Expanding Avenue of Fast Neutron Mediated Mutagenesis for Crop Improvement
Abstract
1. Introduction
2. Fast Neutron Radiation-Induced Mutagenesis
3. Combining FN Mutagenesis with Other Mutagenesis Methods
4. Genetic Stability of Mutants Generated Using Fast Neutron Radiations
5. Fast Neutron Mutagenesis in Polyploidy Plant Species
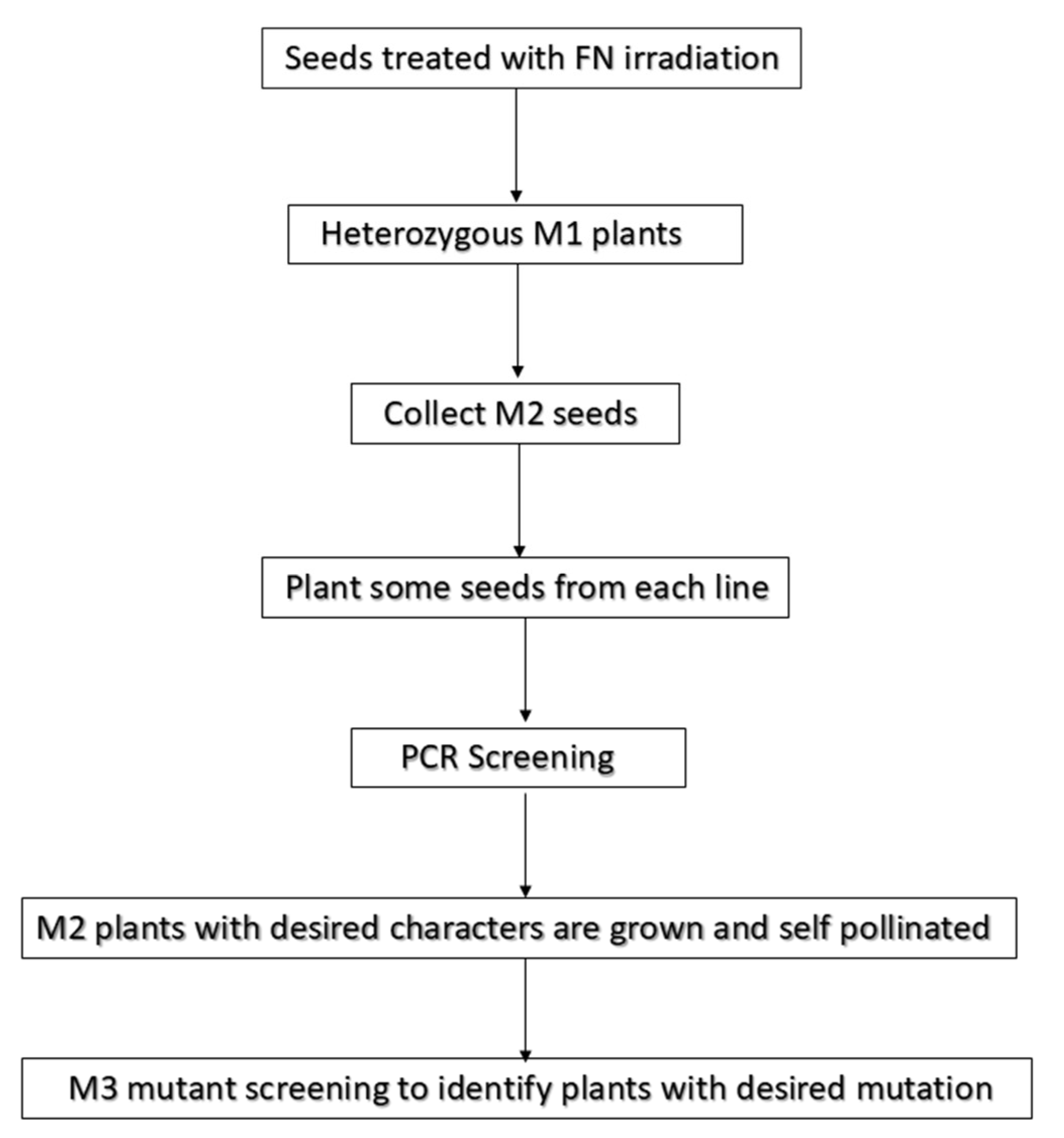
6. Mapping of Mutation Induced by Fast Neutron Radiations
7. PCR-Based Candidate Gene Screening for the Localization of Mutations
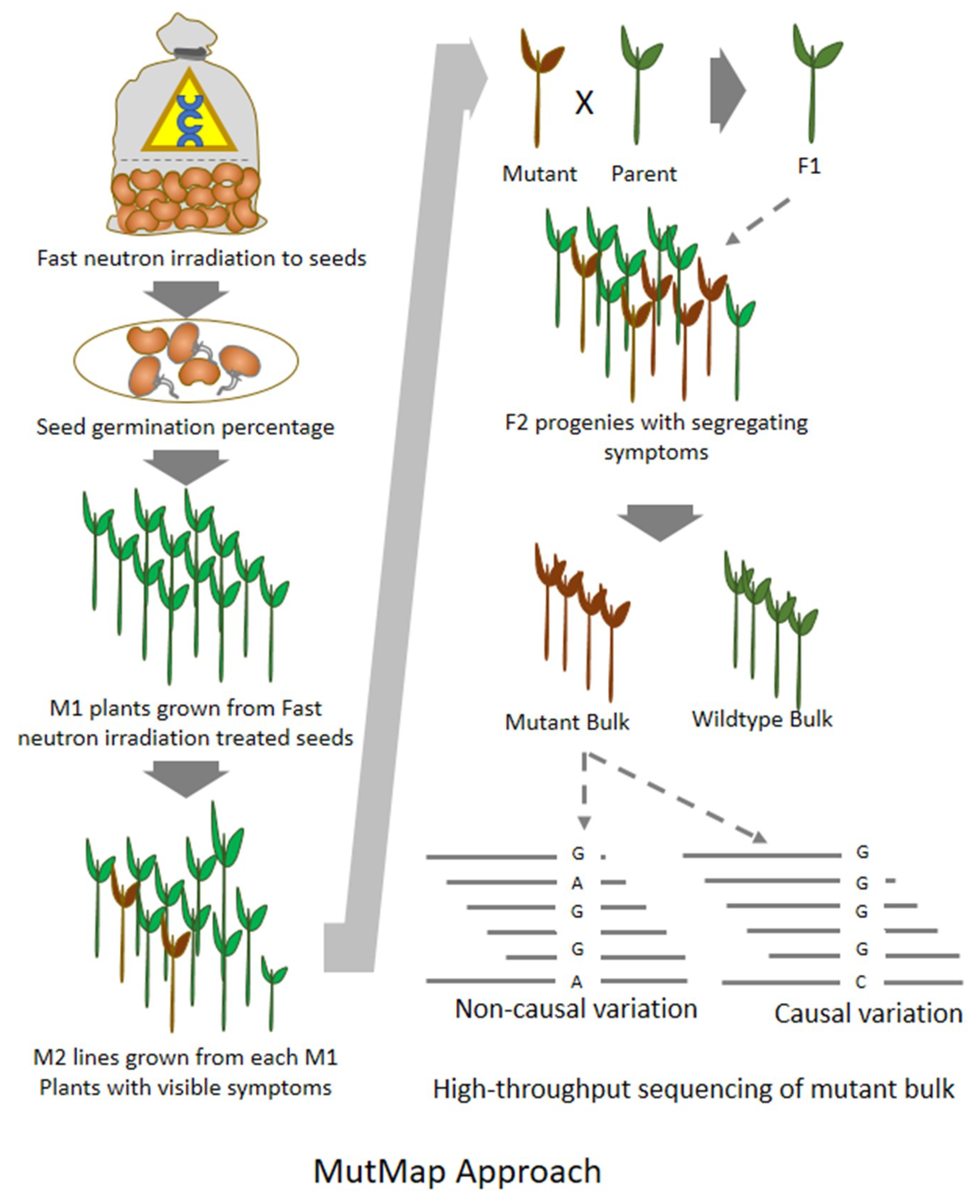
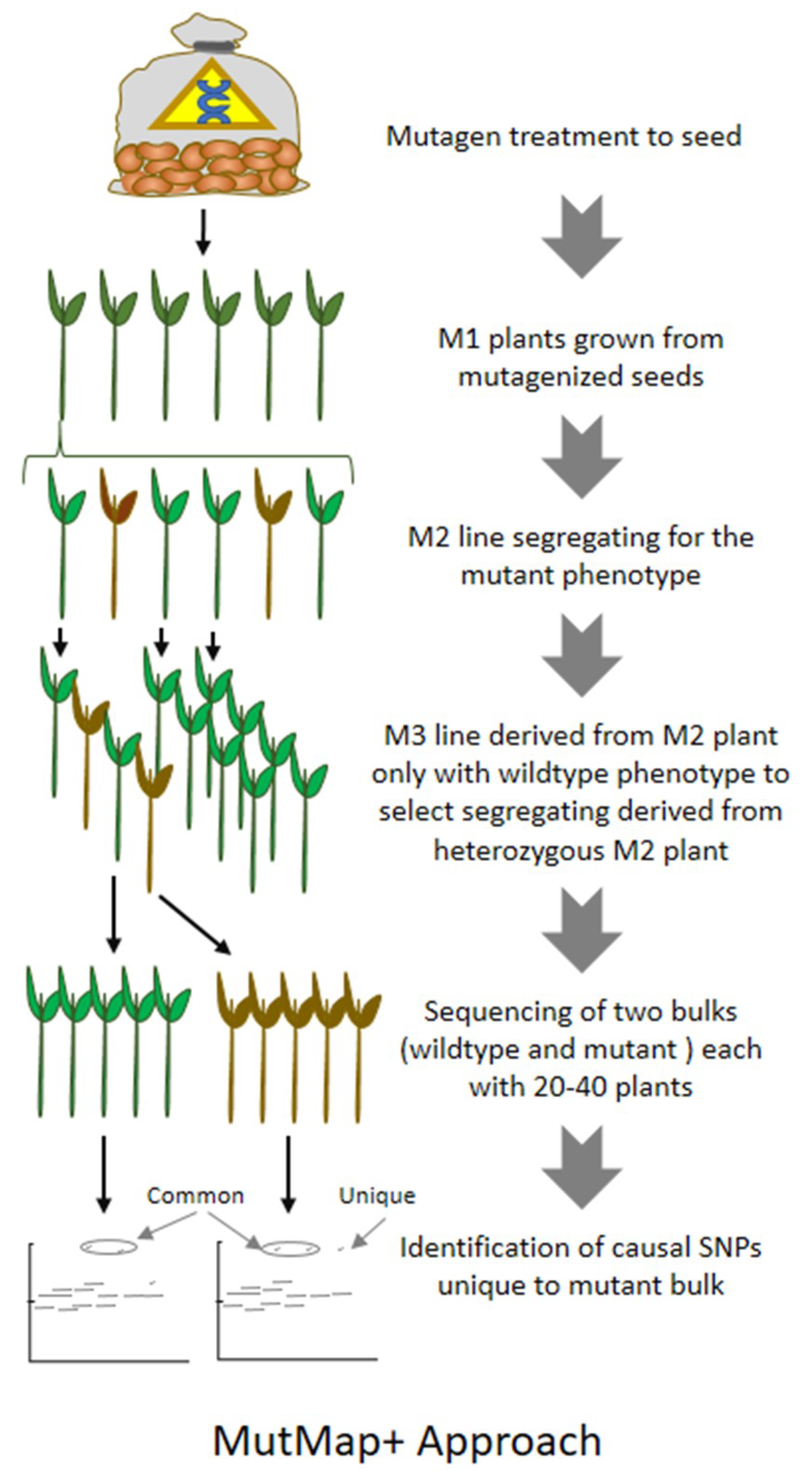
8. MutMap: An Efficient Approach for Mutation Mapping in Small Genomes
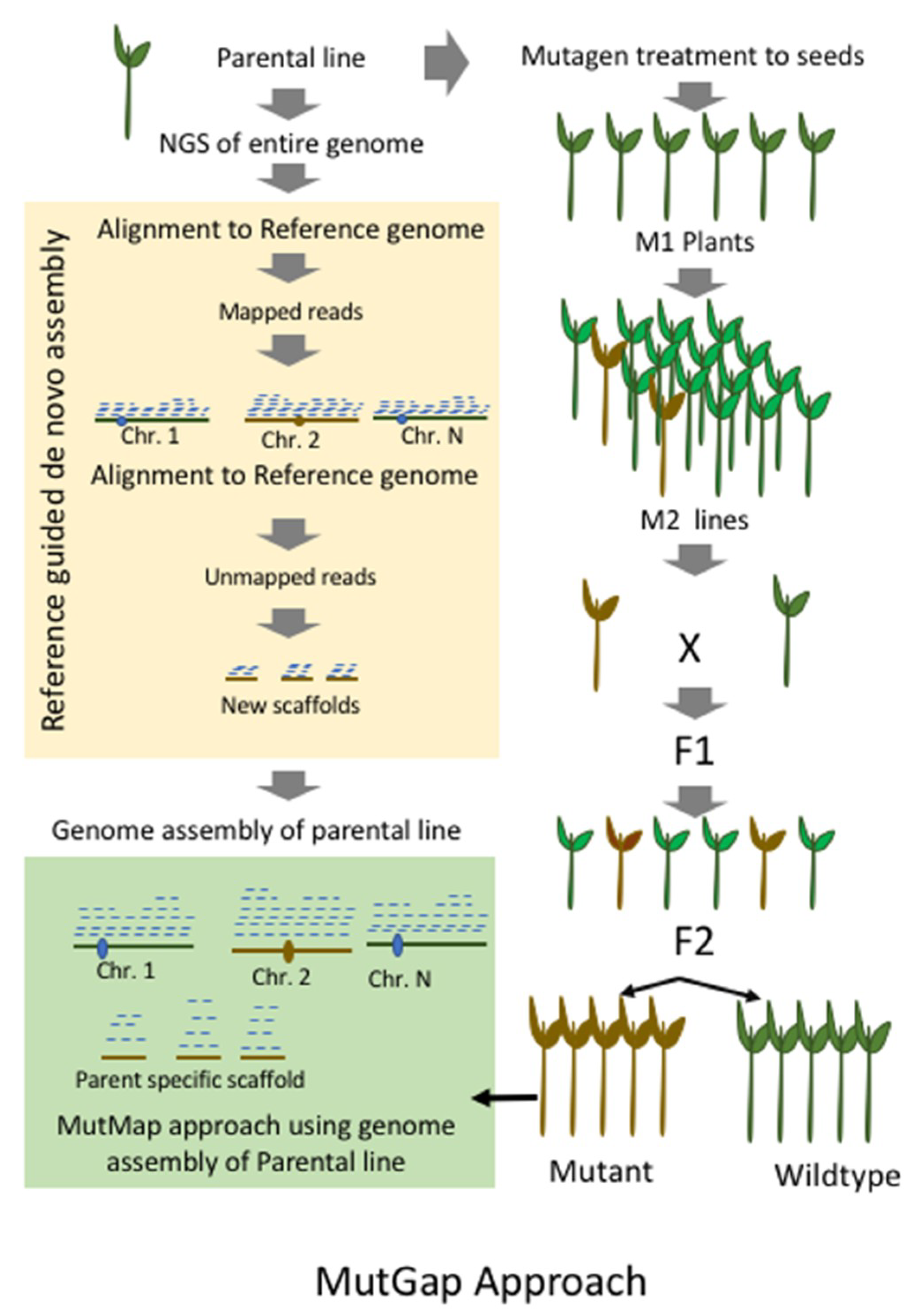
9. MutMap-Gap
10. MutChromSeq: A Method Based on Selected Chromosome Sequencing
11. Exon Capture
12. MutRen-Seq
13. Targeting Induced Local Lesions in Genomes (TILLING) Approaches
14. Whole Genome Sequencing
15. Genotyping by Sequencing-Based Mutation Mapping
16. Challenges for the Efficient Utilization of FN Mutagenesis Approaches for Crop Improvement
17. General Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friedberg, E.C.; Walker, G.C.; Siede, W.; Wood, R.D. DNA Repair Mutagenesis; American Society for Microbiology Press: Washington, DC, USA, 2005. [Google Scholar]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef]
- Ripley, L.S. Mutation. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 534–539. [Google Scholar] [CrossRef]
- Kodym, A.; Afza, R. Physical and chemical mutagenesis. In Plant Functional Genomics; Springer: Berlin/Heidelberg, Germany, 2003; pp. 189–203. [Google Scholar]
- Tadege, M.; Wang, T.L.; Wen, J.; Ratet, P.; Mysore, K.S. Mutagenesis and beyond! Tools for understanding legume biology. Plant Physiol. 2009, 151, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Topping, J.F.; Lindsey, K. Insertional mutagenesis and promoter trapping in plants for the isolation of genes and the study of development. Transgenic Res. 1995, 4, 291–305. [Google Scholar] [CrossRef]
- Wu, J.-L.; Wu, C.; Lei, C.; Baraoidan, M.; Bordeos, A.; Madamba, M.R.S.; Ramos-Pamplona, M.; Mauleon, R.; Portugal, A.; Ulat, V.J. Chemical-and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol. Biol. 2005, 59, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, J.; Alisha, A.; Bhatt, V.; Chandanshive, S.; Kumar, N.; Mir, Z.; Kumar, A.; Yadav, S.K.; Shivaraj, S.; Sonah, H. Mutation Breeding in Tomato: Advances, Applicability and Challenges. Plants 2019, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Tadege, M.; Ratet, P.; Mysore, K.S. Insertional mutagenesis: A Swiss Army knife for functional genomics of Medicago truncatula. Trends Plant Sci. 2005, 10, 229–235. [Google Scholar] [CrossRef]
