Molecular and Morphological Changes Induced by Leonardite-based Biostimulant in Beta vulgaris L.
Abstract
1. Introduction
2. Results
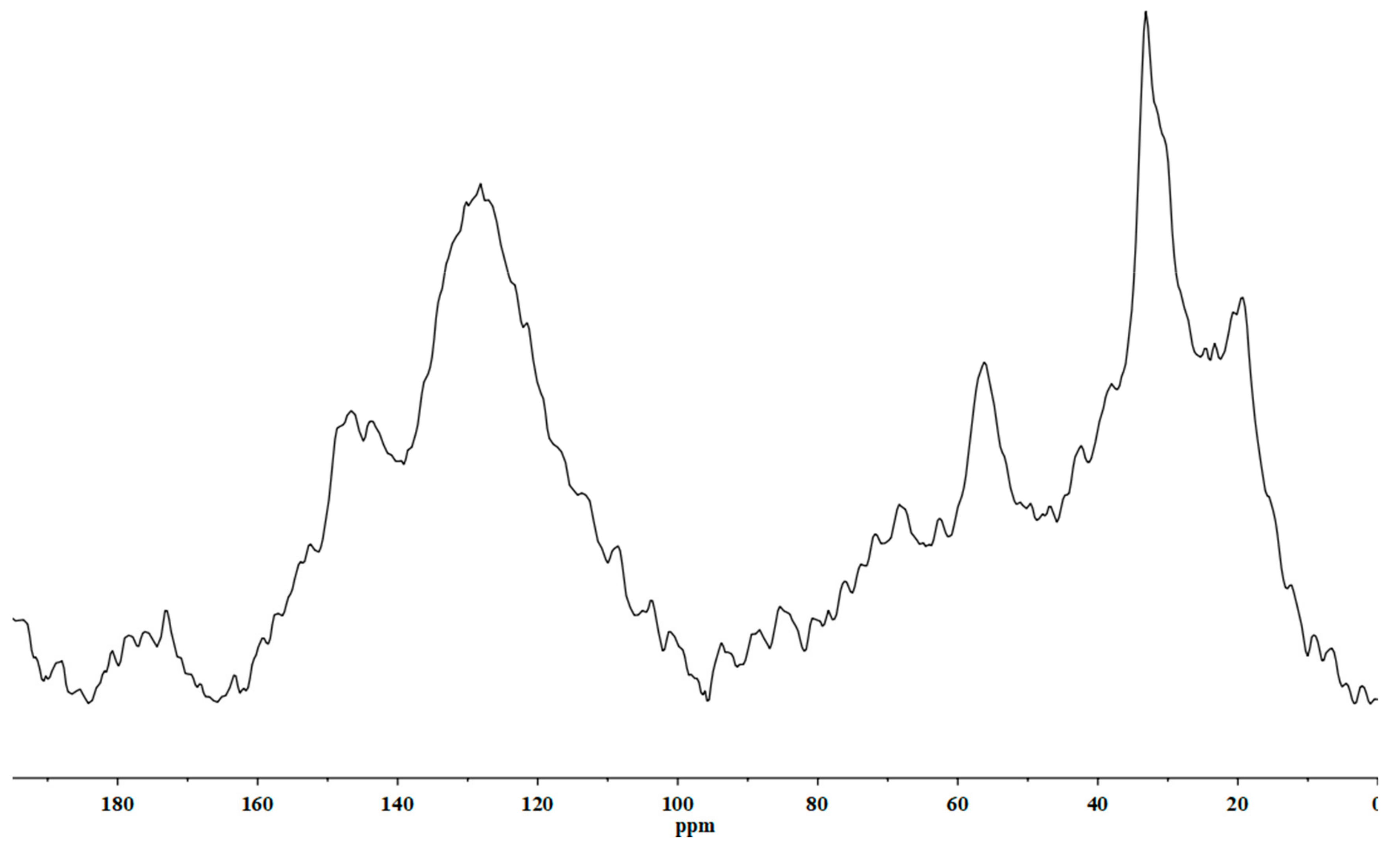
2.1. Chemical Characterization of Biostimulant
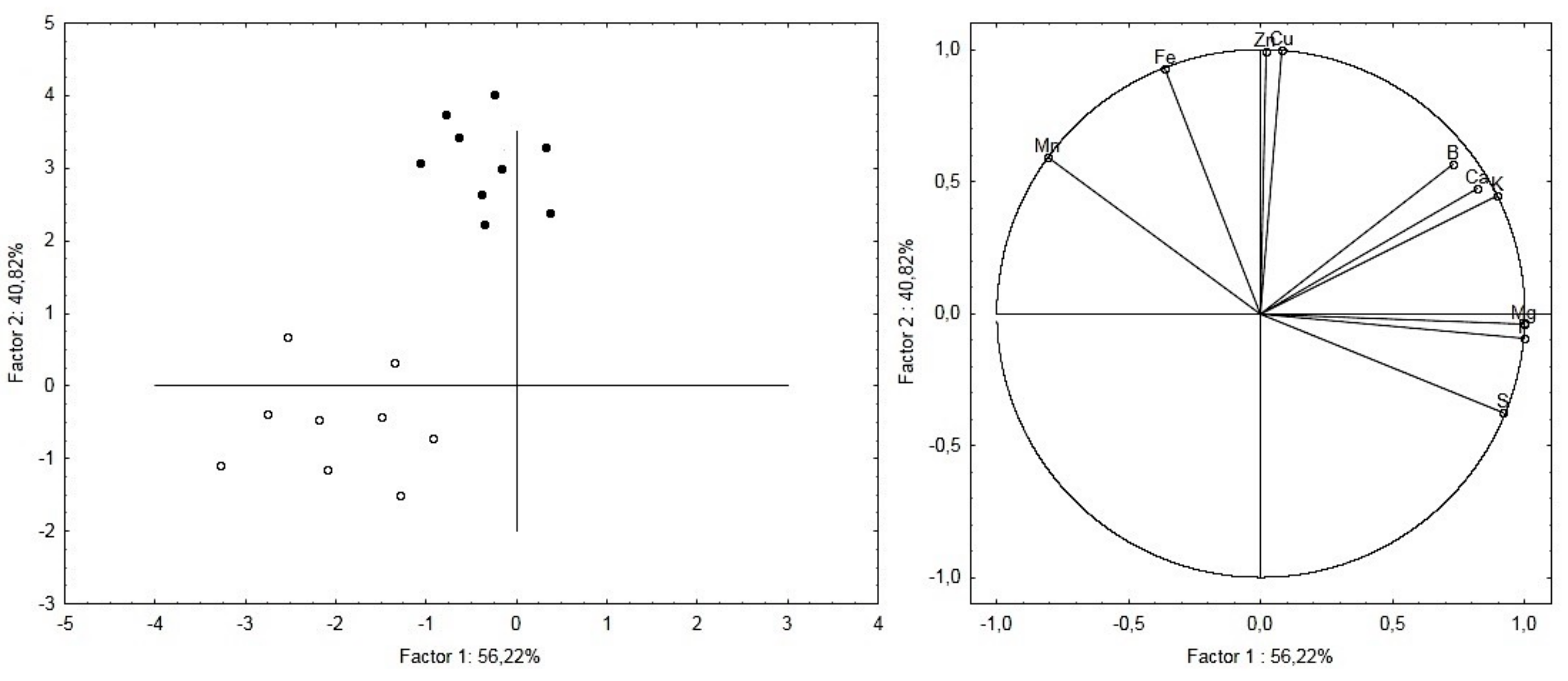
2.2. Ionomic Analysis
