic acid genotypes. Phytochemistry 2003, 62, 691–706. [Google Scholar] [CrossRef]
- Bhati, K.K.; Sharma, S.; Aggarwal, S.; Kaur, M.; Shukla, V.; Kaur, J.; Mantri, S.; Pandey, A.K. Genome-wide identification and expression characterization of ABCC-MRP transporters in hexaploid wheat. Front. Plant Sci. 2015, 6, 488. [Google Scholar] [CrossRef]
- Oltmans, S.; Fehr, W.; Welke, G.; Cianzio, S. Inheritance of low-phytate phosphorus in soybean. Crop Sci. 2004, 44, 433–435. [Google Scholar] [CrossRef]
- Oltmans, S.; Fehr, W.; Welke, G.; Raboy, V.; Peterson, K. Agronomic and seed traits of soybean lines with low-phytate phosphorus. Crop Sci. 2005, 45, 593–598. [Google Scholar] [CrossRef]
- Chappell, A.; Scaboo, A.; Wu, X.; Nguyen, H.; Pantalone, V.; Bilyeu, K. Characterization of the MIPS gene family in Glycine max. Plant Breed. 2006, 125, 493–500. [Google Scholar] [CrossRef]
- Scaboo, A.; Pantalone, V.; Walker, D.; Boerma, H.; West, D.; Walker, F.; Sams, C. Confirmation of molecular markers and agronomic traits associated with seed phytate content in two soybean RIL populations. Crop Sci. 2009, 49, 426–432. [Google Scholar] [CrossRef]
- Hulke, B.; Fehr, W.; Welke, G. Agronomic and seed characteristics of soybean with reduced phytate and palmitate. Crop Sci. 2004, 44, 2027–2031. [Google Scholar] [CrossRef]
- Spear, J.; Fehr, W. Genetic improvement of seedling emergence of soybean lines with low phytate. Crop Sci. 2007, 47, 1354–1360. [Google Scholar] [CrossRef]
- Anderson, B.; Fehr, W. Seed source affects field emergence of low-phytate soybean lines. Crop Sci. 2008, 48, 929–932. [Google Scholar] [CrossRef]
- Campion, B.; Glahn, R.; Tava, A.; Perrone, D.; Doria, E.; Sparvoli, F.; Cecotti, R.; Dani, V.; Nielsen, E. Genetic reduction of antinutrients in common bean (Phaseolus vulgaris L.) seed, increases nutrients and in vitro iron bioavailability without depressing main agronomic traits. Field Crops Res. 2013, 141, 27–37. [Google Scholar] [CrossRef]
- Zhang, W.; Gruszewski, H.; Chevone, B.; Nessler, C. An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol. 2008, 146, 431–440. [Google Scholar] [CrossRef]
- Torabinejad, J.; Donahue, J.; Gunesekera, B.; Allen-Daniels, M.; Gillaspy, G. VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 2009, 150, 951–961. [Google Scholar] [CrossRef]
- Zhao, H.; Frank, T.; Tan, Y.; Zhou, C.; Jabnoune, M.; Arpat, A.B.; Cui, H.; Huang, J.; He, Z.; Poirier, Y.; et al. Disruption of OsSULTR3;3 reduces phytate and phosphorus concentrations and alters the metabolite profile in rice grains. New Phytol. 2016, 211, 926–939. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.C.; Polzin, K.; Labate, J.; Specht, J.; Brummer, E.C.; Olson, T.; Young, N.; Concibido, V.; Wilcox, J.; Tamulonis, J.P.; et al. Genome duplication in soybean (Glycine subgenus soja). Genetics 1996, 144, 329–338. [Google Scholar] [PubMed]