- Hebert, C.G.; Valdes, J.J.; Bentley, W.E. Beyond silencing—Engineering applications of RNA interference and antisense technology for altering cellular phenotype. Curr. Opin. Biotechnol. 2008, 19, 500–505. [Google Scholar] [CrossRef]
- Krysan, P.J.; Young, J.C.; Sussman, M.R. T-DNA as an Insertional Mutagen in Arabidopsis. Plant Cell 1999, 11, 2283–2290. [Google Scholar] [CrossRef]
- Sakai, K.; Suzuki, S.; Nakamura, N.; Okada, S. Induction and subsequent repair of DNA damage by fast neutrons in cultured mammalian cells. Radiat. Res. 1987, 110, 311–320. [Google Scholar] [CrossRef]
- Bewley, D. A comparison of the response of mammalian cells to fast neutrons and charged particle beams. Radiat. Res. 1968, 34, 446–458. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y. Reverse genetics by fast neutron mutagenesis in higher plants. Funct. Integr. Genom. 2002, 2, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, E.; Haughn, G. Reverse genetics techniques: Engineering loss and gain of gene function in plants. Brief. Funct. Genom. 2010, 9, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Chern, M.; Jain, R.; Martin, J.A.; Schackwitz, W.S.; Jiang, L.; Vega-Sánchez, M.E.; Lipzen, A.M.; Barry, K.W.; Schmutz, J. Genome-wide sequencing of 41 rice (Oryza sativa L.) mutated lines reveals diverse mutations induced by fast-neutron irradiation. Mol. Plant 2016, 9, 1078–1081. [Google Scholar] [CrossRef] [PubMed]
- Hendry, J.H. The slower cellular recovery after higher-LET irradiations, including neutrons, focuses on the quality of DNA breaks. Radiat. Res. 1991, 128, S111–S113. [Google Scholar] [CrossRef]
- Li, X.; Song, Y.; Century, K.; Straight, S.; Ronald, P.; Dong, X.; Lassner, M.; Zhang, Y. A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J. 2001, 27, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Belfield, E.J.; Gan, X.; Mithani, A.; Brown, C.; Jiang, C.; Franklin, K.; Alvey, E.; Wibowo, A.; Jung, M.; Bailey, K. Genome-wide analysis of mutations in mutant lineages selected following fast-neutron irradiation mutagenesis of Arabidopsis thaliana. Genome Res. 2012, 22, 1306–1315. [Google Scholar] [CrossRef]
