Nitrogen Dioxide at Ambient Concentrations Induces Nitration and Degradation of PYR/PYL/RCAR Receptors to Stimulate Plant Growth: A Hypothetical Model
Abstract
1. Introduction
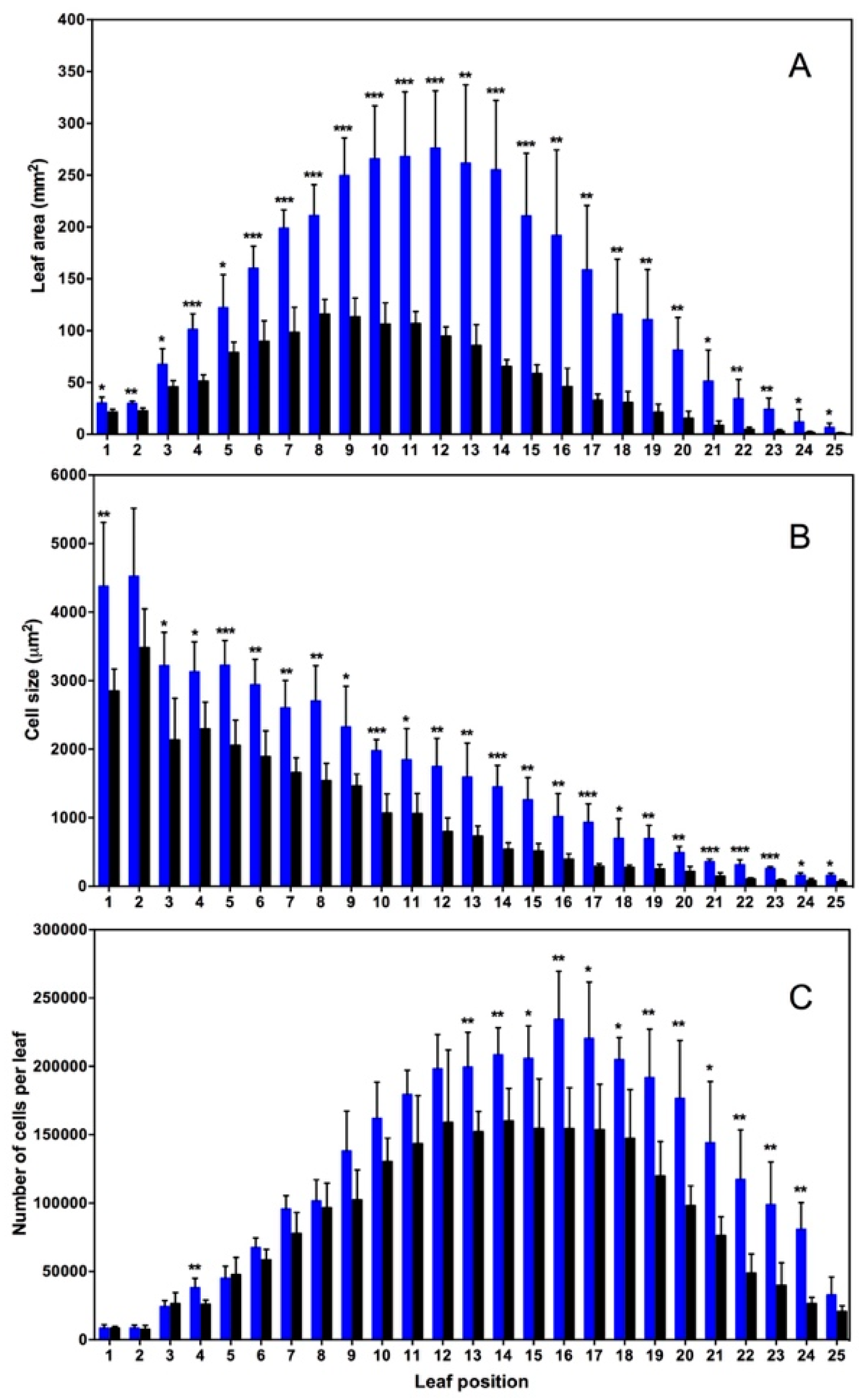
2. Nitrogen Dioxide at Ambient Concentrations of 10–50 ppb Acts as a Positive Plant Growth Signal in Arabidopsis Thaliana
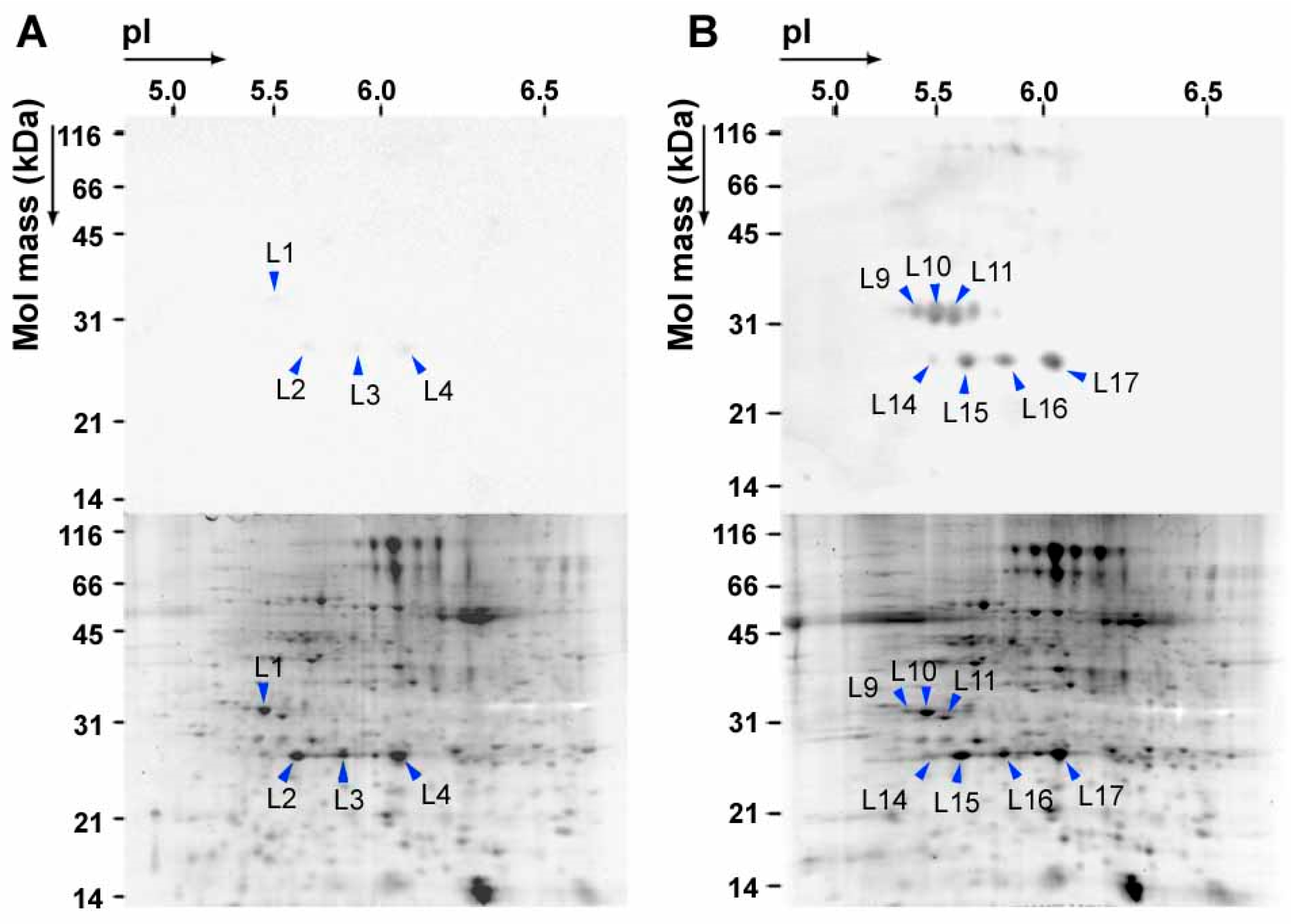
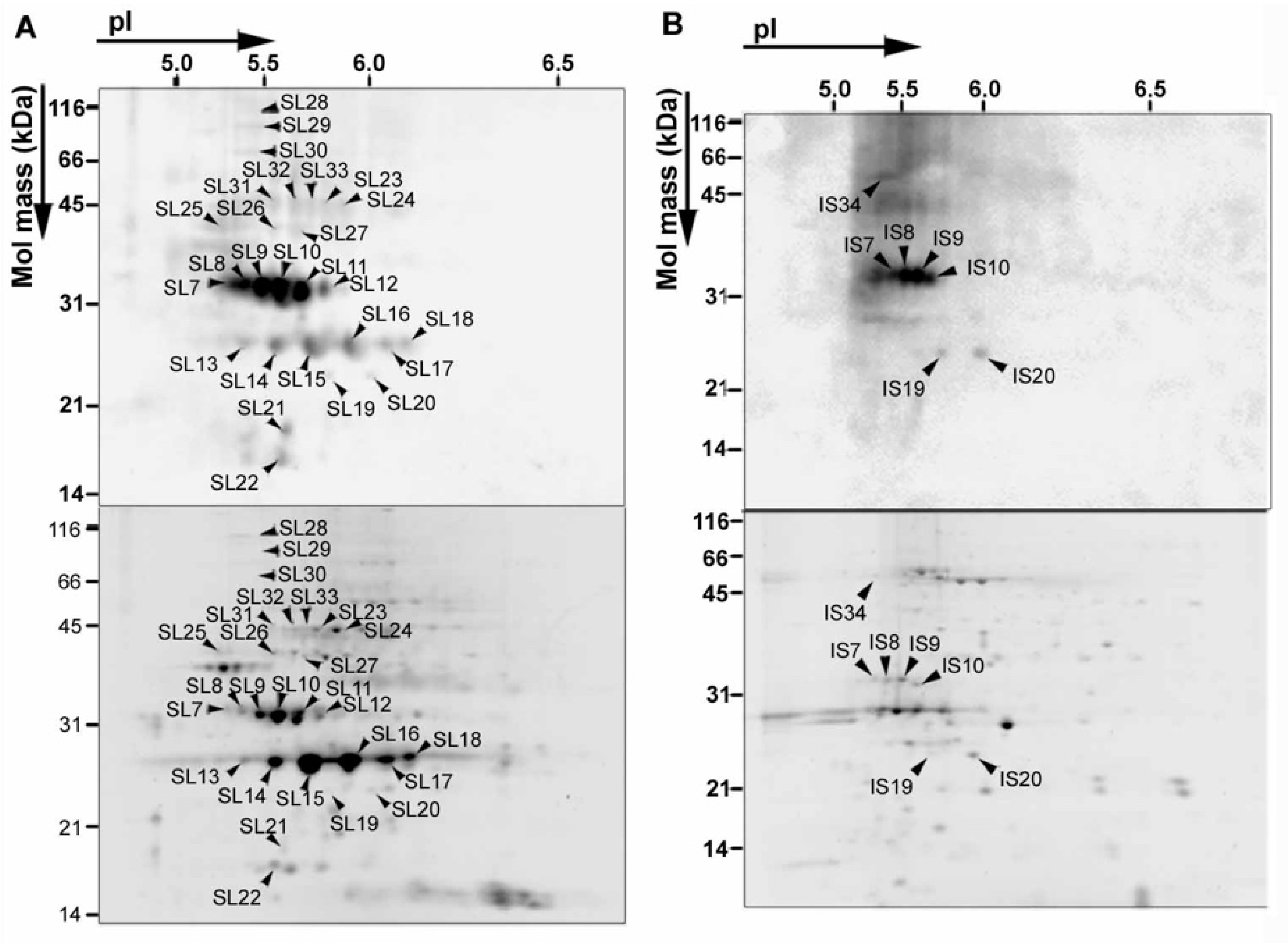
