Mutation Breeding in Tomato: Advances, Applicability and Challenges
Abstract
1. Introduction
2. Induced Mutagenesis
2.1. Chemical Mutagenesis
2.2. Physical Mutagenesis
2.3. Insertional Mutagenesis
2.4. Mutagenesis by Antisense Approach
2.5. Mutagenesis by Genome-Editing Approaches
2.6. Genome Editing by Zinc Finger Nucleases (ZFNs)
2.7. Genome Editing by Transcription Activator-Like Effector Nucleases (TALENs)
2.8. Gene Editing by CRISPR/Cas9
3. Mutation Mapping Approaches
3.1. MutMap Approach
3.2. MutChromSeq Approach
3.3. Whole-Genome Sequencing (WGS)-Based Mapping
4. Tomato Mutant Resources
4.1. Genes that Make Tomatoes (http://zamir.sgn.cornell.edu/mutants/)
4.2. LycoTILL (http://www.agrobios.it/tilling/)
5. The Red Setter TILLING Platform
TOMATOMA (http://tomatoma.nbrp.jp/indexAction.do)
6. Limitations and Challenges for Mutagenesis in Tomato
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Just, D.; Garcia, V.; Fernandez, L.; Bres, C.; Mauxion, J.-P.; Petit, J.; Jorly, J.; Assali, J.; Bournonville, C.; Ferrand, C. Micro-Tom mutants for functional analysis of target genes and discovery of new alleles in tomato. Plant Biotechnol. 2013, 30, 225–231. [Google Scholar] [CrossRef]
- Ranjan, A.; Ichihashi, Y.; Sinha, N.R. The tomato genome: Implications for plant breeding, genomics and evolution. Genome Biol. 2012, 13, 167. [Google Scholar] [CrossRef]
- Consortium, T.G. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar] [CrossRef]
- Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Liang, D.; Liu, J.; Sahoo, G.; Kelliher, T. OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 2018, 4, 530. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019, 2, 46. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018, 37, 1353–1356. [Google Scholar] [CrossRef]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef]
- Lor, V.S.; Starker, C.G.; Voytas, D.F.; Weiss, D.; Olszewski, N.E. Targeted mutagenesis of the tomato PROCERA gene using transcription activator-like effector nucleases. Plant Physiol. 2014, 166, 1288–1291. [Google Scholar] [CrossRef]
- Wisman, E.; Koornneef, M.; Chase, T.; Lifshytz, E.; Ramanna, M.; Zabel, P. Genetic and molecular characterization of an Adh-1 null mutant in tomato. Mol. Gen. Genet. 1991, 226, 120–128. [Google Scholar] [PubMed]
- Menda, N.; Semel, Y.; Peled, D.; Eshed, Y.; Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 2004, 38, 861–872. [Google Scholar] [CrossRef]
- Adamu, A.; Aliyu, H. Morphogical effects of sodium azide on tomato (Lycopersicon esculentum Mill). Sci. World J. 2007, 2, 9–12. [Google Scholar] [CrossRef]
- Kostov, K.; Batchvarova, R.; Slavov, S. Application of chemical mutagenesis to increase the resistance of tomato to Orobanche ramosa L. Bulg. J. Agric. Sci. 2007, 13, 505–513. [Google Scholar]
- Matsukura, C.; Yamaguchi, I.; Inamura, M.; Ban, Y.; Kobayashi, Y.; Yin, Y.-g.; Saito, T.; Kuwata, C.; Imanishi, S.; Nishimura, S. Generation of gamma irradiation-induced mutant lines of the miniature tomato (Solanum lycopersicum L.) cultivar ‘Micro-Tom ’. Plant Biotechnol. 2007, 24, 39–44. [Google Scholar] [CrossRef]