- Yamaji, N.; Ma, J.F. Node-controlled allocation of mineral elements in Poaceae. Curr. Opin. Plant Biol. 2017, 39, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Zuber, H.; Davidian, J.C.; Aubert, G.; Aimé, D.; Belghazi, M.; Lugan, R.; Heintz, D.; Wirtz, M.; Hell, R.; Thompson, R.; et al. The seed composition of Arabidopsis mutants for the group 3 sulfate transporters indicates a role in sulfate translocation within developing seeds. Plant Physiol. 2010, 154, 913–926. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, P.X.; Miao, Z.Q.; Qi, G.F.; Wang, Z.; Yuan, Y.; Ahmad, N.; Cao, M.J.; Hell, R.; Wirtz, M.; et al. SULTR3s function in chloroplast sulfate uptake and affect ABA biosynthesis and the stress response. Plant Physiol. 2019, 180, 593–604. [Google Scholar] [CrossRef]
- Ding, G.; Lei, G.J.; Yamaji, N.; Yokosho, K.; Mitani-Ueno, N.; Huang, S.; Ma, J.F. Vascular cambium-localized AtSPDT mediates xylem-to-phloem transfer of phosphorus for its preferential distribution in Arabidopsis. Mol. Plant 2019. [Google Scholar] [CrossRef]
- Raboy, V.; Cichy, K.; Peterson, K.; Reichman, S.; Sompong, U.; Srinives, P.; Saneoka, H. Barley (Hordeum vulgare L.) Low phytic acid 1-1: An endosperm-specific, filial determinant of seed total phosphorus. J. Hered. 2014, 105, 656–665. [Google Scholar] [CrossRef]
- Zhou, C.; Tan, Y.; Goßner, S.; Li, Y.; Shu, Q.; Engel, K.H. Stability of the metabolite signature resulting from the OsSULTR3;3 mutation in low phytic acid rice (Oryza sativa L.) seeds upon cross-breeding. J. Agric. Food Chem. 2018, 66, 9366–9376. [Google Scholar] [CrossRef]
- Zhou, C.; Tan, Y.; Goßner, S.; Li, Y.; Shu, Q.; Engel, K.H. Impact of crossing parent and environment on the metabolite profiles of progenies generated from a low phytic acid rice (Oryza sativa L.) mutant. J. Agric. Food Chem. 2019, 67, 2396–2407. [Google Scholar] [CrossRef]
- Tong, C.; Chen, Y.; Tan, Y.; Liu, L.; Waters, D.L.E.; Rose, T.J.; Shu, Q.; Bao, J. Analysis of lysophospholipid content in low phytate rice mutants. J. Agric. Food Chem. 2017, 65, 5435–5441. [Google Scholar] [CrossRef]
- Otegui, M.; Capp, R.; Staehelin, L. Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. Plant Cell 2002, 14, 1311–1327. [Google Scholar] [CrossRef] [PubMed]
- Bregitzer, P.; Raboy, V. Effects of four independent low-phytate mutations on barley agronomic performance. Crop Sci. 2006, 46, 1318–1322. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Liao, W.; Chen, S.; Zemetra, R. Bioethanol production using genetically modified and mutant wheat and barley straws. Biomass Bioenergy 2011, 35, 542–548. [Google Scholar] [CrossRef]
- Liu, F.; Chang, X.; Ye, Y.; Xie, W.; Wu, P.; Lian, X. Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant 2011, 4, 1105–1122. [Google Scholar] [CrossRef] [PubMed]
- PFAM Software. Available online: http://pfam.xfam.org/ (accessed on 20 November 2019).
- Kopriva, S.; Chu, C. Are we ready to improve phosphorus homeostasis in rice? J. Exp. Bot. 2018, 69, 3515–3522. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H. Sulfate transport systems in plants: Functional diversity and molecular mechanisms underlying regulatory coordination. J. Exp. Bot. 2019, 70, 4075–4087. [Google Scholar] [CrossRef]
- Rouached, H. Multilevel coordination of phosphate and sulfate homeostasis in plants. Plant Signal. Behav. 2011, 6, 952–955. [Google Scholar] [CrossRef]
- Sacchi, G.A.; Nocito, F.F. Plant sulfate transporters in the low phytic acid network: Some educated guesses. Plants 2019, 8, 616. [Google Scholar] [CrossRef]