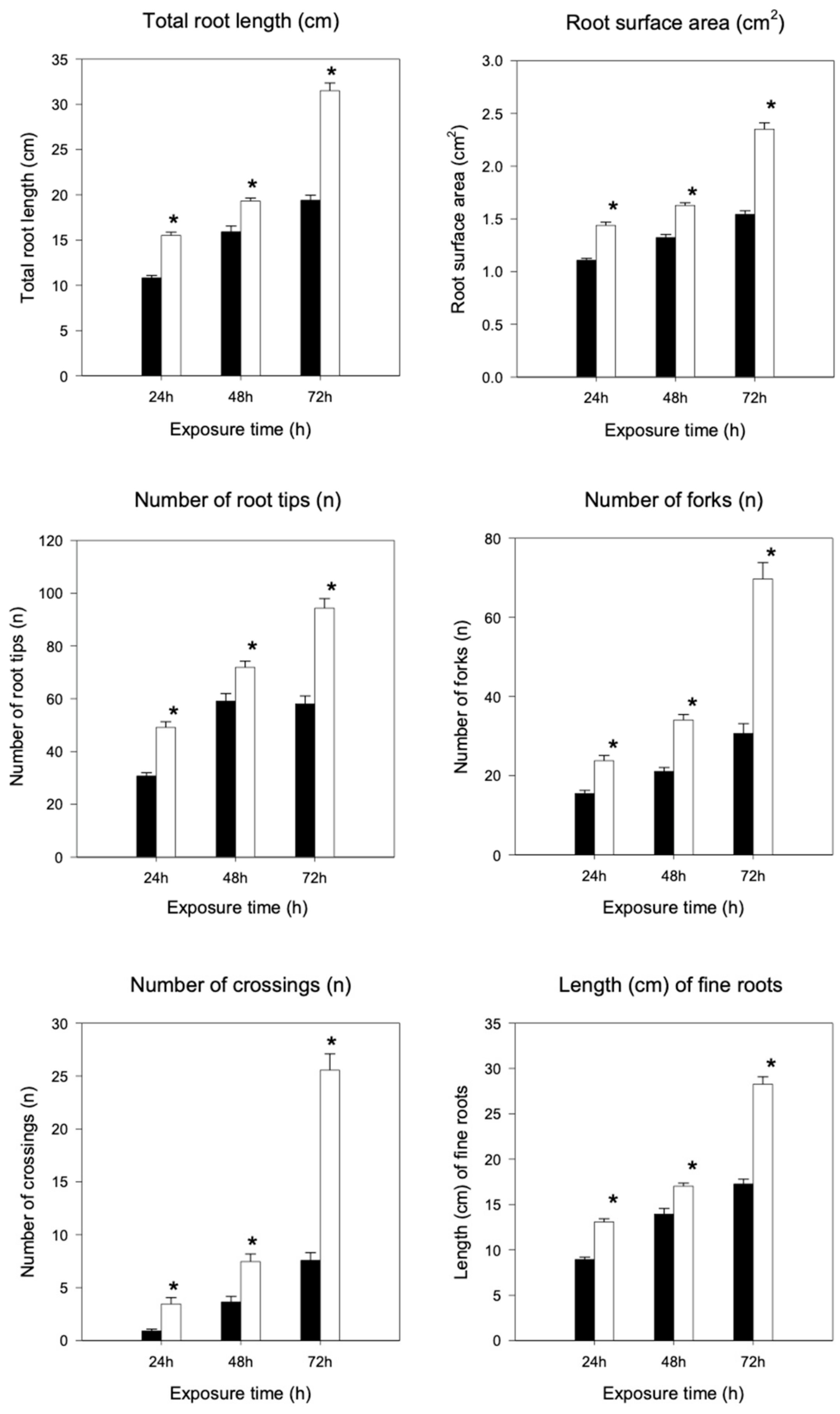
2.3. Root Morphological Analysis
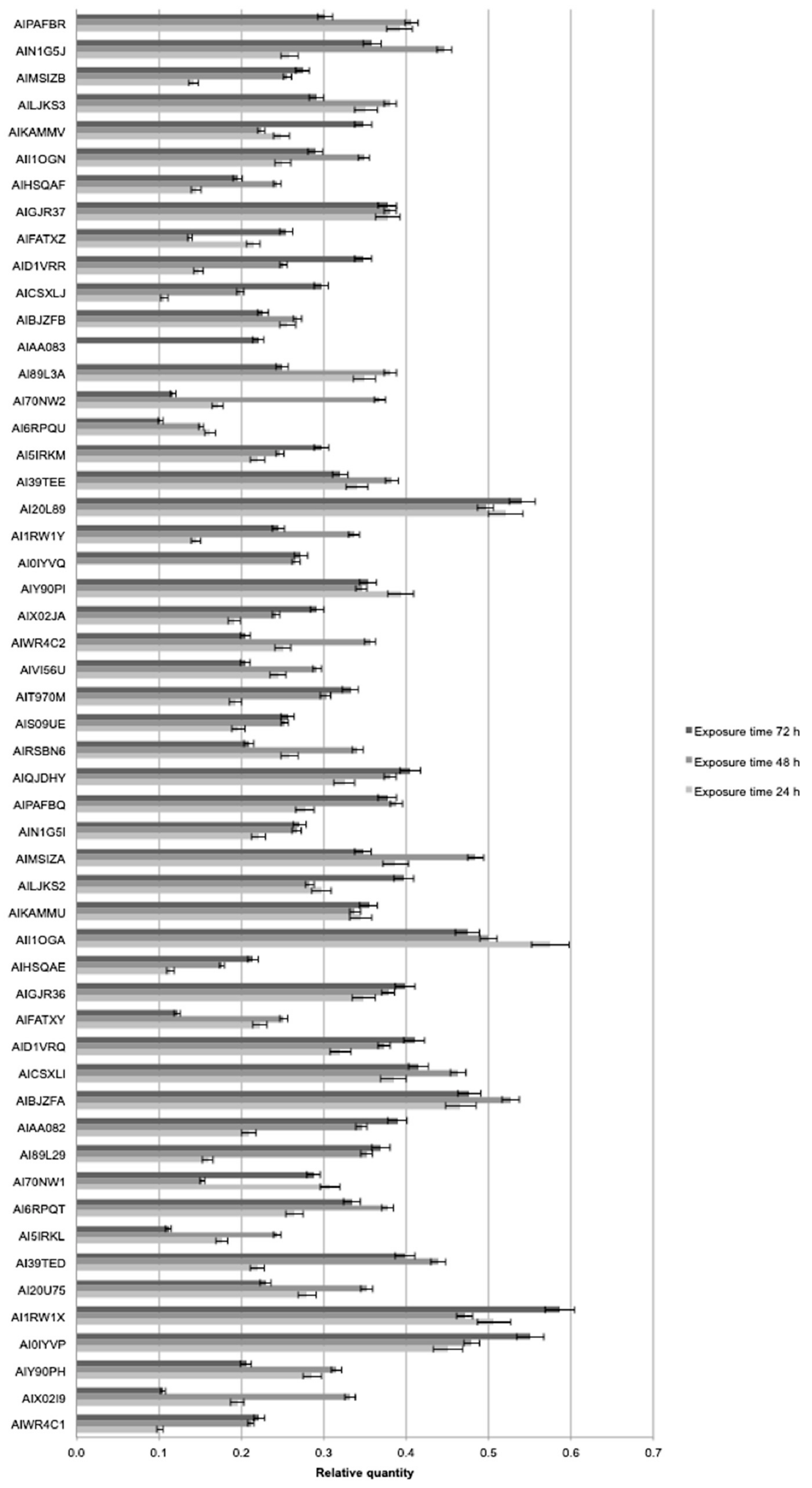
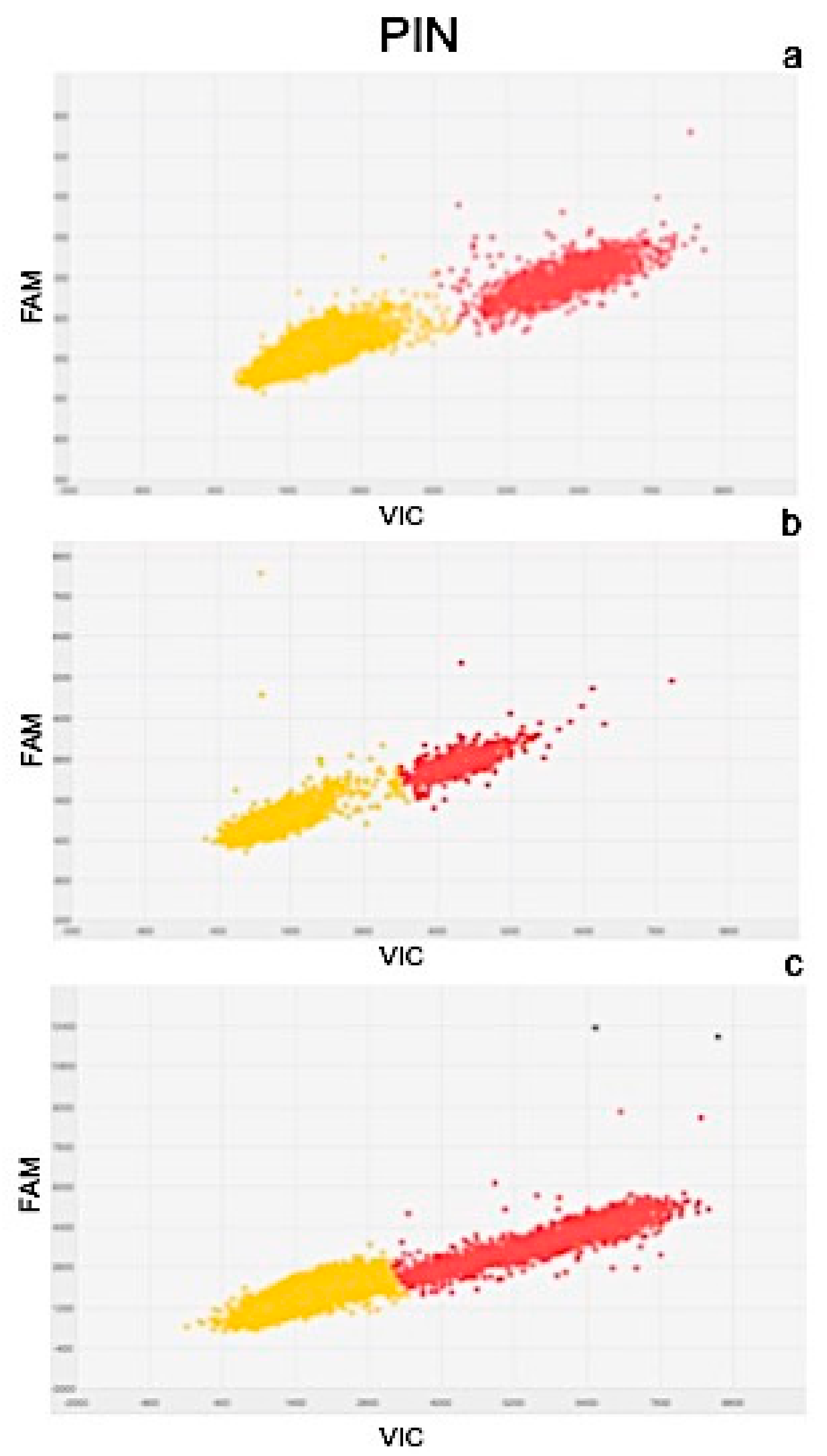
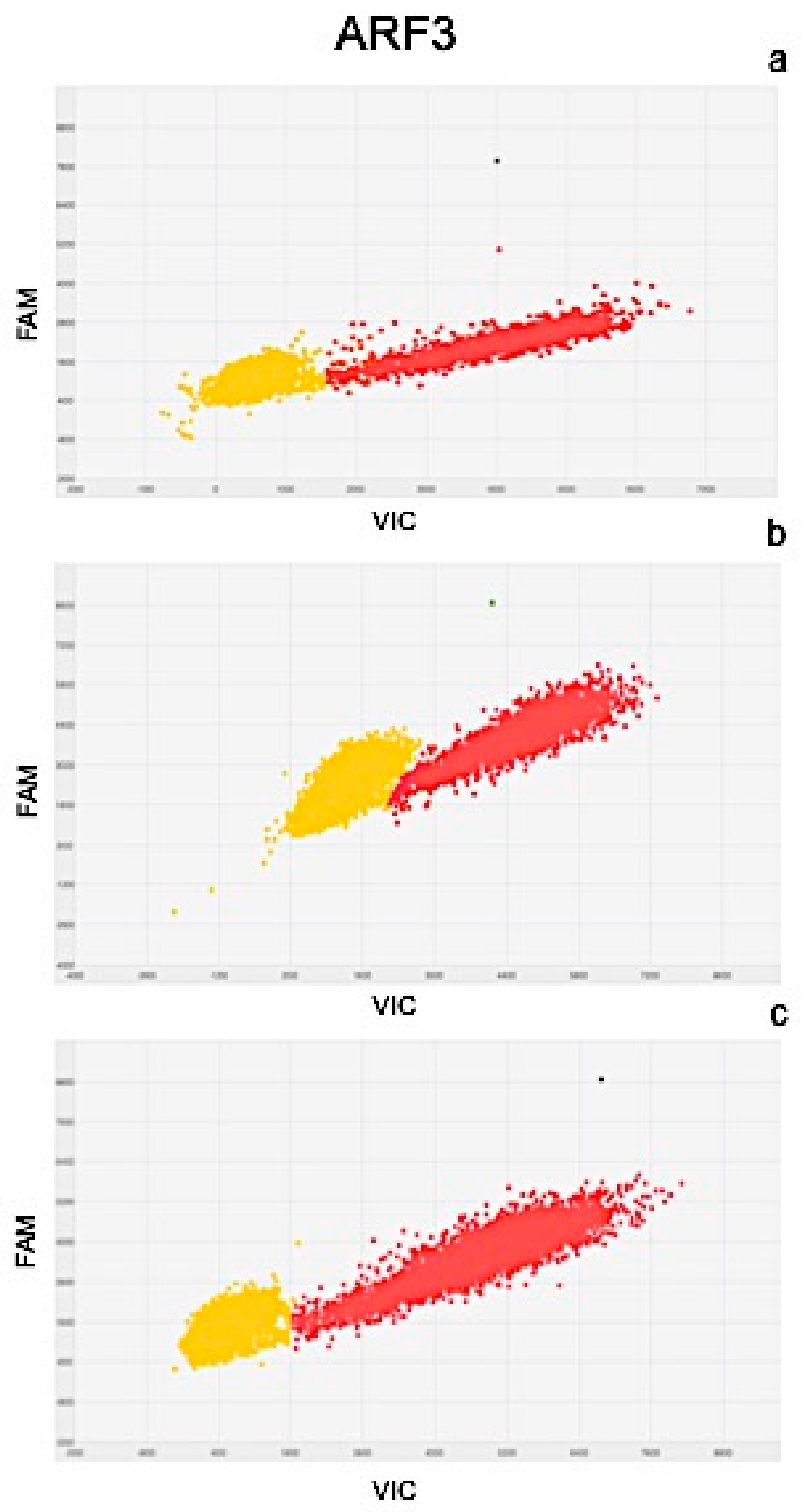
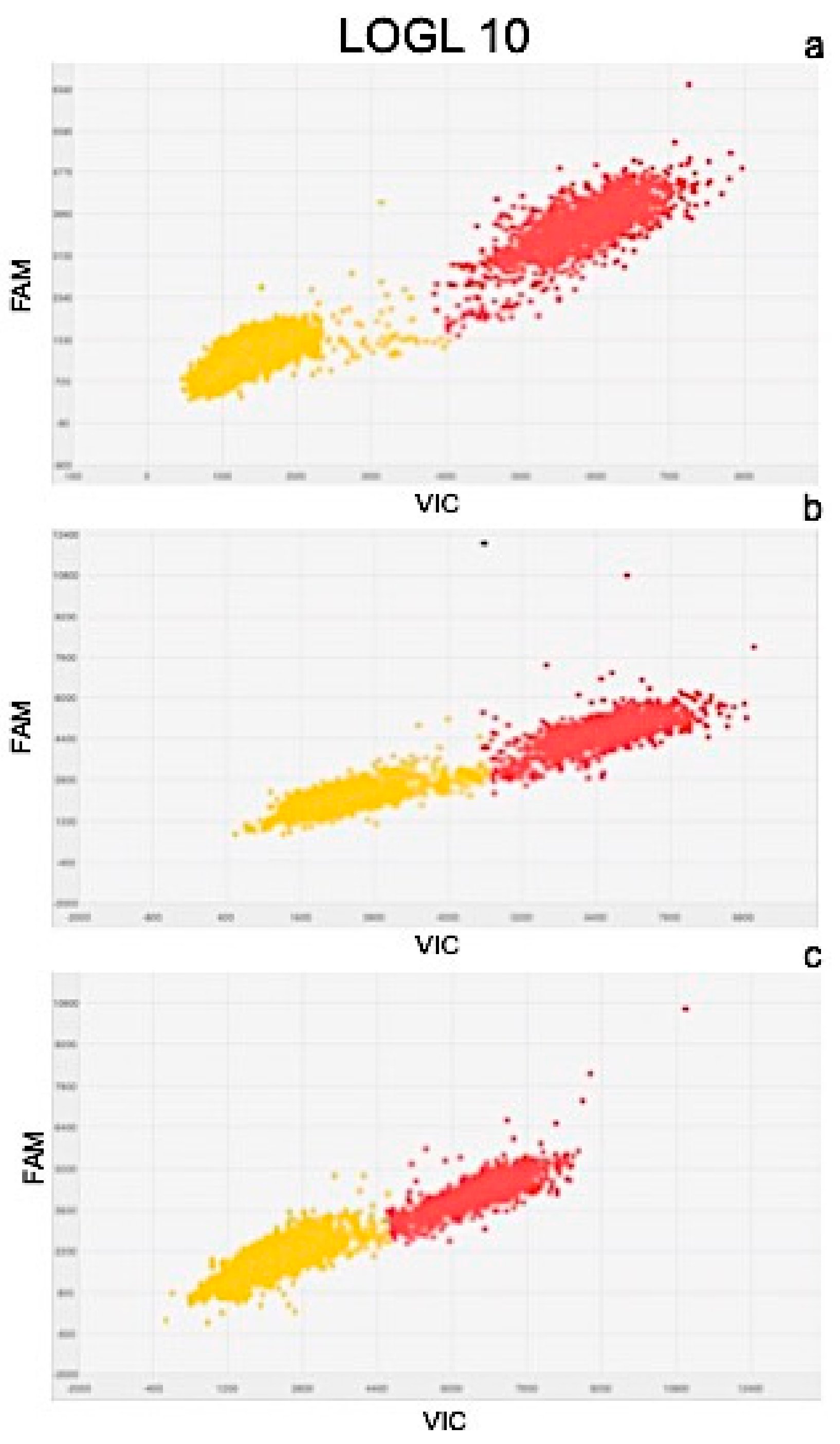
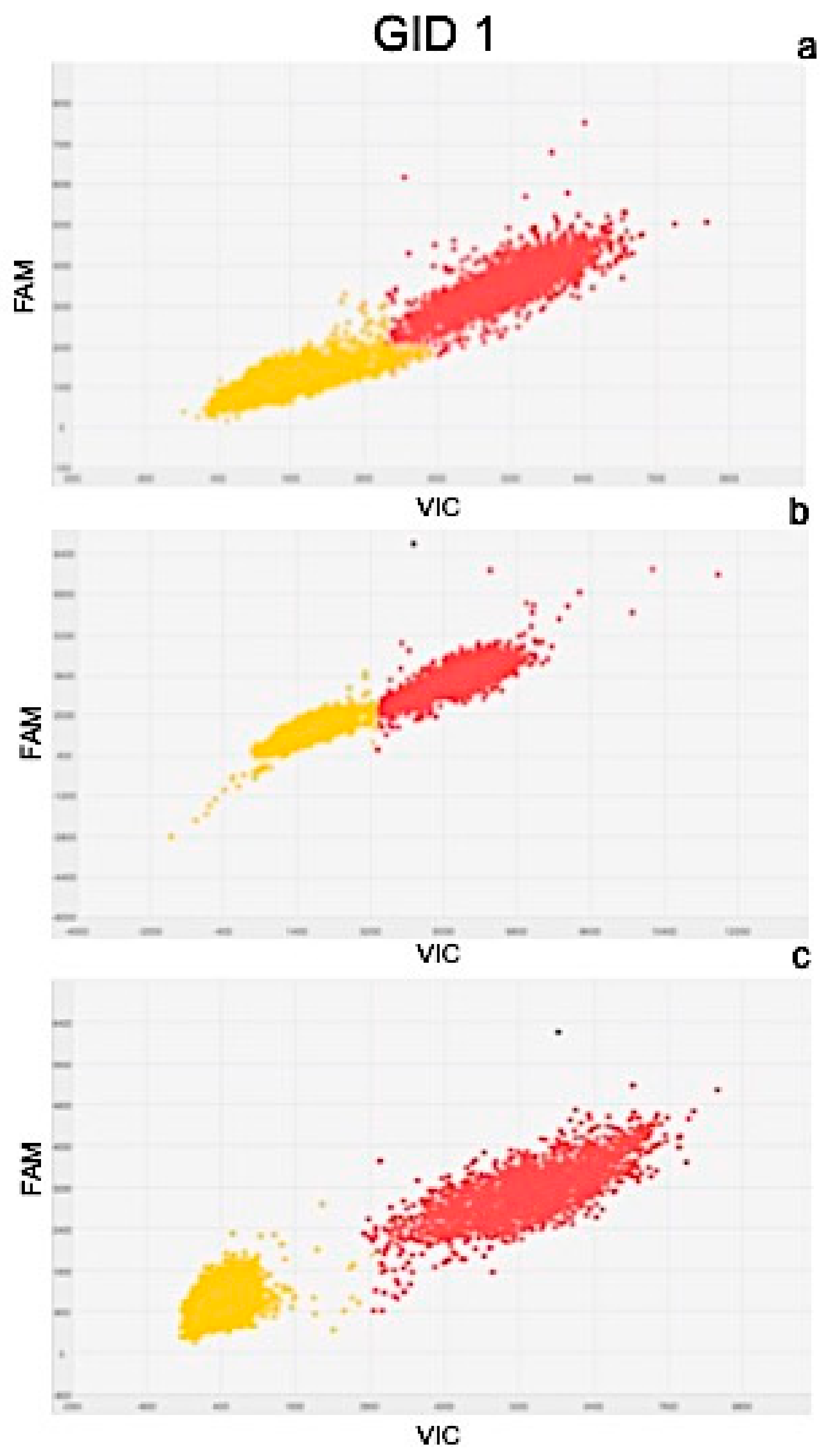
2.4. Molecular Analysis
3. Discussion
4. Materials and Methods
4.1. Chemical Characterization of HS
4.1.1. Elemental Analysis
4.1.2. NMR Analysis
4.2. Experimental Design, Plant Material, and Growing Conditions
4.2.1. Preliminary Investigation
4.2.2. Experimental Design
4.2.3. Plant Material
4.2.4. Growing Conditions
4.3. Ionomic Analysis
4.4. Root Morphological Analysis
4.5. Molecular Analysis
4.5.1. Root RNA Isolation and cDNA Synthesis
4.5.2. Real Time PCR and Digital PCR Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Calvo, P.; Nelson, L.; Kloepper, J.W. Agricultural uses of plant biostimulants. Plant Soil 2014, 383, 3–41. [Google Scholar] [CrossRef]
- European Biostimulant Industry Council. Economic Overview of the Biostimulants Sector in Europe. 2013. Available online: http://www.biostimulants.eu/2013/04/2013-overview-of-the-european-biostimulants-market/17April2013.pdf (accessed on 17 April 2013).