- Minoia, S.; Petrozza, A.; D’Onofrio, O.; Piron, F.; Mosca, G.; Sozio, G.; Cellini, F.; Bendahmane, A.; Carriero, F. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 2010, 3, 69. [Google Scholar] [CrossRef]
- Piron, F.; Nicolaï, M.; Minoïa, S.; Piednoir, E.; Moretti, A.; Salgues, A.; Zamir, D.; Caranta, C.; Bendahmane, A. An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 2010, 5, e11313. [Google Scholar] [CrossRef]
- Watanabe, S.; Mizoguchi, T.; Aoki, K.; Kubo, Y.; Mori, H.; Imanishi, S.; Yamazaki, Y.; Shibata, D.; Ezura, H. Ethylmethanesulfonate (EMS) mutagenesis of Solanum lycopersicum cv. Micro-Tom for large-scale mutant screens. Plant Biotechnol. 2007, 24, 33–38. [Google Scholar] [CrossRef]
- Hammond-Kosack, K.E.; Jones, D.A.; Jones, J.D. Identification of two genes required in tomato for full Cf-9-dependent resistance to Cladosporium fulvum. Plant Cell 1994, 6, 361–374. [Google Scholar]
- Kurowska, M.; Daszkowska-Golec, A.; Gruszka, D.; Marzec, M.; Szurman, M.; Szarejko, I.; Maluszynski, M. TILLING-a shortcut in functional genomics. J. Appl. Genet. 2011, 52, 371. [Google Scholar] [CrossRef]
- Martí, E.; Gisbert, C.; Bishop, G.J.; Dixon, M.S.; García-Martínez, J.L. Genetic and physiological characterization of tomato cv. Micro-Tom. J. Exp. Bot. 2006, 57, 2037–2047. [Google Scholar] [CrossRef]
- Gilchrist, E.; Haughn, G. Reverse genetics techniques: Engineering loss and gain of gene function in plants. Brief. Funct. Genom. 2010, 9, 103–110. [Google Scholar] [CrossRef]
- Li, X.; Song, Y.; Century, K.; Straight, S.; Ronald, P.; Dong, X.; Lassner, M.; Zhang, Y. A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J. 2001, 27, 235–242. [Google Scholar] [CrossRef]
- Lee, H.-K.; Mysore, K.S.; Wen, J. Tnt1 Insertional Mutagenesis in Medicago truncatula. Functional Genomics in Medicago truncatula; Humana Press: New York, NY, USA, 2018; pp. 107–114. [Google Scholar]
- Sun, S.; Chen, Y.; Cheng, J.; Li, Q.; Zhang, Z.; Lan, Z. Isolation, characterization, genomic sequencing, and GFP-marked insertional mutagenesis of a high-performance nitrogen-fixing bacterium, Kosakonia radicincitans GXGL-4A and visualization of bacterial colonization on cucumber roots. Folia Microbiol. 2018, 1–14. [Google Scholar] [CrossRef]
- Cooley, M.B.; Yoder, J.; Goldsbrough, A.; Still, D. Site-selected insertional mutagenesis of tomato with maizeAc andDs elements. Mol. Gen. Genet. 1996, 252, 184–194. [Google Scholar] [CrossRef]
- Cooley, M.B.; Yoder, J.I. Insertional inactivation of the tomato polygalacturonase gene. Plant Mol. Biol. 1998, 38, 521–530. [Google Scholar] [CrossRef]
- Healy, J.; Corr, C.; DeYoung, J.; Baker, B. Linked and unlinked transposition of a genetically marked Dissociation element in transgenic tomato. Genetics 1993, 134, 571–584. [Google Scholar]
- Goldsbrough, A.P.; Tong, Y.; Yoder, J.I. Lc as a non-destructive visual reporter and transposition excision marker gone for tomato. Plant J. 1996, 9, 927–933. [Google Scholar] [CrossRef]
- Mathews, H.; Clendennen, S.K.; Caldwell, C.G.; Liu, X.L.; Connors, K.; Matheis, N.; Schuster, D.K.; Menasco, D.; Wagoner, W.; Lightner, J. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 2003, 15, 1689–1703. [Google Scholar] [CrossRef] [PubMed]