- Bolon, Y.-T.; Haun, W.J.; Xu, W.W.; Grant, D.; Stacey, M.G.; Nelson, R.T.; Gerhardt, D.J.; Jeddeloh, J.A.; Stacey, G.; Muehlbauer, G.J. Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol. 2011, 156, 240–253. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, R. Physical Mutagenesis in Medicago truncatula Using Fast Neutron Bombardment (FNB) for Symbiosis and Developmental Biology Studies. In Functional Genomics in Medicago Truncatula; Springer: Berlin/Heidelberg, Germany, 2018; pp. 61–69. [Google Scholar]
- Wang, J.-S.; Sui, J.-M.; Xie, Y.-D.; Guo, H.-J.; Qiao, L.-X.; Zhao, L.-L.; Yu, S.-L.; Liu, L.-X. Generation of peanut mutants by fast neutron irradiation combined with in vitro culture. J. Radiat. Res. 2015, 56, 437–445. [Google Scholar] [CrossRef]
- Hoffmann, D.; Jiang, Q.; Men, A.; Kinkema, M.; Gresshoff, P.M. Nodulation deficiency caused by fast neutron mutagenesis of the model legume Lotus japonicus. J. Plant Physiol. 2007, 164, 460–469. [Google Scholar] [CrossRef]
- Koornneeff, M.; Dellaert, L.; Van der Veen, J. EMS-and relation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 1982, 93, 109–123. [Google Scholar] [CrossRef]
- Goodhead, D.T. Neutrons are forever! Historical perspectives. Int. J. Radiat. Biol. 2019, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, P.C.; Lawrence, J.H. The physiological effects of neutron rays. Annu. Rev. Physiol. 1942, 4, 25–48. [Google Scholar] [CrossRef]
- Smith, H.H. Radiation in the production of useful mutations. Bot. Rev. 1958, 24, 1–24. [Google Scholar] [CrossRef]
- Dunning, J.; Pegram, G.; Fink, G.; Mitchell, D. Interaction of neutrons with matter. Phys. Rev. 1935, 48, 265. [Google Scholar] [CrossRef]
- Tanaka, K.; Gajendiran, N.; Endo, S.; Komatsu, K.; Hoshi, M.; Kamada, N. Neutron energy-dependent initial DNA damage and chromosomal exchange. J. Radiat. Res. 1999, 40, S36–S44. [Google Scholar] [CrossRef] [PubMed]
- Brunner, H. Radiation induced mutations for plant selection. Appl. Radiat. Isot. 1995, 46, 589–594. [Google Scholar] [CrossRef]
- Rogers, C.; Wen, J.; Chen, R.; Oldroyd, G. Deletion-based reverse genetics in Medicago truncatula. Plant Physiol. 2009, 151, 1077–1086. [Google Scholar] [CrossRef]
- Fujii, T. Relative Biological Effectiveness of 14-MeV Fast Neutrons to Co 60 Gamma-Rays in Einkorn Wheat. In Biological Effects of Neutron and Proton Irradiations. Vol. II. Proceedings of the Symposium on Biological Effects of Neutron Irradiations, Upton, NY, USA, 7–11 December 1963; International Atomic Energy Agency (IAEA): Vienna, Austria, 1964. [Google Scholar]
- Li, X.; Lassner, M.; Zhang, Y. Deleteagene: A fast neutron deletion mutagenesis-based gene knockout system for plants. Int. J. Genom. 2002, 3, 158–160. [Google Scholar] [CrossRef]