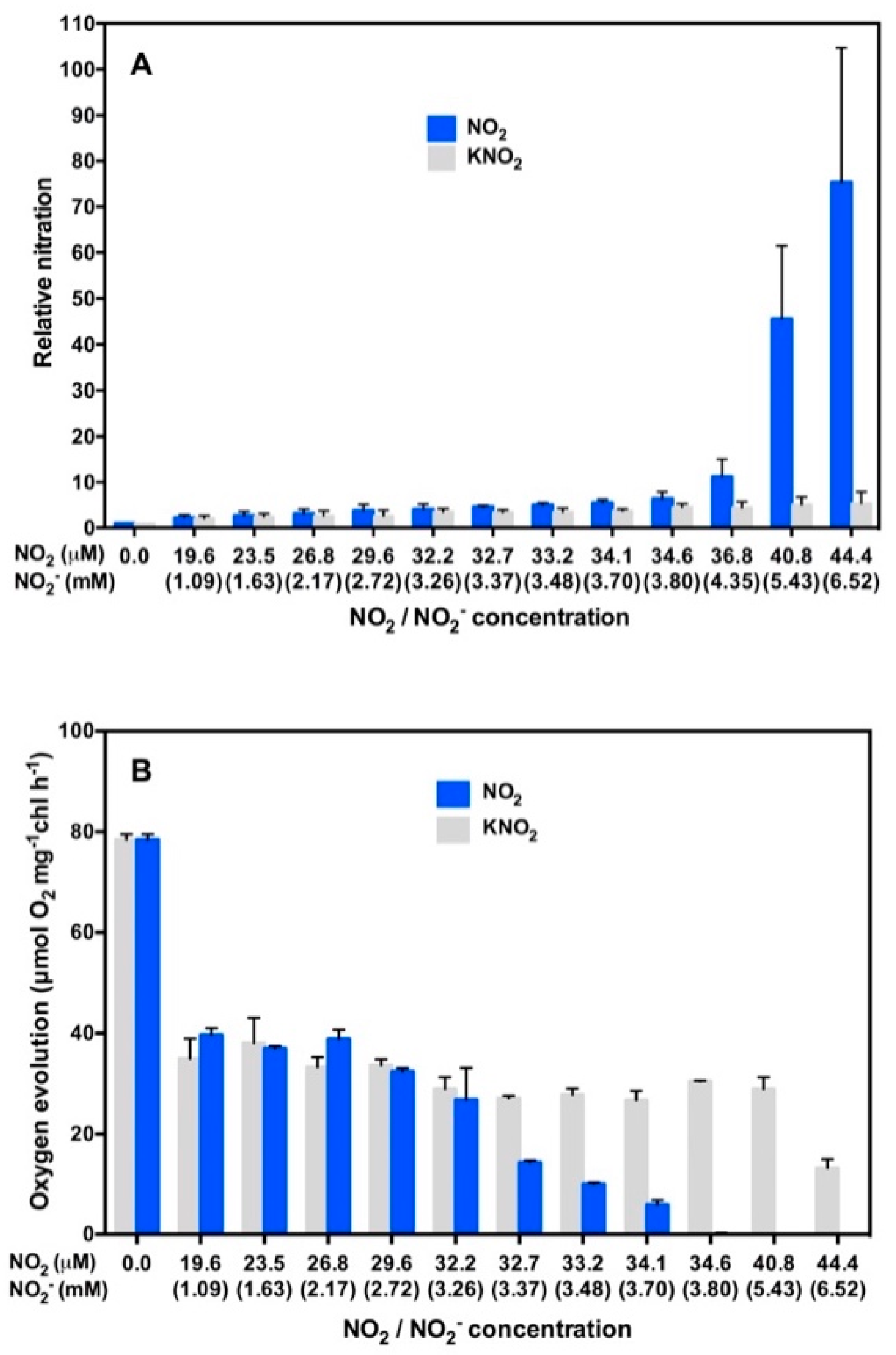
3. NO2 Selectively Nitrates Specific Cellular Proteins in Arabidopsis Leaves
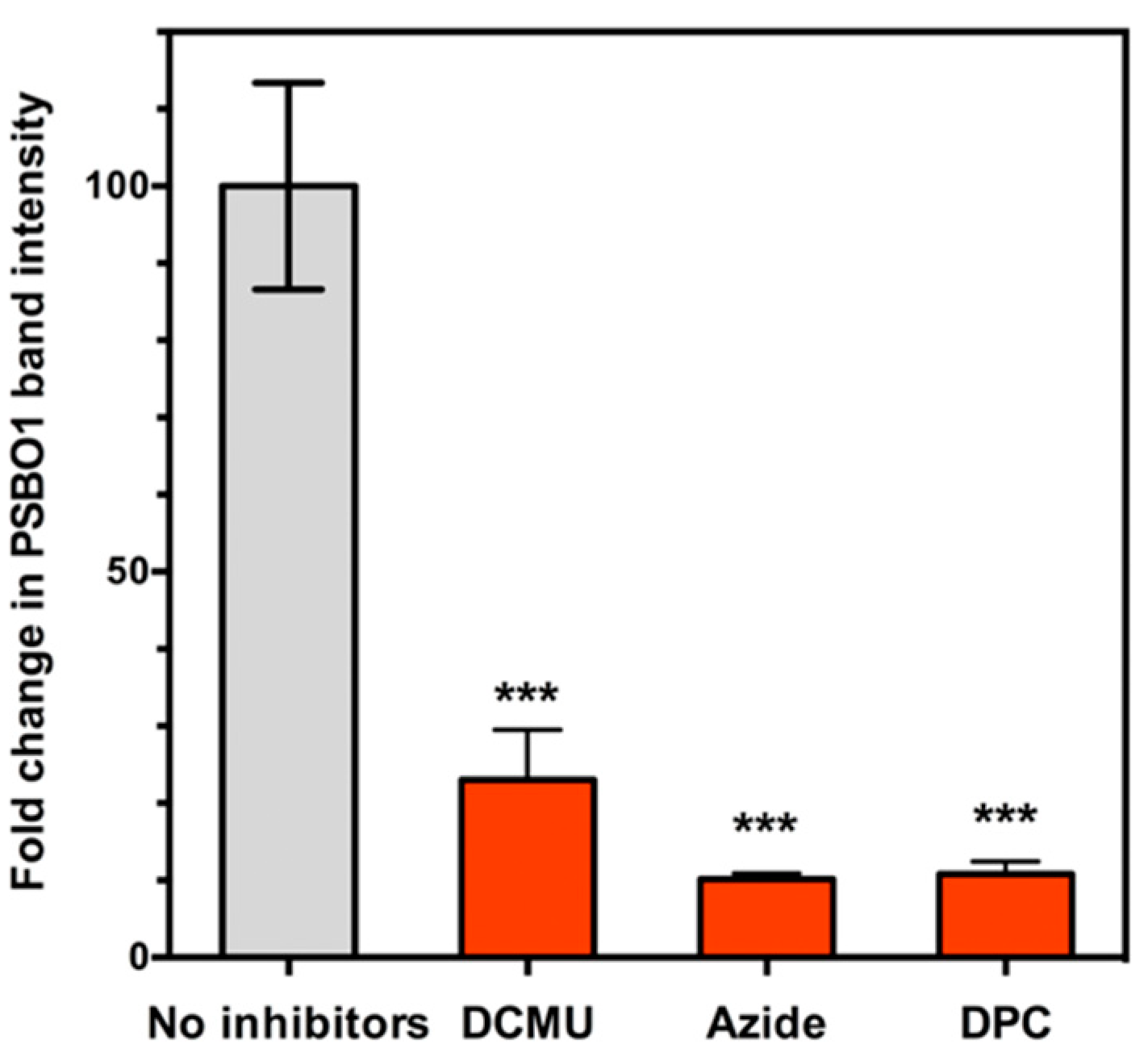
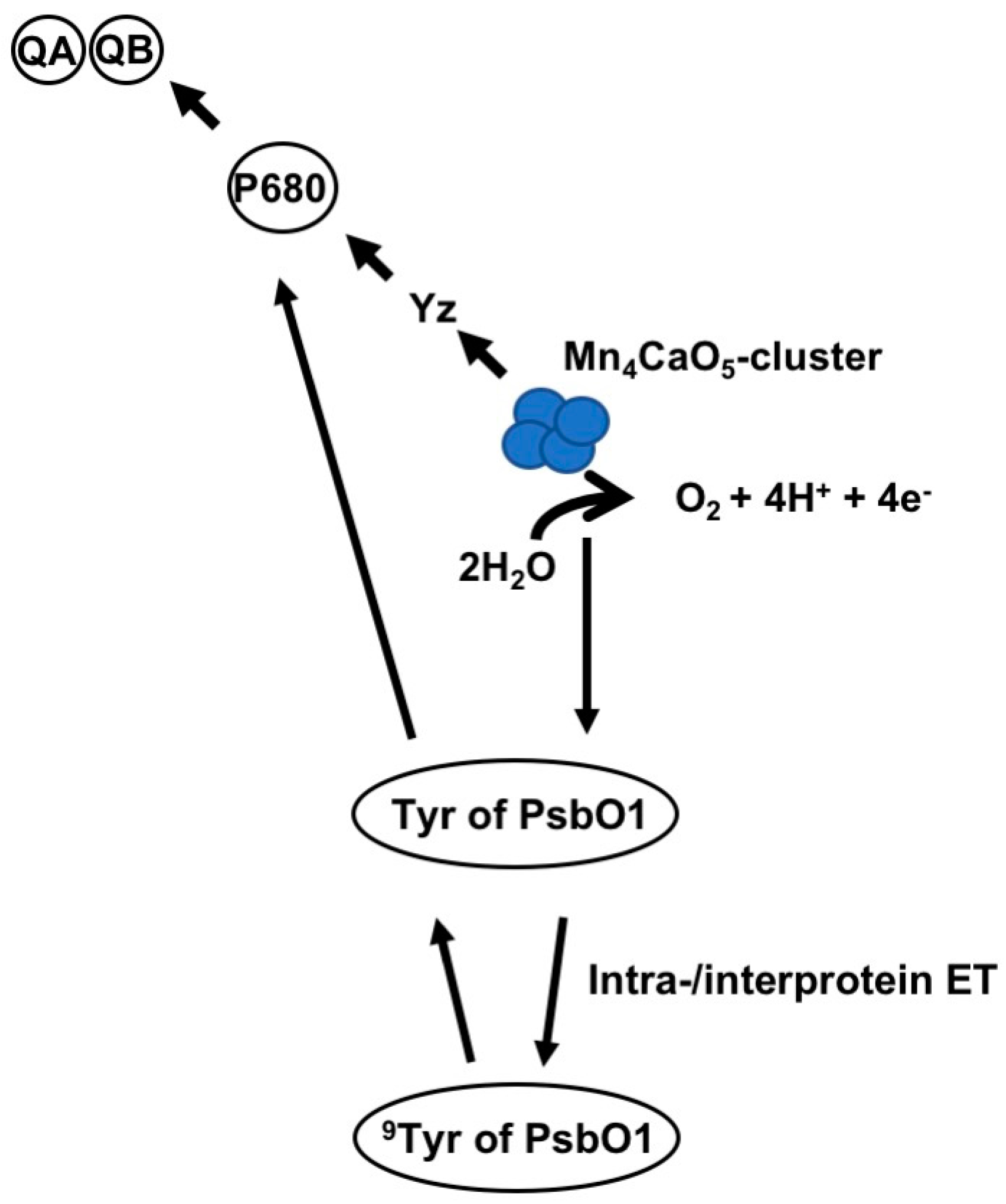
4. PsbO1 May Function as an Electron Element Like Yz in PSII Electron Transport Chain
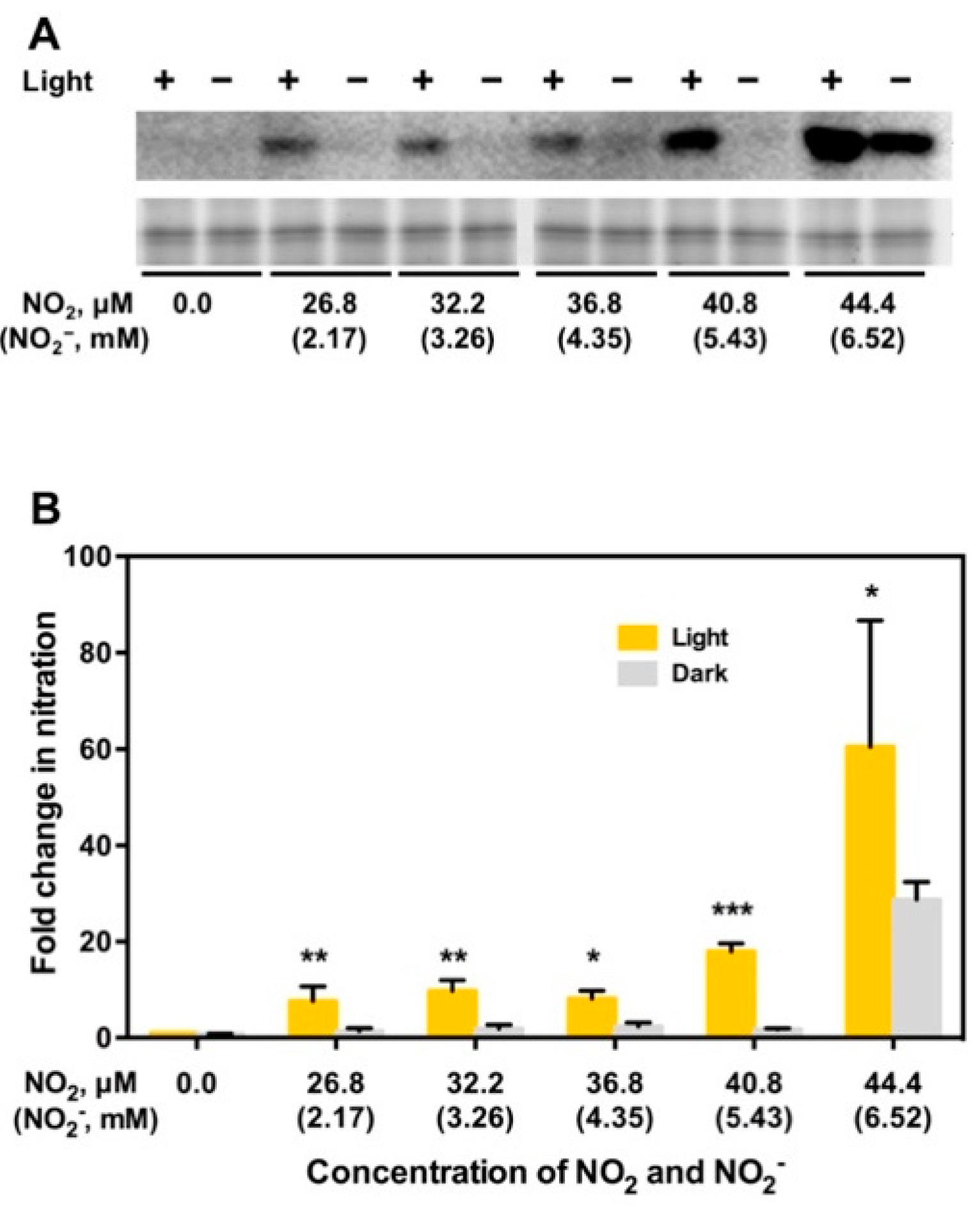
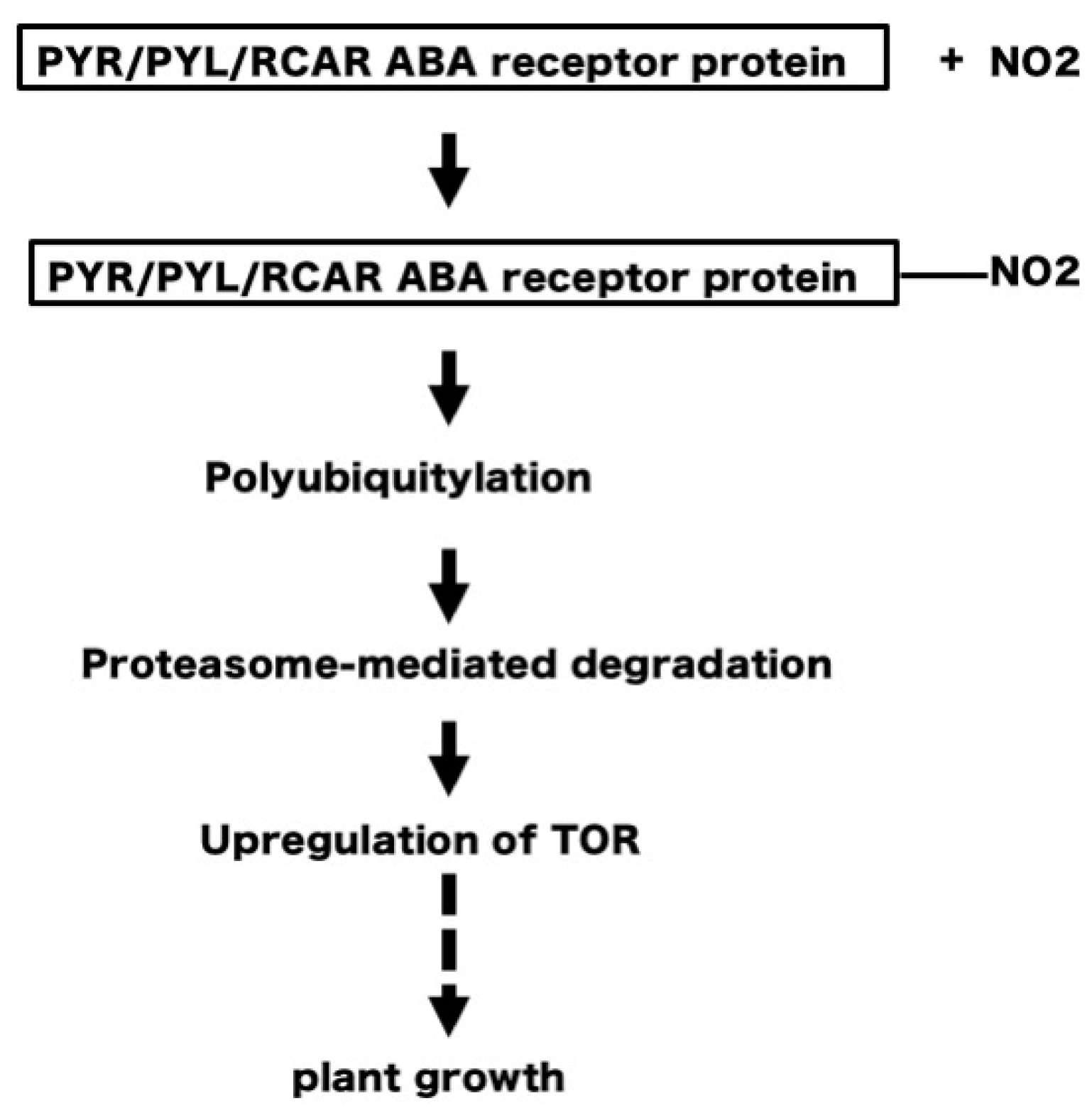