- Roldan, M.V.G.; Périlleux, C.; Morin, H.; Huerga-Fernandez, S.; Latrasse, D.; Benhamed, M.; Bendahmane, A. Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Sci. Rep. 2017, 7, 4402. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, R.; Xiao, J.; Qian, C.; Wang, T.; Li, H.; Ouyang, B.; Ye, Z. A single-base deletion mutation in SlIAA9 gene causes tomato (Solanum lycopersicum) entire mutant. J. Plant Res. 2007, 120, 671–678. [Google Scholar] [CrossRef]
- Uluisik, S.; Chapman, N.H.; Smith, R.; Poole, M.; Adams, G.; Gillis, R.B.; Besong, T.M.; Sheldon, J.; Stiegelmeyer, S.; Perez, L. Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 2016, 34, 950. [Google Scholar] [CrossRef]
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of Sl PL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 2017, 15, 1544–1555. [Google Scholar] [CrossRef] [PubMed]
- Rothan, C.; Diouf, I.; Causse, M. Trait discovery and editing in tomato. Plant J. 2019, 97, 73–90. [Google Scholar] [CrossRef]
- Mao, Y.; Botella, J.R.; Liu, Y.; Zhu, J.-K. Gene Editing in Plants–Progress and Challenges. Natl. Sci. Rev. 2019. [Google Scholar] [CrossRef]
- Jung, C.; Capistrano-Gossmann, G.; Braatz, J.; Sashidhar, N.; Melzer, S. Recent developments in genome editing and applications in plant breeding. Plant Breed. 2018, 137, 1–9. [Google Scholar] [CrossRef]
- Mani, M.; Kandavelou, K.; Dy, F.J.; Durai, S.; Chandrasegaran, S. Design, engineering, and characterization of zinc finger nucleases. Biochem. Biophys. Res. Commun. 2005, 335, 447–457. [Google Scholar] [CrossRef]
- Hilioti, Z.; Ganopoulos, I.; Ajith, S.; Bossis, I.; Tsaftaris, A. A novel arrangement of zinc finger nuclease system for in vivo targeted genome engineering: The tomato LEC1-LIKE4 gene case. Plant Cell Rep. 2016, 35, 2241–2255. [Google Scholar] [CrossRef] [PubMed]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed]
- Gould, K.S. Nature’s Swiss army knife: The diverse protective roles of anthocyanins in leaves. BioMed Res. Int. 2004, 2004, 314–320. [Google Scholar] [CrossRef]
- Qiu, Z.; Wang, H.; Li, D.; Yu, B.; Hui, Q.; Yan, S.; Huang, Z.; Cui, X.; Cao, B. Identification of Candidate HY5-dependent and-independent Regulators of Anthocyanin Biosynthesis in Tomato. Plant Cell Physiol. 2018. [Google Scholar] [CrossRef]
- Jacobs, T.B.; Zhang, N.; Patel, D.; Martin, G.B. Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol. 2017, 174, 2023–2037. [Google Scholar] [CrossRef]
- Shimatani, Z.; Ariizumi, T.; Fujikura, U.; Kondo, A.; Ezura, H.; Nishida, K. Targeted Base Editing with CRISPR-Deaminase in Tomato. In Plant Genome Editing with CRISPR Systems; Humana Press: New York, NY, USA, 2019; pp. 297–307. [Google Scholar]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441. [Google Scholar] [CrossRef]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018. [Google Scholar] [CrossRef]
- Rivero, R.M.; Mestre, T.C.; Mittler, R.; Rubio, F.; GARCIA-SANCHEZ, F.; Martinez, V. The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant Cell Environ. 2014, 37, 1059–1073. [Google Scholar] [CrossRef]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.-H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784. [Google Scholar] [CrossRef]