- Alam, M.Z.; Braun, G.; Norrie, J.; Hodges, D.M. Ascophyllum extract application can promote plant growth and root yield in carrot associated with increased root-zone soil microbial activity. Can. J. Plan. Sci. 2014, 94, 337–348. [Google Scholar] [CrossRef]
- Parrado, J.; Bautista, J.; Romero, E.J.; García-Martínez, A.M.; Friaza, V.; Tejada, M. Production of a carob enzymatic extract: Potential use as a biofertilizer. Bioresour. Technol. 2008, 99, 2312–2318. [Google Scholar] [CrossRef] [PubMed]
- Nardi, S.; Pizzeghello, D.; Schiavon, M.; Ertani, A. Plant biostimulants: Physiological responses induced by protein hydrolyzed-based products and humic substances in plant metabolism. Sci. Agric. 2016, 73, 18–23. [Google Scholar] [CrossRef]
- Canellas, L.P.; Dantas, D.J.; Aguiar, N.O.; Peres, L.E.P.; Zsogon, A.; Olivares, F.L.; Dobbss, L.B.; Facanha, A.R.; Nebbioso, A.; Piccolo, A. Probing the hormonal activity of fractionated molecular humic components in tomato auxin mutants. Ann. Appl. Biol. 2011, 159, 202–211. [Google Scholar] [CrossRef]
- Mora, V.; Baigorri, R.; Bacaicoa, E.; Zamarreno, A.M.; Garcia-Mina, J.M. The humic acid-induced changes in the root concentration of nitric oxide, IAA and ethylene do not explain the changes in root architecture caused by humic acid in cucumber. Environ. Exp. Bot. 2012, 76, 24–32. [Google Scholar] [CrossRef]
- Tahir, M.M.; Khurshid, M.; Khan, M.Z.; Abbasi, M.K.; Kazmi, M.H. Lignite-derived humic acid effect on growth of wheat plants in different soils. Pedosphere 2011, 21, 124–131. [Google Scholar] [CrossRef]
- Jindo, K.; Martim, S.A.; Navarro, E.C.; Aguiar, N.O.; Canellas, L.P. Root growth promotion by humic acids from composted and non-composted urban organic wastes. Plant Soil 2012, 353, 209–220. [Google Scholar] [CrossRef]
- Aydin, A.; Kant, C.; Turan, M. Humic acid application alleviates salinity stress of bean (Phaseolus vulgaris L.) plants decreasing membrane leakage. Afr. J. Agric. Res. 2012, 7, 1073–1086. [Google Scholar] [CrossRef]
- Cimrin, K.M.; Onder, T.; Turan, M.; Burcu, T. Phosphorus and humic acid application alleviate salinity stress of pepper seedling. Afr. J. Biotechnol. 2010, 9, 5845–5851. [Google Scholar]
- García, A.C.; Santos, L.A.; Izquierdo, F.G.; Sperandio, M.V.L.; Castro, R.N.; Berbara, R.L.L. Vermicompost humic acids as an ecological pathway to protect rice plant against oxidative stress. Ecol. Eng. 2012, 47, 203–208. [Google Scholar] [CrossRef]
- O’Donnell, R.W. The auxin-like effects of humic preparations from leonardite. Soil Sci. 1973, 116, 106–112. [Google Scholar] [CrossRef]
- Muscolo, A.; Sidari, M.; Nardi, S. Humic substance: Relationship between structure and activity. Deeper information suggests univocal findings. J. Geochem. Explor. 2013, 129, 57–63. [Google Scholar] [CrossRef]
- Tahiri, A.; Delporte, F.; Muhovski, Y.; Ongena, M.; Thonart, P.; Druart, P. Change in ATP-binding cassette B1/19, glutamine synthetase and alcohol dehydrogenase gene expression during root elongation in Betula pendula Roth and Alnus glutinosa L. Gaertn in response to leachate and leonardite humic substances. Plant Physiol. Biochem. 2016, 98, 25–38. [Google Scholar] [CrossRef]
- Pertuit, A.J.; Dudley, J.B.; Toler, J.E. Leonardite and fertilizer levels influence tomato seedling growth. Hort. Sci. 2001, 36, 913–915. [Google Scholar] [CrossRef]
- Akinremi, O.O.; Janzen, H.H.; Lemke, R.L.; Larney, F.J. Response of canola, wheat and green beans to leonardite additions. Can. J. Soil Sci. 2000, 80, 437–443. [Google Scholar] [CrossRef]
- Biancardi, E.; McGrath, J.M.; Panella, L.W.; Lewellen, R.T.; Stevanato, P. Sugar Beet. In Handbook of Plant Breeding, Tuber and Root Crops; Bradshaw, J., Ed.; Springer: New York, NY, USA, 2010; Volume 4, pp. 173–219. [Google Scholar]
- Norouzi, P.; Stevanato, P.; Mahmoudi, S.B.; Fasahat, P.; Biancardi, E. Molecular progress in sugar beet breeding for resistance to biotic stresses in sub-arid conditions—Current status and perspectives. J. Crop Sci. Biotechnol. 2017, 20, 99–105. [Google Scholar] [CrossRef]
- Barone, V.; Baglieri, A.; Stevanato, P.; Broccanello, C.; Bertoldo, G.; Bertaggia, M.; Cagnin, M.; Pizzeghello, D.; Moliterni, V.M.C.; Mandolino, G.; et al. Root morphological and molecular responses induced by microalgae extracts in sugar beet (Beta vulgaris L.). J. Appl. Phycol. 2018, 30, 1061–1071. [Google Scholar] [CrossRef]
- Stevanato, P.; Broccanello, C.; Moliterni, V.; Mandolino, G.; Barone, V.; Lucini, L.; Bertoldo, G.; Bertaggia, M.; Cagnin, M.; Pizzeghello, D.; et al. Innovative approaches to evaluate sugar beet responses to changes in sulfate availability. Front. Plant Sci. 2018, 9, 14. [Google Scholar] [CrossRef]
- Kögel-Knabner, I. 13C and 15N NMR spectroscopy as a tool in soil organic matter studies. Geoderma 1997, 80, 243–270. [Google Scholar] [CrossRef]
- Hatcher, P.G.; VanderHart, D.L.; Earl, W.L. Use of solid-state 13C NMR in structural studies of humic acids and humin from Holocene sediments. Org. Geochem. 1980, 2, 87–92. [Google Scholar] [CrossRef]
- Canellas, L.P.; Olivares, F.L.; Aguiar, N.O.; Jones, D.L.; Nebbioso, A.; Mazzei, P.; Piccolo, A. Humic and fulvic acids as biostimulants in horticulture. Sci. Hortic. 2015, 196, 15–27. [Google Scholar] [CrossRef]
- Preston, C.M.; Ripmeester, J.A. Application of solution and solid state 13C NMR to four organic soils, their humic acids, fulvic acids, humins and hydrolysis residues. Can. J. Spectrosc. 1982, 27, 99–105. [Google Scholar]
- Chilom, G.; Baglieri, A.; Johnson-Edler, C.A.; Rice, J.A. Hierarchical self-assembling properties of natural organic matter’s components. Org. Geochem. 2013, 57, 119–126. [Google Scholar] [CrossRef]
- Thorn, K.A.; Folan, D.W.; MacCarthy, P. Characterization of the International Humic Substances Society Standard and Reference Fulvic and Humic Acids by Solution State Carbon-13 (13C) and Hydrogen-1 (1H) Nuclear Magnetic Resonance Spectrometry; Water-Resources Investigations Report; U.S. Geological Survey: Denver, CO, USA, 1989; p. 93.