5. NO2 May Induce Tyrosine Nitration of PYR/PYL/RCAR ABA Receptors Leading to Degradation of the Receptors and Upregulation of TOR, to Stimulate Plant Growth
6. Future Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aneja, V.P.; Roelle, P.A.; Murray, G.C.; Southerland, J.; Erisman, J.W.; Fowler, D.; Asman, W.A.H.; Naveen Patni, H. Atmospheric nitrogen compounds II: Emissions, transport, transformation, deposition and assessment. Atmos. Environ. 2001, 35, 1903–1911. [Google Scholar] [CrossRef]
- Oswald, R.; Behrendt, T.; Ermel, M.; Wu, D.; Su, H.; Cheng, Y.; Breuninger, C.; Moravek, A.; Mougin, E.; Delon, C.; et al. HONO emissions from soil bacteria as a major source of atmospheric reactive nitrogen. Science 2013, 6151, 1233–1235. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency Home Page Integrated Science Assessment for Oxides of Nitrogen—Health Criteria (2016 Final Report). Available online: https://cfpub.epa.gov/ncea/isa/recordisplay.cfm?deid=310879 (accessed on 1 January 2019).
- World Health Organization. Home Page. Available online: http://www.who.int/mediacentre/factsheets/fs313/en/ (accessed on 1 January 2019).
- NationMaster Home Page. Available online: http://www.nationmaster.com/index.php (accessed on 1 January 2019).