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Toki, S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 2015, 467, 76–82. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Qin, L.; Liu, X.; He, Y.; Wang, J.; Chen, L.; Lu, G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 2016, 6, 24765. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Park, S.J.; Van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [CrossRef]
- de Toledo Thomazella, D.P.; Brail, Q.; Dahlbeck, D.; Staskawicz, B.J. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv 2016, 064824. [Google Scholar]
- Ueta, R.; Abe, C.; Watanabe, T.; Sugano, S.S.; Ishihara, R.; Ezura, H.; Osakabe, Y.; Osakabe, K. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 2017, 7, 507. [Google Scholar] [CrossRef] [PubMed]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162. [Google Scholar] [CrossRef]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.; Salts, Y.; Barg, R. Tomato facultative parthenocarpy results from Sl AGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 2017, 15, 634–647. [Google Scholar] [CrossRef]
- Nonaka, S.; Arai, C.; Takayama, M.; Matsukura, C.; Ezura, H. Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 2017, 7, 7057. [Google Scholar] [CrossRef]
- Yu, Q.-h.; Wang, B.; Li, N.; Tang, Y.; Yang, S.; Yang, T.; Xu, J.; Guo, C.; Yan, P.; Wang, Q. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 2017, 7, 11874. [Google Scholar] [CrossRef]
- Li, R.; Li, R.; Li, X.; Fu, D.; Zhu, B.; Tian, H.; Luo, Y.; Zhu, H. Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 415–427. [Google Scholar] [CrossRef]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174. [Google Scholar] [CrossRef]
- Garcia, V.; Bres, C.; Just, D.; Fernandez, L.; Tai, F.W.J.; Mauxion, J.-P.; Le Paslier, M.-C.; Bérard, A.; Brunel, D.; Aoki, K. Rapid identification of causal mutations in tomato EMS populations via mapping-by-sequencing. Nat. Protoc. 2016, 11, 2401. [Google Scholar] [CrossRef]
- Sánchez-Martín, J.; Steuernagel, B.; Ghosh, S.; Herren, G.; Hurni, S.; Adamski, N.; Vrána, J.; Kubaláková, M.; Krattinger, S.G.; Wicker, T. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 2016, 17, 221. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Fukuoka, H.; Matsunaga, H.; Kobayashi, Y.; Kobayashi, I.; Hirakawa, H.; Isobe, S.; Tabata, S. Genome-wide association studies using single nucleotide polymorphism markers developed by re-sequencing of the genomes of cultivated tomato. DNA Res. 2013, 20, 593–603. [Google Scholar] [CrossRef]
- Koornneef, M.; Bosma, T.; Hanhart, C.; Van der Veen, J.; Zeevaart, J. The isolation and characterization of gibberellin-deficient mutants in tomato. Theor. Appl. Genet. 1990, 80, 852–857. [Google Scholar] [CrossRef]
- Koornneef, M.; Bade, J.; Hanhart, C.; Horsman, K.; Schel, J.; Soppe, W.; Verkerk, R.; Zabel, P. Characterization and mapping of a gene controlling shoot regeneration in tomato. Plant J. 1993, 3, 131–141. [Google Scholar] [CrossRef]