- Batchelor, A.; Phillips, R.J.; Searle, A. A comparison of the mutagenic effectiveness of chronic neutron-and γ-irradiation of mouse spermatogonia. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 1966, 3, 218–229. [Google Scholar] [CrossRef]
- Chakraborty, N.; Sarkar, G.M.; Lahiri, S.C. Effect of physical irradiation and chemical mutagen treatment on methane production by methanogenic bacteria. World J. Microbiol. Biotechnol. 2003, 19, 145–150. [Google Scholar] [CrossRef]
- Ayaki, T.; Fujikawa, K.; Ryo, H.; Itoh, T.; Kondo, S. Induced rates of mitotic crossing over and possible mitotic gene conversion per wing anlage cell in Drosophila melanogaster by X rays and fission neutrons. Genetics 1990, 126, 157–166. [Google Scholar] [PubMed]
- Unrau, P. The relative biological effectiveness of 14.5-MeV neutrons for the induction of gene conversion and mutation in yeast. Radiat. Res. 1986, 107, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Smittle, B.; LaBrecque, G.; Carroll, E. Comparative effectiveness of fast neutrons and gamma rays in producing sterility in house flies. J. Econ. Entomol. 1971, 64, 1030–1032. [Google Scholar] [CrossRef] [PubMed]
- Tahmisian, T.N.; Vogel, H.H., Jr. Relative Biological Effectiveness of Fast Neutrons, Gamma Rays, X-Rays on Grasshopper Nymph Ovarioles. (Melanoplus differentialis). Proc. Soc. Exp. Biol. Med. 1953, 84, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Erdman, H.E. Fast-neutron Effects on Productivity of Young and Old Flour Beetles, Tribolium Castaneum. Herbst, and Alterations at Different Temperatures and after Exposure of Either or Both Sexes. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1965, 9, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Xu, Y.; Sun, Y.; Liu, J.; Wang, X. Effect of fast neutron irradiation on production of cellulase from Trichoderma viride. J. Jilin Agric. Univ. 2011, 33, 177–180. [Google Scholar]
- Gragg, R. Response of Chinese Hamster Ovary Cells to Fast Neutron Radiotherapy Beams. [Gamma Radiation]; Texas University: Houston, TX, USA, 1974. [Google Scholar]
- Nakamura, N.; Okada, S.; Suzuki, S.; Ito, A. Mutations induced by γ-rays and fast neutrons in cultured mammalian cells. Mutat. Res. 1982, 104, 383–387. [Google Scholar] [CrossRef]
- Liu, S.; Xu, J.; Chen, W.; Fu, H.; Ma, L.Y.; Xu, H.; Xinnian, L.; Wu, M.; Ma, F. Enhancement of lipid productivity in green microalgae Chlorella sp. via fast neutron irradiation. Biomass Bioenergy 2016, 91, 196–203. [Google Scholar] [CrossRef]