- Klepper, L. Nitric oxide (NO) and nitrogen dioxide (NO2) emissions from herbicide-treated soybean plants. Atmos. Environ. 1979, 13, 537–542. [Google Scholar] [CrossRef]
- Dean, J.V.; Harper, J.E. Nitric oxide and nitrous oxide production by soybean and winged bean during the in vivo nitrate reductase assay. Plant Physiol. 1986, 82, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Wildt, J.; Kley, D.; Rockel, A.; Rockel, P.; Segschneider, H.J. Emission of NO from several higher plant species. J. Geochem. Res. 1997, 102, 5919–5927. [Google Scholar] [CrossRef]
- Hari, P.; Raivonen, M.; Vesala, T.; Munger, J.W.; Pilegaard, K.; Kulmala, M. Atmospheric science: Ultraviolet light and leaf emission of NO(x). Nature 2003, 422, 134. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, T.; Sasakawa, H. Transformation of atmospheric NO2 absorbed in spinach leaves. Plant Cell Physiol. 1979, 20, 263–266. [Google Scholar]
- Yoneyama, T.; Sasakawa, H.; Ishizuka, S.; Totsuka, T. Absorption of atmospheric NO2 by plants and soils. Soil Sci. Plant Nutr. 1979, 25, 255–265. [Google Scholar]
- Breuninger, C.; Meixner, F.X.; Kesselmeier, J. Field investigations of nitrogen dioxide (NO2) exchange between plants and the atmosphere. Atmos. Chem. Phys. 2013, 13, 773–790. [Google Scholar] [CrossRef]
- Wellburn, A.R. Why are atmospheric oxides of nitrogen usually phytotoxic and not alternative fertilizers? New Phytol. 1990, 115, 395–429. [Google Scholar] [CrossRef]
- Chauhan, A.J.; Inskip, H.M.; Linaker, C.H.; Smith, S.; Schreiber, J.; Johnston, S.L.; Holgate, S.T. Personal exposure to nitrogen dioxide (NO2) and the severity of virus-induced asthma in children. Lancet 2003, 9373, 1939–1944. [Google Scholar] [CrossRef]
- Capron, T.M.; Mansfield, T.M. Inhibition of growth in tomato by air polluted with nitrogen oxides. J. Exp. Bot. 1977, 28, 112–116. [Google Scholar] [CrossRef]
- Sandhu, R.; Gupta, G. Effects of nitrogen dioxide on growth and yield of black turtle bean (Phaseolus vulgaris L.) cv. ‘Domino’. Environ. Pollut. 1989, 59, 337–344. [Google Scholar] [CrossRef]
- Saxe, H. Relative sensitivity of greenhouse pot plants to long-term exposures of NO- and NO2-containing air. Environ. Pollut. 1994, 85, 283–290. [Google Scholar] [CrossRef]
- Morikawa, H.; Higaki, A.; Nohno, M.; Takahashi, M.; Kamada, M.; Nakata, M.; Toyohara, G.; Okamura, Y.; Matsui, K.; Kitani, S.; et al. More than a 600-fold variation in nitrogen dioxide assimilation among 217 plant taxa. Plant Cell Environ. 1998, 21, 180–190. [Google Scholar] [CrossRef]
- von Liebig, J. Extrait d’une note sur la nitrification. Ann. Chem. Phys. 1827, 35, 329–333. [Google Scholar]
- Takahashi, M.; Higaki, A.; Nohno, M.; Kamada, M.; Okamura, Y.; Matsui, K.; Kitani, S.; Morikawa, H. Differential assimilation of nitrogen dioxide by 70 taxa of roadside trees at an urban pollution level. Chemosphere 2005, 61, 633–639. [Google Scholar] [CrossRef]
- Takahashi, M.; Sasaki, Y.; Ida, S.; Morikawa, H. Enrichment of nitrite reductase gene improves the ability of Arabidopsis thaliana plants to assimilate nitrogen dioxide. Plant Physiol. 2001, 126, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, H.; Takahashi, M.; Kawamura, Y. Metabolism and genetics of atmospheric nitrogen dioxide control using pollutant-philic plants. In Phytoremediation Transformation and Control of Contaminants; McCutcheon, S.C., Schnoor, J.L., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2003; pp. 765–786. ISBN 0-471-39435-1. [Google Scholar]
- Takahashi, M.; Morikawa, H. Air-pollutant-philic plants for air remediation. J. Environ. Prot. 2012, 3, 1346–1352. [Google Scholar] [CrossRef][Green Version]
- Takahashi, M.; Nakagawa, M.; Sakamoto, A.; Ohsumi, C.; Matsubara, T.; Morikawa, H. Atmospheric nitrogen dioxide gas is a plant vitalization signal to increase plant size and the contents of cell constituents. New Phytol. 2005, 168, 149–154. [Google Scholar] [CrossRef]
- Takahashi, M.; Furuhashi, T.; Ishikawa, N.; Horiguchi, G.; Sakamoto, A.; Tsukaya, H.; Morikawa, H. Nitrogen dioxide regulates organ growth by controlling cell proliferation and enlargement in Arabidopsis. New Phytol. 2014, 201, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Morikawa, H. Nitrogen dioxide is a positive regulator of plant growth. Plant Signal. Behav. 2014, 9, e28033. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.E.H.; Shigeto, J.; Sakamoto, A.; Takahashi, M.; Morikawa, H. Atmospheric nitrogen dioxide at ambient levels stimulates growth and development of horticultural plants. Botany 2008, 86, 213–217. [Google Scholar] [CrossRef]
- Takahashi, M.; Morikawa, H. Differential responses of Arabidopsis thaliana accessions to atmospheric nitrogen dioxide at ambient concentrations. Plant Signal. Behav. 2014, 9, e28563. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Morikawa, H. Kinematic evidence that atmospheric nitrogen dioxide increases the rates of cell proliferation and enlargement to stimulate leaf expansion in Arabidopsis. Plant Signal. Behav. 2015, 10, e1022011. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takahashi, M.; Morikawa, H. Nitrogen dioxide accelerates flowering without changing the number of leaves at flowering in Arabidopsis thaliana. Plant Signal. Behav. 2014, 9, e970433. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Sakamoto, A.; Ezura, H.; Morikawa, H. Prolonged exposure to atmospheric nitrogen dioxide increases fruit yield of tomato plants. Plant Biotechnol. 2011, 8, 485–487. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Xu, Q.; Zhou, B.; Ma, C.; Xu, X.; Xu, J.; Jiang, Y.; Liu, C.; Li, G.; Herbert, S.J.; Hao, L. Salicylic acid-altering Arabidopsis mutants response to NO2 exposure. Environ. Contam. Toxicol. 2010, 84, 106–111. [Google Scholar] [CrossRef]
- Kotchoni, S.O.; Larrimore, K.E.; Mukherjee, M.; Kempinski, C.F.; Barth, C. Alterations in the endogenous ascorbic acid content affect flowering time in Arabidopsis. Plant Physiol. 2009, 149, 803–815. [Google Scholar] [CrossRef]
- Adam, S.E.H.; Abdel-Banat, B.M.A.; Sakamoto, A.; Takahashi, M.; Morikawa, H. Effect of Atmospheric Nitrogen Dioxide on Mulukhiya (Corchorus olitorius) Growth and Flowering. Am. J. Plant Physiol. 2008, 3, 180–184. [Google Scholar] [CrossRef]
- He, Y.; Tang, R.H.; Hao, Y.; Stevens, R.D.; Cook, C.W.; Ahn, S.M.; Jing, L.; Yang, Z.; Chen, L.; Guo, F.; et al. Nitric oxide represses the Arabidopsis floral transition. Science 2004, 305, 1968–1971. [Google Scholar] [CrossRef] [PubMed]