- Van Tuinen, A.; Hanhart, C.J.; Kerckhoffs, L.H.J.; Nagatani, A.; Boylan, M.T.; Quail, P.H.; Kendrick, R.E.; Koornneef, M. Analysis of phytochrome-deficient yellow-green-2 and aurea mutants of tomato. Plant J. 1996, 9, 173–182. [Google Scholar] [CrossRef]
- Thompson, A.J.; Tor, M.; Barry, C.S.; Vrebalov, J.; Orfila, C.; Jarvis, M.C.; Giovannoni, J.J.; Grierson, D.; Seymour, G.B. Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol. 1999, 120, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Ronen, G.; Carmel-Goren, L.; Zamir, D.; Hirschberg, J. An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. USA 2000, 97, 11102–11107. [Google Scholar] [CrossRef] [PubMed]
- Barry, C.S.; McQuinn, R.P.; Thompson, A.J.; Seymour, G.B.; Grierson, D.; Giovannoni, J.J. Ethylene insensitivity conferred by the Green-ripe and Never-ripe 2 ripening mutants of tomato. Plant Physiol. 2005, 138, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948. [Google Scholar] [CrossRef]
- Barry, C.S.; McQuinn, R.P.; Chung, M.-Y.; Besuden, A.; Giovannoni, J.J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008, 147, 179–187. [Google Scholar] [CrossRef]
- Kang, J.-H.; Liu, G.; Shi, F.; Jones, A.D.; Beaudry, R.M.; Howe, G.A. The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol. 2010, 154, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Barry, C.; Aldridge, G.M.; Herzog, G.; Ma, Q.; McQuinn, R.P.; Hirschberg, J.; Giovannoni, J.J. Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent 2 locus of tomato. Plant Physiol. 2012, 159, 1086–1098. [Google Scholar] [CrossRef]
- Nadakuduti, S.S.; Pollard, M.R.; Kosma, D.K.; Allen, C.; Ohlrogge, J.; Barry, C. Pleiotropic phenotypes of the sticky peel (pe) mutant provide new insight into the role of CUTIN DEFICIENT 2 in epidermal cell function in tomato. Plant Physiol. 2012, 159, 945–960. [Google Scholar] [CrossRef] [PubMed]
- Petit, J. Identification et Validation Fonctionnelle de gènes Candidats Contrôlant la Composition de la Cuticule chez le Fruit de tomate. Université Sciences et Technologies-Bordeaux I, 2013. Available online: https://www.theses.fr/2013BOR15225 (accessed on 17 December 2013).
- Ariizumi, T.; Kishimoto, S.; Kakami, R.; Maoka, T.; Hirakawa, H.; Suzuki, Y.; Ozeki, Y.; Shirasawa, K.; Bernillon, S.; Okabe, Y. Identification of the carotenoid modifying gene PALE YELLOW PETAL 1 as an essential factor in xanthophyll esterification and yellow flower pigmentation in tomato (Solanum lycopersicum). Plant J. 2014, 79, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-H.; McRoberts, J.; Shi, F.; Moreno, J.; Jones, D.; Howe, G.A. The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 2014, 164, 1161–1174. [Google Scholar] [CrossRef]
- Neuman, H.; Galpaz, N.; Cunningham Jr, F.X.; Zamir, D.; Hirschberg, J. The tomato mutation nxd1 reveals a gene necessary for neoxanthin biosynthesis and demonstrates that violaxanthin is a sufficient precursor for abscisic acid biosynthesis. Plant J. 2014, 78, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Ezura, H. Micro-Tom Tomato as an Alternative Plant Model System: Mutant Collection and Efficient Transformation. In Plant Signal Transduction; Humana Press: New York, NY, USA, 2016; pp. 47–55. [Google Scholar]