- Mehandjiev, A.; Kosturkova, G.; Mihov, M. Enrichment of Pisum sativum gene resources through combined use of physical and chemical mutagens. Israel J. Plant Sci. 2001, 49, 280–284. [Google Scholar] [CrossRef]
- Abdel Haliem, E.; Abdullah, H.; AL-Huqail, A.A. Oxidative damage and mutagenic potency of fast neutron and UV-B radiation in pollen mother cells and seed yield of Vicia faba L. BioMed Res. Int. 2013, 2013, 824656. [Google Scholar] [CrossRef] [PubMed]
- Menda, N.; Semel, Y.; Peled, D.; Eshed, Y.; Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 2004, 38, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Bolon, Y.-T.; Stec, A.O.; Michno, J.-M.; Roessler, J.; Bhaskar, P.B.; Ries, L.; Dobbels, A.A.; Campbell, B.W.; Young, N.P.; Anderson, J.E. Genome resilience and prevalence of segmental duplications following fast neutron irradiation of soybean. Genetics 2014, 198, 967–981. [Google Scholar] [CrossRef] [PubMed]
- KA, S.; Larik, H. Persistence of Chromosomal Aberrations in Mutated Populations of Triticum aestivum. Cytologia 1982, 47, 247–256. [Google Scholar]
- Bruggemann, E.; Handwerger, K.; Essex, C.; Storz, G. Analysis of fast neutron-generated mutants at the Arabidopsis thaliana HY4 locus. Plant J. 1996, 10, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, S.; Swaminathan, M. Polyploidy and radiosensitivity in wheat and barley. Genetica 1960, 31, 449–480. [Google Scholar] [CrossRef]
- Fitzgerald, T.L.; Powell, J.J.; Stiller, J.; Weese, T.L.; Abe, T.; Zhao, G.; Jia, J.; McIntyre, C.L.; Li, Z.; Manners, J.M. An assessment of heavy ion irradiation mutagenesis for reverse genetics in wheat (Triticum aestivum L.). PLoS ONE 2015, 10, e0117369. [Google Scholar] [CrossRef] [PubMed]
- Chopra, V. Mutagenesis: Investigating the process and processing the outcome for crop improvement. Curr. Sci. Bangalore 2005, 89, 353. [Google Scholar]
- Krishnaswami, R. The Relationship between Response to Radiations and Nature of Polyploidy in Some Crop Plants. Caryologia 1968, 21, 303–310. [Google Scholar] [CrossRef]
- Chai, J.-S.; Kim, J.-H.; Yang, T.-G.; Lyu, J.-I.; Lee, H.-Y.; Yang, D.-C.; Bae, C.-H. Characteristics of tobacco and rice plants irradiated with neutron beam. Korean J. Plant Resour. 2005, 18, 359–366. [Google Scholar]
- Xiao, J.; Jia, X.; Wang, H.; Zhao, R.; Fang, Y.; Gao, R.; Wu, Z.; Cao, A.; Wang, J.; Xue, Z. A fast-neutron induced chromosome fragment deletion of 3BS in wheat landrace Wangshuibai increased its susceptibility to Fusarium head blight. Chromosome Res. 2011, 19, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Hanafy, M.S.; Mohamed, H.A. Effect of irradiation of wheat grains with fast neutrons on the grain yield and other characteristics of the plants. Appl. Radiat. Isot. 2014, 86, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174. [Google Scholar] [CrossRef] [PubMed]