- Leshem, Y.Y.; Haramaty, E. The characterization and contrasting effects of the nitric oxide free radical in vegetative stress and senescence of Pisum sativum Linn. foliage. J. Plant Physiol. 1996, 148, 258–263. [Google Scholar] [CrossRef]
- Jin, C.W.; Du, S.T.; Zhang, Y.S.; Tang, C.X.; Lin, X.Y. Atmospheric nitric oxide stimulates plant growth and improves the quality of spinach (Spinaciaoleracea). Ann. Appl. Biol. 2009, 155, 113–120. [Google Scholar] [CrossRef]
- Morikawa, H.; Takahashi, M.; Sakamoto, A.; Matsubara, T.; Arimura, G.-I.; Kawamura, Y.; Fukunaga, K.; Fujita, K.; Sakurai, N.; Hirata, T.; et al. Formation of unidentified nitrogen in plants: An implication for a novel nitrogen metabolism. Planta 2004, 219, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, T.; Tsukaya, H.; Uchimiya, H. Two independent and polarized processes of cell elongation regulate leaf blade. Development 1996, 122, 1589–1600. [Google Scholar] [PubMed]
- Callos, J.D.; Medford, J.I. Organ positions and pattern formation in the shoot apex. Plant J. 1994, 6, 1–7. [Google Scholar] [CrossRef]
- Potters, G.; Pasternak, T.P.; Guisez, Y.; Palme, K.J.; Jansen, M.A. Stress-induced morphogenic responses: Growing out of trouble? Trends Plant Sci. 2007, 3, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Granier, C.; Tardieu, F. Multi-scale phenotyping of leaf expansion in response to environmental changes: The whole is more than the sum of parts. Plant Cell Environ. 2009, 32, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Kawade, K.; Horiguchi, G.; Tsukaya, H. Non-cell-autonomously coordinated organ size regulation in leaf development. Development 2010, 137, 4221–4227. [Google Scholar] [CrossRef]
- Ischiropoulos, H. Protein tyrosine nitration: An update. Arch. Biochem. Biophys. 2009, 484, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Radi, R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Nat. Acad. Sci. USA 2004, 101, 4003–4008. [Google Scholar] [CrossRef] [PubMed]
- Rubbo, H.; Radi, R. Protein and lipid nitration: Role in redox signaling and injury. Biochim. Biophys. Acta 2008, 1780, 1318–1324. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Chaki, M.; Leterrier, M.; Barroso, J.B. Protein tyrosine nitration: A new challenge in plants. Plant Signal. Behav. 2009, 4, 920–923. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; David, A.; Baluška, F.; Bhatla, S.C. Rapid auxin-induced nitric oxide accumulation and subsequent tyrosine nitration of proteins during adventitious root formation in sunflower hypocotyls. Plant Signal. Behav. 2013, 8, e23196. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ara, J.; Przedborski, S.; Naini, A.B.; Jackson-Lewis, V.; Trifiletti, R.R.; Horwitz, J.; Ischiropoulos, H. Inactivation of tyrosine hydroxylase by nitration following exposure to peroxynitrite and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP). Proc. Natl. Acad. Sci. USA 1998, 95, 7659–7663. [Google Scholar] [CrossRef]
- Souza, J.M.; Peluffo, G.; Radi, R. Protein tyrosine nitration–Functional alteration or just a biomarker? Free Radic. Biol. Med. 2008, 45, 357–366. [Google Scholar] [CrossRef]
- Álvarez, C.; Lozano-Juste, J.; Romero, L.C.; García, I.; Gotor, C.; León, J. Inhibition of Arabidopsis O-acetylserine(thiol)lyase A1 by tyrosine nitration. J. Biol. Chem. 2011, 286, 578–586. [Google Scholar] [CrossRef]
- Abello, N.; Kerstjens, H.A.M.; Postma, D.S.; Bischoff, R. Protein tyrosine nitration: Selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 2009, 8, 3222–3238. [Google Scholar] [CrossRef]
- Koeck, T.; Fu, X.; Hazen, S.L.; Crabb, J.W.; Stuehr, D.J.; Aulark, K.S. Rapid and selective oxygen-regulated protein tyrosine denitration and nitration in mitochondria. J. Biol. Chem. 2004, 279, 27257–27262. [Google Scholar] [CrossRef]
- Morot-Gaudry-Talarmain, Y.; Rockel, P.; Moureaux, T.; Quilleré, I.; Leydecker, M.T.; Kaiser, W.M.; Morot-Gaudry, J.F. Nitrite accumulation and nitric oxide emission in relation to cellular signaling in nitrite reductase antisense tobacco. Planta 2002, 215, 708–715. [Google Scholar] [PubMed]
- Cecconi, D.; Orzetti, S.; Vandelle, E.; Rinalducci, S.; Zolla, L.; Delledonne, M. Protein nitration during defense response in Arabidopsis thaliana. Electrophoresis 2009, 30, 2460–2468. [Google Scholar] [CrossRef]
- Takahashi, M.; Shigeto, J.; Sakamoto, A.; Izumi, S.; Asada, K.; Morikawa, H. Dual selective nitration in Arabidopsis: Almost exclusive nitration of PsbO and PsbP, and highly susceptible nitration of four non-PSII proteins, including peroxiredoxin II E. Electrophoresis 2015, 36, 2569–2578. [Google Scholar] [CrossRef] [PubMed]
- Nelson, N.; Yocum, C.F. Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 2006, 57, 521–565. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.M.; Chow, W.S.; De Las Rivas, J. Dynamic flexibility in the structure and function of photosystem II in higher plant thylakoid membranes: The grana enigma. Photosynth. Res. 2008, 98, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Suorsa, M.; Sirpiö, S.; Allahverdiyeva, Y.; Paakkarinen, V.; Mamedov, F.; Styring, S.; Aro, E.M. PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. J. Biol. Chem. 2006, 281, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Shigeto, J.; Shibata, T.; Sakamoto, A.; Izumi, S.; Morikawa, H. Differential abilities of nitrogen dioxide and nitrite to nitrate proteins in thylakoid membranes isolated from Arabidopsis leaves. Plant Signal. Behav. 2016, 11, e1237329. [Google Scholar] [CrossRef] [PubMed]
- Huie, R.E. The reaction kinetics of NO2. Toxicology 1994, 89, 193–216. [Google Scholar] [CrossRef]
- Kawamura, Y.; Takahashi, M.; Arimura, G.; Isayama, T.; Irifune, K.; Goshima, N.; Morikawa, H. Determination of levels of NO3−, NO2− and NH4+ ions in leaves of various plants by capillary electrophoresis. Plant Cell Physiol. 1996, 37, 878–880. [Google Scholar] [CrossRef]
- Takahashi, M.; Shigeto, J.; Sakamoto, A.; Morikawa, H. Light-triggered selective nitration of PsbO1 in isolated Arabidopsis thylakoid membranes is inhibited by photosynthetic electron transport inhibitors. Plant Signal. Behav. 2016, 11, e1263413. [Google Scholar] [CrossRef]
- Peltier, J.B.; Friso, G.; Kalume, D.E.; Roepstorff, P.; Nilsson, F.; Adamska, I.; Van Wijk, K.J. Proteomics of the chloroplast: Systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 2000, 12, 319–341. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Noguchi, T. Infrared detection of a proton released from tyrosine YD to the bulk upon its photo-oxidation in photosystem II. Biochemistry 2015, 54, 5045–5053. [Google Scholar] [CrossRef] [PubMed]