- Ariizumi, T.; Aoki, K.; Ezura, H. Systematic development of tomato bioresources in Japan. Interdiscip. Bio Cent. 2011, 3, 1–7. [Google Scholar] [CrossRef][Green Version]
- Saito, T.; Ariizumi, T.; Okabe, Y.; Asamizu, E.; Hiwasa-Tanase, K.; Fukuda, N.; Mizoguchi, T.; Yamazaki, Y.; Aoki, K.; Ezura, H. TOMATOMA: A novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 2011, 52, 283–296. [Google Scholar] [CrossRef] [PubMed]
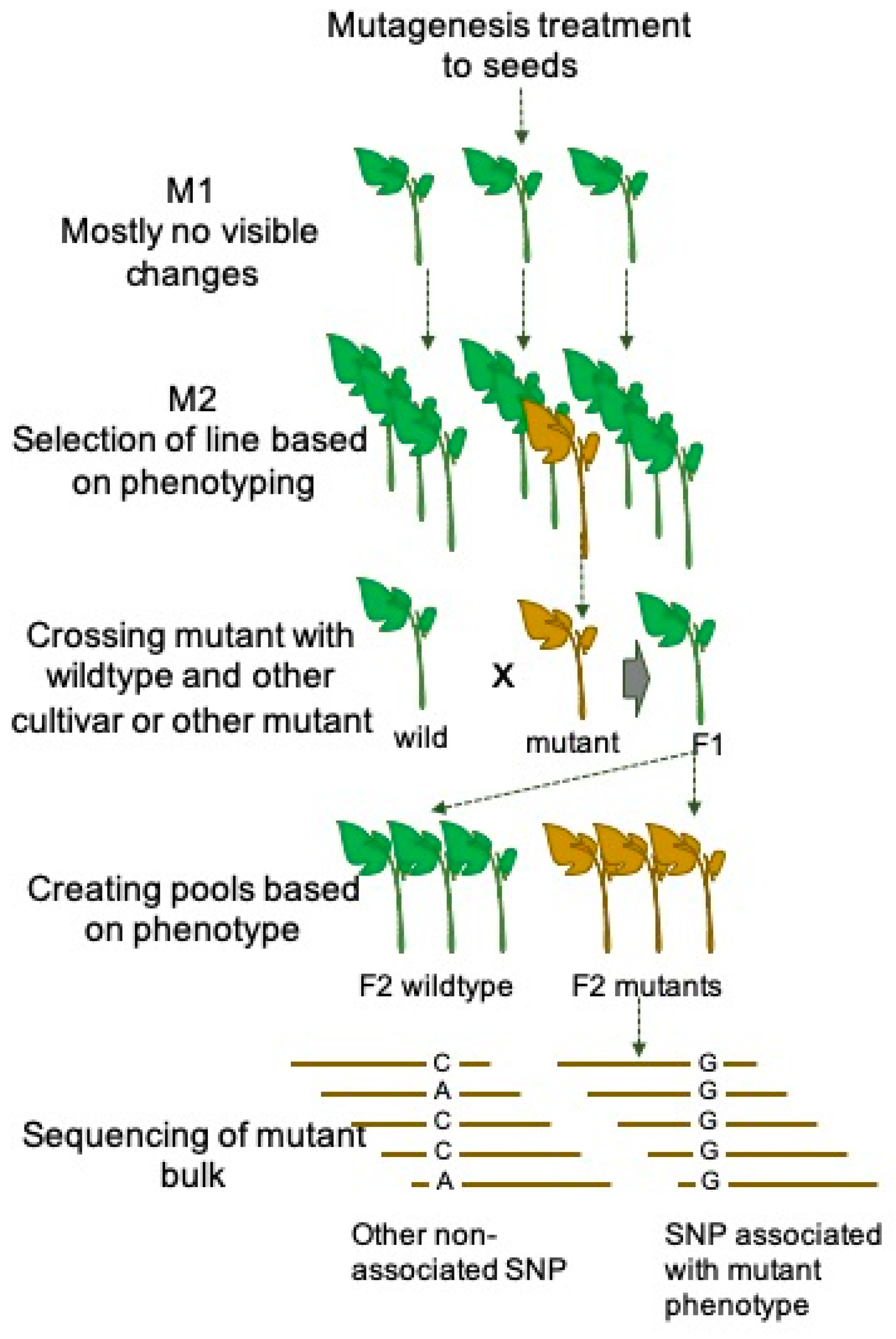
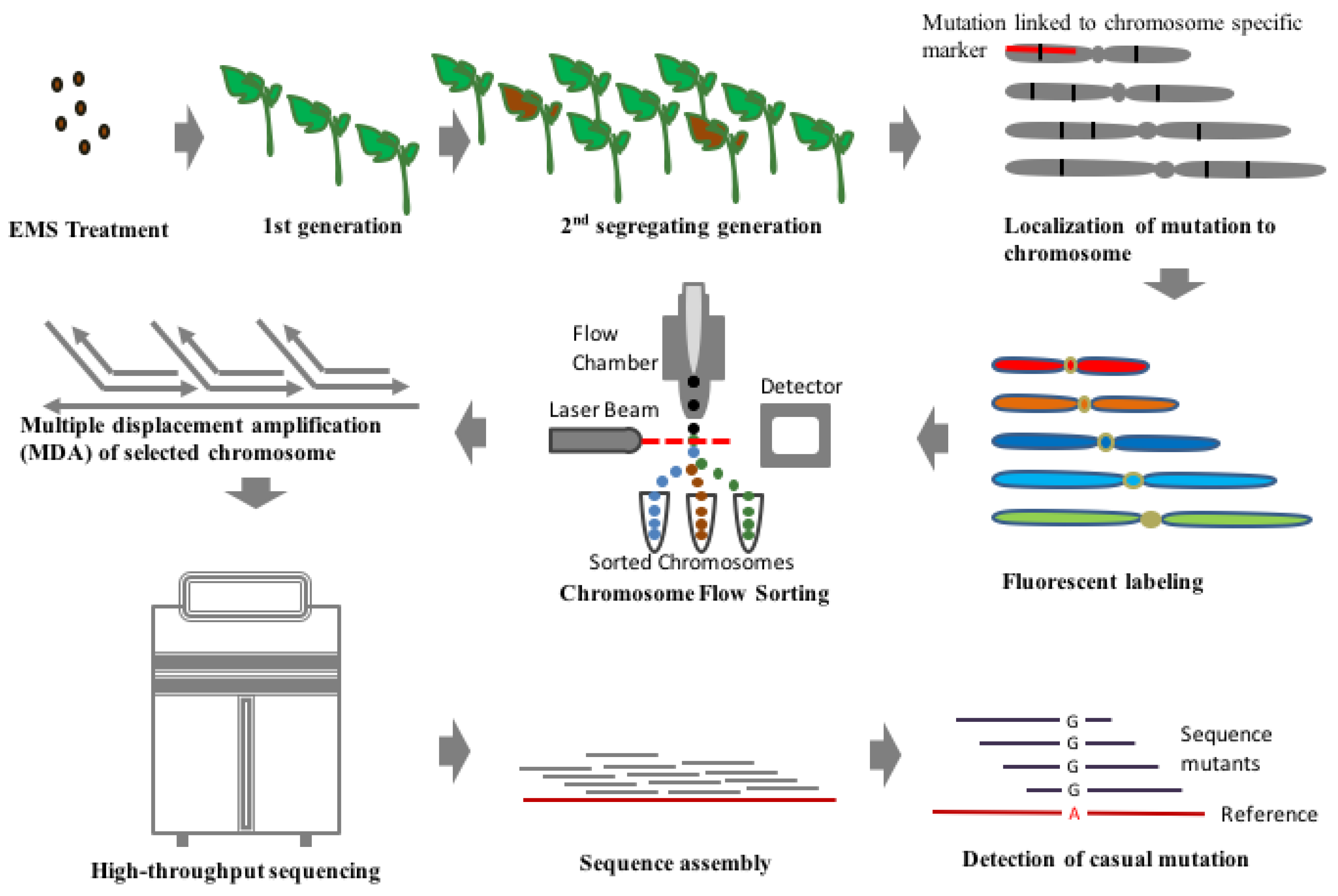
| Tomato Cultivar | Mutagen | Concentration/Dose | Number of Mutants | Reason | Reference |
|---|---|---|---|---|---|
| Moneymaker | EMS (ethyl methane sulfonate) | 60 mM | NA | Isolated Adh1 null mutant of tomato | [9] |
| M82 | EMS | 0.5% | 2552 | For functional genomic studies | [10] |
| M82 | Fast neutron | 15 Gy | 865 | For functional genomic studies | [10] |
| Lycopersicon esculentum Mill. | Sodium azide | 4 mM | 31.07% | To improve the variety | [11] |
| Lycopersicon esculentum Mill. | EMS | 1.5% | 16 | For resistance to Orobanche ramosa L. | [12] |
| Micro-Tom | Gamma-ray irradiation | 300 Gy | 6347 | For functional genomics studies | [13] |
| Red Setter | EMS | 1% | 4500 | To develop Red Setter TILLING platform | [14] |
| Red Setter | EMS | 0.7% | 8500 | To develop Red Setter TILLING platform | [14] |
| M82 | EMS | 4759 | For resistance to Potyvirus | [15] | |
| Micro-Tom | EMS | 1% | NA | For forward and reverse genetic studies | [1] |
| Tomato Variety/Cultivar | Insertional Mutagen | Target Gene/s | Transformation Method; Vector | Reference |
|---|---|---|---|---|
| cv. VF36 | Dissociation transposable element with Ac3 | NA | Agrobacterium-mediated; pBH2 | [26] |
| cv. VFNT Cherry (LA 1221) | Maize transposable element Ds1 | polygalacturonase (PG) and dihydroflavonol 4-reductase (DHFR) genes | Agrobacterium-mediated; pMON200 | [24] |
| VFNT Cherry and New Yorker | Chimeric Ds element | Lc | Agrobacterium-mediated; pAL69 and pAL144 | [27] |
| VF36 (LA490) and VFNT Cherry (LA1221) | Ac transposase and a chimeric Ds element | PG | Agrobacterium-mediated; pVCY1601 (VFNT Cherry) | [25] |
| cv. Micro-Tom | Activation-tagging technology | ant1 | Agrobacterium-mediated; pSKI015 | [28] |
| LA0315 and LA3899 | Rider transposon | SlMBP21 | Agrobacterium-mediated; pGEM-T Easy vector | [29] |
| Tomato Variety/Cultivar | Target Gene | Cas9 Promoter | Transformation Method; Vector | Effect | Reference |
|---|---|---|---|---|---|
| cv. M82 | ARGONAUTE7 (SlAGO7) | U6 promoter | Agrobacterium tumefaciens-mediated; pAGM4723 | First leaves of mutant plants having leaflets without petioles and later-formed leaves lacking laminae | [7] |
| Solanum lycopersicum var. M82 | SHORT-ROOT (SHR) and SCARECROW (SCR) | 35S promoter | A. rhizogenes transformation; pK7m24GW vector | Demonstrates that SHORT-ROOT and SCARECROW gene function is conserved between Arabidopsis and tomato | [45] |
| cv. M82 | SlCLV3 | 35S promoter | Agrobacterium-mediated; pSC-A-amp/kan vector | Increased fruit size | [46] |
| cv. Micro-Tom | ANT1 | 35S promoter | A. tumefaciens-mediated; pCAMBIA1300 | Resulted in intensely purple plant tissue | [38] |