- Yugandhar, P.; Sun, Y.; Liu, L.; Negi, M.; Nallamothu, V.; Sun, S.; Neelamraju, S.; Rai, V.; Jain, A. Characterization of the loss-of-function mutant NH101 for yield under phosphate deficiency from EMS-induced mutants of rice variety Nagina22. Plan. Physiol. Biochem. 2018, 130, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, J.; Steuernagel, B.; Ghosh, S.; Herren, G.; Hurni, S.; Adamski, N.; Vrána, J.; Kubaláková, M.; Krattinger, S.G.; Wicker, T. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 2016, 17, 221. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Gaikwad, K. From genomes to GENE-omes: Exome sequencing concept and applications in crop improvement. Front. Plant Sci. 2017, 8, 2164. [Google Scholar] [CrossRef] [PubMed]
- Tadele, Z.; Chikelu, M.; Till, B.J. TILLING for mutations in model plants and crops. In Molecular Techniques in Crop Improvement; Springer: Berlin/Heidelberg, Germany, 2010; pp. 307–332. [Google Scholar]
- Sevanthi, A.M.; Kandwal, P.; Kale, P.B.; Prakash, C.; Ramkumar, M.; Yadav, N.; Mahato, A.K.; Sureshkumar, V.; Behera, M.; Deshmukh, R.K. Whole genome characterization of a few EMS-induced mutants of upland rice variety Nagina 22 reveals a staggeringly high frequency of SNPs which show high phenotypic plasticity towards the wild-type. Front. Plant Sci. 2018, 9, 1179. [Google Scholar] [CrossRef]
- Wachsman, G.; Modliszewski, J.L.; Valdes, M.; Benfey, P.N. A simple pipeline for mapping point mutations. Plant Physiol. 2017, 174, 1307–1313. [Google Scholar] [CrossRef]
- Sonah, H.; Bastien, M.; Iquira, E.; Tardivel, A.; Légaré, G.; Boyle, B.; Normandeau, É.; Laroche, J.; Larose, S.; Jean, M. An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS ONE 2013, 8, e54603. [Google Scholar] [CrossRef]
- Kamolsukyeunyong, W.; Ruengphayak, S.; Chumwong, P.; Kusumawati, L.; Chaichoompu, E.; Jamboonsri, W.; Saensuk, C.; Phoonsiri, K.; Toojinda, T.; Vanavichit, A. Identification of spontaneous mutation for broad-spectrum brown planthopper resistance in a large, long-term fast neutron mutagenized rice population. Rice 2019, 12, 16. [Google Scholar] [PubMed]