- Allard, B. A comparative study on the chemical composition of humic acids from forest soil, agricultural soil and lignite deposit: Bound lipid, carbohydrate and amino acid distributions. Geoderma 2006, 130, 77–96. [Google Scholar] [CrossRef]
- Piccolo, A.; Conte, P.; Spaccini, R.; Mbagwu, J.S.C. Influence of land use on the characteristics of humic substances in some tropical soils of Nigeria. Eur. J. Soil Sci. 2005, 56, 343–352. [Google Scholar] [CrossRef]
- García, A.C.; de Souza, L.G.A.; Pereira, M.G.; Castro, R.N.; García-Mina, J.M.; Zonta, E.; Lisboa, F.J.G.; Berbara, R.L.L. Structure-property-function relationship in humic substances to explain the biological activity in plants. Sci. Rep. 2016, 6, 20798. [Google Scholar] [CrossRef]
- Vaccaro, S.; Muscolo, A.; Pizzeghello, D.; Spaccini, R.; Piccolo, A.; Nardi, S. Effect of a compost and its water-soluble fractions on key enzymes of nitrogen metabolism in maize seedlings. J. Agric. Food Chem. 2009, 57, 11267–11276. [Google Scholar] [CrossRef]
- Spaccini, R.; Piccolo, A.; Contea, P.; Haberhauer, G.; Gerzabek, M.H. Increased soil organic carbon sequestration through hydrophobic protection by humic substances. Soil Biol. Biochem. 2002, 34, 1839–1851. [Google Scholar] [CrossRef]
- Canellas, L.P.; Olivares, F.L. Physiological Responses to Humic Substances as Plant Growth Promoter. Chem. Biol. Technol. Agric. 2014, 1–3. [Google Scholar] [CrossRef]
- Sessi, E.; Nardi, S.; Gessa, C. Effects of low and high molecular weight humic substances from two different soils on nitrogen assimilation pathway in maize seedlings. Humic Subst. Environ. 2000, 2, 39–46. [Google Scholar]
- Nardi, S.; Muscolo, A.; Vaccaro, S.; Baiano, S.; Spaccini, R.; Piccolo, A. Relationship between molecular characteristics of soil humic fractions and glycolytic pathway and krebs cycle in maize seedlings. Soil Biol. Biochem. 2007, 39, 3138–3146. [Google Scholar] [CrossRef]
- De Dorlodot, S.; Forster, B.; Pagès, L.; Price, A.; Tuberosa, R.; Draye, X. Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 2007, 12, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Lucini, L.; Rouphael, Y.; Cardarelli, M.; Canaguier, R.; Kumar, P.; Colla, G. The effect of a plant-derived biostimulant on metabolic profiling and crop performance of lettuce grown under saline conditions. Sci. Hortic. 2015, 182, 124–133. [Google Scholar] [CrossRef]
- Stevanato, P.; Trebbi, D.; Saccomani, M. Root traits and yield in sugar beet: Identification of AFLP markers associated with root elongation rate. Euphytica 2010, 173, 289–298. [Google Scholar] [CrossRef]
- Rani Debi, B.; Taketa, S.; Ichii, M. Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (Oryza sativa). J. Plant Physiol. 2005, 162, 507–515. [Google Scholar] [CrossRef]
- Zhou, D.; Yin, K.; Xu, Z.; Xue, H. Effect of polar auxin transport on rice root development. Acta Bot. Sin. 2003, 45, 1421–1427. [Google Scholar]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef]
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef]
- Ganguly, A.; Lee, S.H.; Cho, M.; Lee, O.R.; Yoo, H.; Cho, H.T. Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol. 2010, 153, 1046–1061. [Google Scholar] [CrossRef] [PubMed]
- Millier, M.J.; Stamp, L.K.; Hessian, P.A. Digital-PCR for gene expression: Impact from inherent tissue RNA degradation. Sci. Rep. 2017, 7, 17235. [Google Scholar] [CrossRef]
- Povero, G.; Mejia, J.F.; Di Tommaso, D.; Piaggesi, A.; Warrior, P. A systematic approach to discover and characterize natural plant biostimulants. Front. Plant Sci. 2016, 7, 435. [Google Scholar] [CrossRef]
- Sanders, R.; Mason, D.J.; Foy, C.A.; Huggett, J.F. Evaluation of digital PCR for absolute RNA quantification. PLoS ONE 2013, 8, e75296. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, P.; Cortimiglia, C.; Picozzi, C.; Minozzi, G.; Malvisi, M.; Luini, M.; Castiglioni, B. Development of a droplet digital polymerase chain reaction for rapid and simultaneous identification of common foodborne pathogens in soft cheese. Front. Microbiol. 2016, 7, 1725. [Google Scholar] [CrossRef]
- Alikian, M.; Whale, A.S.; Akiki, S.; Piechocki, K.; Torrado, C.; Myint, T.; Cowen, S.; Griffiths, M.; Reid, A.G.; Apperley, J.; et al. RT-qPCR and RT-digital PCR: A comparison of different platforms for the evaluation of residual disease in chronic myeloid leukemia. Clin. Chem. 2017, 63, 525–531. [Google Scholar] [CrossRef]
- Kinz, E.; Leiherer, A.; Lang, A.H.; Drexel, H.; Muendlein, A. Accurate quantitation of JAK2 V617F allele burden by array-based digital PCR. Int. J. Lab. Hemat. 2015, 37, 217–224. [Google Scholar] [CrossRef]
- Tan, K.C.; Ipcho, S.V.; Trengove, R.D.; Oliver, R.P.; Solomon, P.S. Assessing the impact of transciptomics, proteomics and metabolomics on fungal phytopathology. Mol. Plant Pathol. 2009, 10, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Morey, J.S.; Ryan, J.C.; Van Dolah, F.M. Microarray validation: Factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online 2006, 8, 175–193. [Google Scholar] [CrossRef]