| cv. Ailsa Craig | Ripening regulator, RIN | PcUbi4-2 | Agrobacterium tumefaciens-mediated; pUC19_AtU6 oligo | Incomplete-ripening fruits were produced and confirming the important role of RIN in ripening | [47] |
| cv. Micro-Tom | SlPDS and SlPIF4 | CaMV 35S and AtUBQ promoter | Agrobacterium tumefaciens-mediated; AtU6-sgRNA-AtUBQ-Cas9 and AtU6-sgRNA-2 × CaMV 35S-Cas9 | Clear albino phenotypes for psd mutants | [48] |
| cv. M82 | SlBOP | 35S promoter | Agrobacterium tumefaciens-mediated; pSC-A-amp/kan vector | Causes pleiotropic defects, most notably simplification of inflorescences into single flowers, resembling tmf mutants | [49] |
| cv. Micro-Tom | DELLA and ETR1 | AtU6 promoter | Agrobacterium tumefaciens-mediated; pZK_FFCas9 and pUC19_AtU6oligo | Generated marker-free plants with homozygous heritable DNA substitutions | [43] |
| S. lycopersicum line FL8000 | SlDMR6-1 | 2 × 35S promoter | A. tumefaciens-mediated; pPZP200 | Confers broad-spectrum disease resistance | [50,51] |
| cv. M82 (LA3475) and single flower truss (sft-7187) | SELF-PRUNING 5G (SP5G) | 35S promoter | Agrobacterium tumefaciens–mediated; Level 1 vector, pICH47751 and pICH47761and level 2 vector pAGM4723 | Rapid flowering and enhance the compact determinate growth habit of field tomatoes | [52] |
| GCR758, a derivative of tomato cultivar Moneymaker | slmlo1 | U6 promoter | A. tumefaciens-mediated; pAGM4723 | Developed transgene-free powdery mildew resistant tomato variety, ‘Tomelo’ | [53] |
| cv. Ailsa Craig | SlMAPK3 | Ubi-H promoter | Agrobacterium -mediated cotyledon transformation; pYLCRISPR/Cas9 vector | Suggests that SlMAPK3 is involved in drought response in tomato plants | [54] |
| cv. M82 | SlAGL6 | Agrobacterium tumefaciens-mediated; pRCS binary vector | Mutant is capable of fruit production under heat stress conditions | [55] | |
| cv. Micro-Tom | SlGAD2 and SlGAD3 | AtU6 promoter | Agrobacterium tumefaciens-mediated; pZD_ AtU6_Hpger_Cas9_NPTII and pDeCas9_Kan | Increased GABA accumulation by 7 to 15 fold | [56] |
| cv. M82 | ALC gene | 35S promoter | Agrobacterium tumefaciens-mediated; pCAM1301 | Recessive homozygous breeding elites with the character of long-shelf life were generated | [57] |
| cv. Micro-Tom and Ailsa Craig | SlIAA9 | 2 × CaMV35S promoter | Agrobacterium tumefaciens-mediated; pEgP526-2A-GFBSD2 | Morphological changes in leaf shape and seedless fruit | [51] |
| cv. Ailsa Craig (AC) and cv. Micro-Tom (MT) | Phytoene desaturase | Ubi promoter | Agrobacterium-mediated; pYLCRISPR/Cas9-slyPDS and -GABA vector | GABA accumulation enhance in both leaves and fruits | [58] |
| Tomato Cultivar | Mutant Trait | Mapping Method | Mutation (Position) | Reference |
|---|---|---|---|---|
| Moneymaker | gib-2 | Linkage mapping | Chr 1 | [63] |
| gib-1 | Chr 6 | |||
| gib-3 | Chr 7 | |||
| VF 11 and K93 | Rg-1 | Classical and RFLP (restriction fragment length polymorphism) mapping | Chr 3 (51 cM) | [64] |
| phytochrome A (phy A)-deficient fri mutants | fri | Classical map | Chr 10 (29 cM) | [65] |
| phyB1-deficient tri mutants | tri, hp-2 | Classical and RFLP mapping | Chr 1 (33 cM) | [65] |
| Ailsa Craig and Liberto | Cnr (colorless nonripening) | RFLP and Linkage Analysis | Chr 2 (4.1-9.2 cM) | [66] |