- Fork, D.C.; Urbach, W. Evidence for the localization of plastocyanin in the electron-transport chain of photosynthesis. Proc. Natl. Acad. Sci. USA 1965, 53, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, K.; Mano, J.; Asada, K. Photoproduction of the azidyl radical from the azide anion on the oxidizing side of photosystem II and suppression of photooxidation of tyrosine Z by the azidyl radical. Plant Cell Physiol. 1995, 36, 121–129. [Google Scholar] [CrossRef]
- Kovacs, L.; Hegde, U.; Padhye, S.; Bernat, G.; Demeter, S. Effect of potassium-(picrate)-(18-crown-6) on the photosynthetic electron transport. Z. Naturforsch. 1996, 51, 539–547. [Google Scholar] [CrossRef]
- Takahashi, M.; Shigeto, J.; Sakamoto, A.; Morikawa, H. Selective nitration of PsbO1 inhibits oxygen evolution from isolated Arabidopsis thylakoid membranes. Plant Signal. Behav. 2017, 12, e1304342. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, J. Changes in spinach thylakoid activity due to nitrite ions. Photosynth. Res. 1987, 12, 255–263. [Google Scholar] [CrossRef]
- Wincencjusz, H.; Yocum, C.F.; Van Gorkom, H.J. Activating anions that replace Cl− in the O2-evolving complex of photosystem II slow the kinetics of the terminal step in water oxidation and destabilize the S2 and S3 states. Biochemistry 1999, 38, 3719–3725. [Google Scholar] [CrossRef]
- Pokhrel, R.; Brudvig, G.W. Investigation of the inhibitory effect of nitrite on Photosystem II. Biochemistry 2013, 52, 3781–3789. [Google Scholar] [CrossRef]
- Takahashi, M.; Morikawa, H. A novel role for PsbO1 in photosynthetic electron transport as suggested by its light-triggered selective nitration in Arabidopsis thaliana. Plant Signal. Behav. 2018, 13, e1513298. [Google Scholar] [CrossRef]
- Su, X.; Ma, J.; Wei, X.; Cao, P.; Zhu, D.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 2017, 357, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Umena, Y.; Kawakami, K.; Shen, J.-R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, K.V.; Brudvi, G.W. Electron paramagnetic resonance distance measurements in photosynthetic reaction centers. In Distance Measurements in Biological Systems by EPR; Berliner, L.J., Eaton, G.R., Eaton, S.S., Eds.; Springer: London, UK, 2000; Volume 19, pp. 513–567. ISBN 978-1-4757-0575-1. [Google Scholar]
- Pigolev, A.V.; Klimov, V.V. The green alga Chlamydomonas reinhardtii as a tool for in vivo study of site-directed mutations in PsbO protein of photosystem II. Biochemistry (Moscow) 2015, 80, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Kimura, S. Long-range electron transfer over 4 nm governed by an inelastic hopping mechanism in self-assembled monolayers of helical peptides. J. Am. Chem. Soc. 2003, 125, 8732–8733. [Google Scholar] [CrossRef] [PubMed]
- Arikuma, Y.; Nakayama, H.; Morita, T.; Kimura, S. Ultra-long-range electron transfer through a self-assembled monolayer on gold composed of 120-Å-long α-helices. Langmuir 2011, 27, 1530–1535. [Google Scholar] [CrossRef] [PubMed]
- Mclendon, G.; Hake, R. Interprotein electron transfer. Chem. Rev. 1992, 92, 481–490. [Google Scholar] [CrossRef]
- Miyashita, O.; Okamura, M.Y.; Onuchic, J.N. Interprotein electron transfer from cytochrome c2 to photosynthetic reaction center: Tunneling across an aqueous interface. Proc. Natl. Acad. Sci. USA 2005, 102, 3558–3563. [Google Scholar] [CrossRef] [PubMed]
- Kolbert, Z.; Feigl, G.; Bordé, Á.; Molnár, Á.; Erdei, L. Protein tyrosine nitration in plants: Present knowledge, computational prediction and future perspectives. Plant Physiol. Biochem. 2017, 113, 56–63. [Google Scholar] [CrossRef]
- Salem, M.A.; Li, Y.; Bajdzienko, K.; Fisahn, J.; Watanabe, M.; Hoefgen, R.; Schöttler, M.A.; Giavalisco, P. RAPTOR controls developmental growth transitions by altering the hormonal and metabolic balance. Plant Physiol. 2018, 177, 565–593. [Google Scholar] [CrossRef]
- Chen, G.H.; Liu, M.J.; Xiong, Y.; Sheen, J.; Wu, S.H. TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 2018, 115, 12823–12828. [Google Scholar] [CrossRef]
- Schepetilnikov, M.; Ryabova, L.A. Recent discoveries on the role of TOR (Target of Rapamycin) signaling in translation in plants. Plant Physiol. 2018, 176, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.C.; Liu, X.; Fu, L.; Hou, Y.J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell. 2018, 69, 100–112.e6. [Google Scholar] [CrossRef] [PubMed]
- Castillo, M.C.; Lozano-Juste, J.; González-Guzmán, M.; Rodriguez, L.; Rodriguez, P.L.; León, J. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci. Signal. 2015, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Karve, A.; Brandon, D.; Moore, B.D. Function of Arabidopsis hexokinase-like1 as a negative regulator of plant growth. J. Exp. Bot. 2009, 60, 4137–4149. [Google Scholar] [CrossRef] [PubMed]
- John, F.; Roffler, S.; Wicker, T.; Ringli, C. Plant TOR signaling components. Plant Signal. Behav. 2011, 6, 1700–1705. [Google Scholar] [CrossRef]
- Antoniuk-Pablant, A.; Sherman, B.D.; Kodis, G.; Gervaldo, M.; Moore, T.A.; Moore, A.L.; Gust, D.; Megiatto, J.D., Jr. Mimicking the electron transfer chain in photosystem II with a molecular triad thermodynamically capable of water oxidation. Proc. Natl. Acad. Sci. USA 2012, 109, 15578–15583. [Google Scholar]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, M.; Morikawa, H. Nitrogen Dioxide at Ambient Concentrations Induces Nitration and Degradation of PYR/PYL/RCAR Receptors to Stimulate Plant Growth: A Hypothetical Model. Plants 2019, 8, 198. https://doi.org/10.3390/plants8070198
Takahashi M, Morikawa H. Nitrogen Dioxide at Ambient Concentrations Induces Nitration and Degradation of PYR/PYL/RCAR Receptors to Stimulate Plant Growth: A Hypothetical Model. Plants. 2019; 8(7):198. https://doi.org/10.3390/plants8070198
Chicago/Turabian StyleTakahashi, Misa, and Hiromichi Morikawa. 2019. "Nitrogen Dioxide at Ambient Concentrations Induces Nitration and Degradation of PYR/PYL/RCAR Receptors to Stimulate Plant Growth: A Hypothetical Model" Plants 8, no. 7: 198. https://doi.org/10.3390/plants8070198
APA StyleTakahashi, M., & Morikawa, H. (2019). Nitrogen Dioxide at Ambient Concentrations Induces Nitration and Degradation of PYR/PYL/RCAR Receptors to Stimulate Plant Growth: A Hypothetical Model. Plants, 8(7), 198. https://doi.org/10.3390/plants8070198