- Gray, W.M. Hormonal Regulation of Plant Growth and Development. PLoS Biol. 2004, 2, 311. [Google Scholar] [CrossRef] [PubMed]
- Torrey, J.G. Endogenous and Exogenous Influences on the Regulation of Lateral Root Formation. In New Root Formation in Plants and Cuttings; Jackson, M.B., Ed.; Nijhoff: Dordrecht, The Netherlands, 1986; pp. 31–66. [Google Scholar]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Harberd, N.P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 2003, 421, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Blasco-Escámez, D.; Lozano-Elena, F.; Fàbregas, N.; Caño-Delgado, A.I. The primary root of Sorghum bicolor (L. Moench) as a model system to study brassinosteroid signaling in crops. In Brassinosteroids; Humana Press: New York, NY, USA, 2017; pp. 181–192. [Google Scholar]
- Stevanato, P.; Fedito, P.; Trebbi, D.; Cagnin, M.; Saccomani, M.; Cacco, G. Effect of sulfate availability on root traits and microRNA395 expression in sugar beet. Biol. Plant. 2015, 59, 491–496. [Google Scholar] [CrossRef]
- Baglieri, A.; Cadili, V.; Monterumici, C.M.; Gennari, M.; Tabasso, S.; Montoneri, E.; Nardi, S.; Negre, M. Fertilization of bean plants with tomato plants hydrolysates. Effect on biomass production, chlorophyll content and N assimilation. Sci. Hortic. 2014, 176, 194–199. [Google Scholar] [CrossRef]
- Stevanato, P.; Broccanello, C.; Biscarini, F.; Del Corvo, M.; Sablok, G.; Panella, L.; Stella, A.; Concheri, G. High-throughput RAD-SNP genotyping for characterization of sugar beet genotypes. Plant Mol. Biol. Rep. 2014, 32, 691–696. [Google Scholar] [CrossRef]
- Arnon, D.I.; Hoagland, D.R. Crop production in artificial culture solution and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci. 1940, 50, 463–483. [Google Scholar]
| Element | Method | Percentage (%) |
|---|---|---|
| C | Combustion analysis | 51.1 |
| N | 0.88 | |
| S | 0.75 | |
| K | ICP analysis | 0.15 |
| P | 0.06 | |
| Ca | 0.90 | |
| Mg | 0.15 | |
| Na | 1.27 | |
| Fe | 2.11 | |
| Al | 1.02 |
| C Functional Group | Method | Percentage (%) |
|---|---|---|
| Alkyl-C | NMR analysis | 36.1 |
| N and O-alkyl-C | 20.9 | |
| Aromatic-C | 39.7 | |
| Carboxyl-C | 3.3 |
| Element | Untreated Roots | Treated Roots | t-Test | ||||
|---|---|---|---|---|---|---|---|
| B | 2.9 | ± | 0.5 | 5.2 | ± | 0.2 | p < 0.001 |
| Ca | 151.8 | ± | 10.8 | 234.5 | ± | 13.9 | p < 0.01 |
| Cu | 1.1 | ± | 0.02 | 2.9 | ± | 0.1 | p < 0.001 |
| Fe | 52 | ± | 6.2 | 112 | ± | 4.6 | p < 0.001 |
| K | 2014.4 | ± | 192.4 | 3959.3 | ± | 234.3 | p < 0.001 |
| Mg | 182.2 | ± | 7.8 | 545.9 | ± | 11.6 | p < 0.001 |
| Mn | 6.7 | ± | 0.5 | 7.2 | ± | 1.2 | p < 0.05 |
| P | 554.2 | ± | 34.1 | 937.8 | ± | 43 | p < 0.01 |
| S | 277.8 | ± | 12.4 | 332.8 | ± | 26.1 | p < 0.05 |
| Zn | 20.4 | ± | 3.4 | 51.2 | ± | 9.8 | p < 0.001 |
| Effect | df | SS | MS | F | p |
|---|---|---|---|---|---|
| Treatment | 1 | 1421 | 1421 | 115.2 | ** |
| Exposure | 2 | 2723 | 1362 | 231.3 | ** |
| Gene | 52 | 4523 | 87 | 6.9 | * |
| Gene Expression Values (copies μL−1) | Exposure Time | |||||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||||
| PIN | 102.06 | ± 3.2 | 80.9 | ± 4.7 | 129.07 | ± 6.3 |
| ARF3 | 105.09 | ± 2.5 | 90.58 | ± 3.1 | 132.19 | ± 3.5 |
| LOGL 10 | 131.95 | ± 1.9 | 119.34 | ± 2.4 | 108.98 | ± 2.0 |
| GID1 | 121.02 | ± 4.1 | 110.12 | ± 1.5 | 123.02 | ± 4.2 |
| BRI1 | 109.98 | ± 1.2 | 128.87 | ± 2.3 | 133.02 | ± 3.4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barone, V.; Bertoldo, G.; Magro, F.; Broccanello, C.; Puglisi, I.; Baglieri, A.; Cagnin, M.; Concheri, G.; Squartini, A.; Pizzeghello, D.; et al. Molecular and Morphological Changes Induced by Leonardite-based Biostimulant in Beta vulgaris L. Plants 2019, 8, 181. https://doi.org/10.3390/plants8060181
Barone V, Bertoldo G, Magro F, Broccanello C, Puglisi I, Baglieri A, Cagnin M, Concheri G, Squartini A, Pizzeghello D, et al. Molecular and Morphological Changes Induced by Leonardite-based Biostimulant in Beta vulgaris L. Plants. 2019; 8(6):181. https://doi.org/10.3390/plants8060181
Chicago/Turabian StyleBarone, Valeria, Giovanni Bertoldo, Francesco Magro, Chiara Broccanello, Ivana Puglisi, Andrea Baglieri, Massimo Cagnin, Giuseppe Concheri, Andrea Squartini, Diego Pizzeghello, and et al. 2019. "Molecular and Morphological Changes Induced by Leonardite-based Biostimulant in Beta vulgaris L." Plants 8, no. 6: 181. https://doi.org/10.3390/plants8060181
APA StyleBarone, V., Bertoldo, G., Magro, F., Broccanello, C., Puglisi, I., Baglieri, A., Cagnin, M., Concheri, G., Squartini, A., Pizzeghello, D., Nardi, S., & Stevanato, P. (2019). Molecular and Morphological Changes Induced by Leonardite-based Biostimulant in Beta vulgaris L. Plants, 8(6), 181. https://doi.org/10.3390/plants8060181