| LA3179 and LA348 | Beta (B) and old-gold (og) | Map-based cloning | Chr 6 | [67] |
| LA2453, LA2455, and LA483 | Green-ripe (Gr) and Never-ripe 2 (Nr-2) | Positional cloning | Chr 1 (2 cM) | [68] |
| Liberto and Ailsa Craig | Colorless non-ripening (Cnr) | Positional cloning | Chr 2 (13 kb) | [69] |
| LA3534, LA4074 and LA4076 | green-flesh (gf) | Positional cloning | Chr 8 (45 cM) | [70] |
| Castlemart | Od-2 | Genetic mapping | Chr 11 (16 cM) | [71] |
| Solanum lycopersicum | lutescent 2 (l2) | Linkage mapping | Chr 10L (7.5 cM) | [72] |
| S. lycopersicum (pe/pe) (LA2467) and S. pimpinellifolium (PE/PE) (LA1589) | pe | Genetic mapping | Chr 1 (424 kb) | [73] |
| P15C12 Micro-Tom glossy mutant × dwarf mutant from the M82 cultivar | P15C12 | Genetic mapping | Chr 11 (4.84 Mb) | [74] |
| Micro-Tom | pyp1 | Candidate gene approach with map-based cloning | Chr 1 (3.4 cM) | [75] |
| af (LA1049) × IL5-2 (LA4055) | Af | Map-Based Cloning | Chr 5 | [76] |
| M82 | Nxd1 | Map-based cloning | Chr 12 (40-42 Mbp) | [77] |
| Information Resource | Genes that Make Tomatoes | LycoTILL | TOMATOMA |
|---|---|---|---|
| Genetic background | Inbred variety M82 | cv. Red Setter | Micro-Tom |
| Mutagens | EMS and fast-neutrons | EMS | EMA and gamma-rays |
| Mutagen dosage | 0.5% EMS and 12 Gy, 15 Gy Fast-neutron irradiation | 0.7% and 1% EMS | 0.3, 0.5, 1 and 1.5% EMS |
| Total M2/M3 families included | 6000 EMS and 7000 fast neutron M2 families | 6677 M2 and 5872 M3 families | 4371 EMS and 6422 gamma-ray irradiated families |
| Total mutants catalogued | 3417 | - | 1048 |
| Total categories | 15 primary and 48 secondary categories | 17 classes and 52 sub-classes | 15 major and 48 sub-categories |
| Managed by | Solanaceae resource | Metapontum Agrobios | National Bioresource Project Tomato (NBRP) |
| Seed request | Seeds obtained by sending email for the list of mutant codes | By signing of a Material Transfer Agreement (MTA) document | By sending 2 copies of MTA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaudhary, J.; Alisha, A.; Bhatt, V.; Chandanshive, S.; Kumar, N.; Mir, Z.; Kumar, A.; Yadav, S.K.; Shivaraj, S.M.; Sonah, H.; et al. Mutation Breeding in Tomato: Advances, Applicability and Challenges. Plants 2019, 8, 128. https://doi.org/10.3390/plants8050128
Chaudhary J, Alisha A, Bhatt V, Chandanshive S, Kumar N, Mir Z, Kumar A, Yadav SK, Shivaraj SM, Sonah H, et al. Mutation Breeding in Tomato: Advances, Applicability and Challenges. Plants. 2019; 8(5):128. https://doi.org/10.3390/plants8050128
Chicago/Turabian StyleChaudhary, Juhi, Alisha Alisha, Vacha Bhatt, Sonali Chandanshive, Nirbhay Kumar, Zahoor Mir, Ashwini Kumar, Satish K. Yadav, S. M. Shivaraj, Humira Sonah, and et al. 2019. "Mutation Breeding in Tomato: Advances, Applicability and Challenges" Plants 8, no. 5: 128. https://doi.org/10.3390/plants8050128
APA StyleChaudhary, J., Alisha, A., Bhatt, V., Chandanshive, S., Kumar, N., Mir, Z., Kumar, A., Yadav, S. K., Shivaraj, S. M., Sonah, H., & Deshmukh, R. (2019). Mutation Breeding in Tomato: Advances, Applicability and Challenges. Plants, 8(5), 128. https://doi.org/10.3390/plants8050128