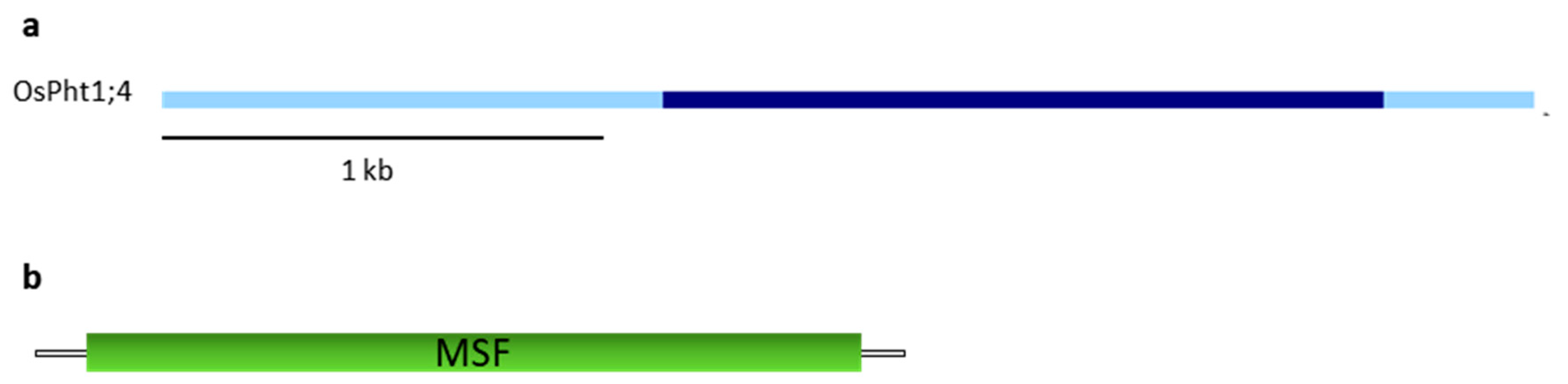
| Class of Transporters | Species | Gene | Phytozome/Genbank/Ensembl Accession Number | Origin of Mutation | Mutation | Reference |
|---|---|---|---|---|---|---|
| MRP | Zea mays | ZmMRP4/ZmABCC4 | EF586878 | EMS | lpa1-1 | [37] |
| lpa1-241 | [38] | |||||
| lpa1-7 | [39] | |||||
| T-DNA insertion | lpa1-mum1 | [24] | ||||
| Embryo specific:RNAi | Ole::MRP4 Glb::MRP4 | [24] | ||||
| Oryza sativa | OsMRP5/OsABCC13 | LOC_Os03g04920 | γ rays + sodium azide | Os-lpa-XS110-2 | [40] | |
| Os-lpa-XS110-3 | [41] | |||||
| T-DNA insertion | 4A-02500 | [41] | ||||
| Embryo specific amiRNA | Ami-MRP5 | [42] | ||||
| Triticum aestivum | TaABCC13-4B | TraesCS4B02G343800 | Constitutive RNAi | TaABCC13 RNAi | [33] | |
| TaABCC13-4D | TraesCS4D02G339000 | |||||
| TaABCC13-5A* | TraesCS5A02G512500 | |||||
| Glycine max | GmMRP3/GmABCC1 | Glyma.03G167800 | EMS | CX1834 | [29,30,31,43] | |
| GmMRP19/GmABCC2 | Glyma.19G169000 | |||||
| GmMRP13/GmABCC3 | Glyma.13G127500 | no reported mutant | no reported mutant | [32] | ||
| Phaseolus vulgaris | PvMRP1/PvABCC1 | Phvul.001G165500 | EMS | lpa1 | [32,44] | |
| lpa12 | [35] | |||||
| PvMRP2/PvABCC2 | Phvul.007G153800 | no reported mutant | no reported mutant | [32] | ||
| SULTR | Oryza sativa | OsSULTR3;3 | LOC_Os04g55800 | γ rays | Oslpa-MH86-1 Os-lpa-Z9B-1 | [28] [19] |
| OsSULTR3;4 | LOC_Os06g05160 | retrotransposon Tos-17 insertion | spdt-1, spdt-2, spdt-3 | [20] | ||
| Hordeum vulgare | Hvst | HORVU2Hr1G113050 | sodium azide | lpa1-1(M422) | [45] [19] | |
| Pht | Oryza sativa | OsPht1;4 | LOC_Os04g10750 | retrotransposon Tos-17 insertion | ospt4-1 (NE1260) ospt4-2 (SHIP_ZSF6267) RNAi | [21] [22] |
| SULTR Group. | Species | Gene Name | Phytozome Accession Number |
|---|---|---|---|
| SULTR3;3 | Zea mays | ZmSULTR3;3 | GRMZM2G395114 |
| Phaseolus vulgaris | PvSULTR3;3 | Phvul.002G095300 | |
| Glycine max | GmSULTR3;3a | Glyma.20G017100 | |
| GmSULTR3;3b | Glyma.07G218900 | ||
| SULTR3;4 | Zea mays | ZmSULTR3;4 | GRMZM2G444801 |
| Phaseolus vulgaris | PvSULTR3;4a | Phvul.005G171800 | |
| PvSULTR3;4b | Phvul.010G151000 | ||
| Glycine max | GmSULTR3;4a | Glyma.07G006500 | |
| GmSULTR3;4b | Glyma.08G207100 | ||
| GmSULTR3;4c | Glyma.13G360000 | ||
| GmSULTR3;4d | Glyma.15G014000 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cominelli, E.; Pilu, R.; Sparvoli, F. Phytic Acid and Transporters: What Can We Learn from low phytic acid Mutants? Plants 2020, 9, 69. https://doi.org/10.3390/plants9010069
Cominelli E, Pilu R, Sparvoli F. Phytic Acid and Transporters: What Can We Learn from low phytic acid Mutants? Plants. 2020; 9(1):69. https://doi.org/10.3390/plants9010069
Chicago/Turabian StyleCominelli, Eleonora, Roberto Pilu, and Francesca Sparvoli. 2020. "Phytic Acid and Transporters: What Can We Learn from low phytic acid Mutants?" Plants 9, no. 1: 69. https://doi.org/10.3390/plants9010069
APA StyleCominelli, E., Pilu, R., & Sparvoli, F. (2020). Phytic Acid and Transporters: What Can We Learn from low phytic acid Mutants? Plants, 9(1), 69. https://doi.org/10.3390/plants9010069