- Sikora, P.; Chawade, A.; Larsson, M.; Olsson, J.; Olsson, O. Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int. J. Plant Genom. 2011, 2011, 314829. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.E.; Michno, J.-M.; Kono, T.J.; Stec, A.O.; Campbell, B.W.; Curtin, S.J.; Stupar, R.M. Genomic variation and DNA repair associated with soybean transgenesis: A comparison to cultivars and mutagenized plants. BMC Biotechnol. 2016, 16, 41. [Google Scholar] [CrossRef] [PubMed]
- Novak, F.; Brunner, H. Plant breeding: Induced mutation technology for crop improvement. IAEA Bull. 1992, 4, 25–33. [Google Scholar]
- Zhong-Hua, W.; Xin-chen, Z.; Yu-lin, J. Development and characterization of rice mutants for functional genomics studies and breeding. Rice Sci. 2014, 21, 60182–60188. [Google Scholar]
- Campbell, B.W.; Hofstad, A.N.; Sreekanta, S.; Fu, F.; Kono, T.J.; O’Rourke, J.A.; Vance, C.P.; Muehlbauer, G.J.; Stupar, R.M. Fast neutron-induced structural rearrangements at a soybean NAP1 locus result in gnarled trichomes. Theor. Appl. Genet. 2016, 129, 1725–1738. [Google Scholar] [CrossRef] [PubMed]
| Method | Advantages | Disadvantages | References |
|---|---|---|---|
| Chemical mutagenesis | Identification of multiple alleles of genes. Heritable. | Isolation of mutated gene is difficult | [6,15] |
| Insertional mutagenesis | Chemically and physically Stable through multiple generations. T-DNA can be used to study specific stages of cellular differentiation or cell fate. | Transactivation properties. Require transformation | [6,11,16] |
| Fast neutron mutagenesis | Saturate the genome. Do not require transformation or tissue culture. Complete knockout of gene | Screening is time taking. Can delete multiple genes at a time | [15,17] |
| Organism | Objective | References |
|---|---|---|
| Mouse | Effect of chronic neutron- and γ-irradiation on spermatogonia | [35] |
| Methanogenic bacteria (TDM, TRM, and SSM) | Effect of physical irradiation (y irradiation, neutron bombardment) and chemical mutagen (acridine orange and colchicine) on methane production | [36] |
| Drosophila melanogaster | Induced rates of mitotic crossing-over | [37] |
| Yeast | Relative biological effectiveness of 14.5- MeV neutrons for the induction of gene conversion | [38] |
| Musca domestica L | To compare effect of fast neutrons and gamma rays in producing sterility | [39] |
| Melanoplus differentialis | To study fast neutrons, gamma rays, X-rays relative biological effectiveness on nymph ovarioles | [40] |
| Tribolium Castaneum | Effect of fast neutron on productivity of young and old flour beetles | [41] |
| Trichoderma viride | Effect of FN on production of cellulose | [42] |
| Chinese hamster | Response of ovary cells to fast neutron | [43] |
| Mammalian cells | To study mutations induced by γ-rays and fast neutrons | [44] |
| Chlorella sp. | Enhancement of lipid productivity | [45] |
| Species | Mutagen | Dose | Number of Mutants | Reason | References |
|---|---|---|---|---|---|
| Soybean cv M92-220 | FN | 4–32 Gray units | 23,000 | Phenotypic screening and associated genomic characterization | [21] |
| Arabidopsis thaliana | FN | 60 Gy | 300 | Screening of elongated hypocotyl mutants | [20] |
| Rice | FN | 20 Gy | 2418 | For genome-wide profiling of mutations | [17] |
| Peanut | FN + in vitro culture | 9.7–18.0 Gy | 19 | Somatic embryogenesis combined with plant regeneration | [23] |
| Medicago truncatula | Fast neutron bombardment (FNB) | 30–40 Gy | 1000 | creened for symbiotic nitrogen fixationSymbiotic nitrogen fixation mutant lines | [22] |
| Vicia faba L. | Fast neutron (FN) and UV-B | 280–320 nm 30–40 Gy | To study combined effect of oxidative stress and mutagenic potential | [47] | |
| Lotus japonicas | FN | 8 Grey (Gy) | 58 | Non-nodulation mutant called FNN5-2 | [24] |
| Lycopersicon esculentum (M82) | FN | 15 Gy | 865 | Functional genomic studies | [48] |
| Pisum sativum | FN + EMS | 10 Gy + 0.2% EMS | 14% | Mutation genetics and breeding study | Mehandjiev, Kosturkova and Mihov [46] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumawat, S.; Rana, N.; Bansal, R.; Vishwakarma, G.; Mehetre, S.T.; Das, B.K.; Kumar, M.; Yadav, S.K.; Sonah, H.; Sharma, T.R.; et al. Expanding Avenue of Fast Neutron Mediated Mutagenesis for Crop Improvement. Plants 2019, 8, 164. https://doi.org/10.3390/plants8060164
Kumawat S, Rana N, Bansal R, Vishwakarma G, Mehetre ST, Das BK, Kumar M, Yadav SK, Sonah H, Sharma TR, et al. Expanding Avenue of Fast Neutron Mediated Mutagenesis for Crop Improvement. Plants. 2019; 8(6):164. https://doi.org/10.3390/plants8060164
Chicago/Turabian StyleKumawat, Surbhi, Nitika Rana, Ruchi Bansal, Gautam Vishwakarma, Sayaji T. Mehetre, Bikram Kishore Das, Manish Kumar, Satish Kumar Yadav, Humira Sonah, Tilak Raj Sharma, and et al. 2019. "Expanding Avenue of Fast Neutron Mediated Mutagenesis for Crop Improvement" Plants 8, no. 6: 164. https://doi.org/10.3390/plants8060164
APA StyleKumawat, S., Rana, N., Bansal, R., Vishwakarma, G., Mehetre, S. T., Das, B. K., Kumar, M., Yadav, S. K., Sonah, H., Sharma, T. R., & Deshmukh, R. (2019). Expanding Avenue of Fast Neutron Mediated Mutagenesis for Crop Improvement. Plants, 8(6), 164. https://doi.org/10.3390/plants8060164







