Conserved Cu-MicroRNAs in Arabidopsis thaliana Function in Copper Economy under Deficiency
Abstract
1. Introduction
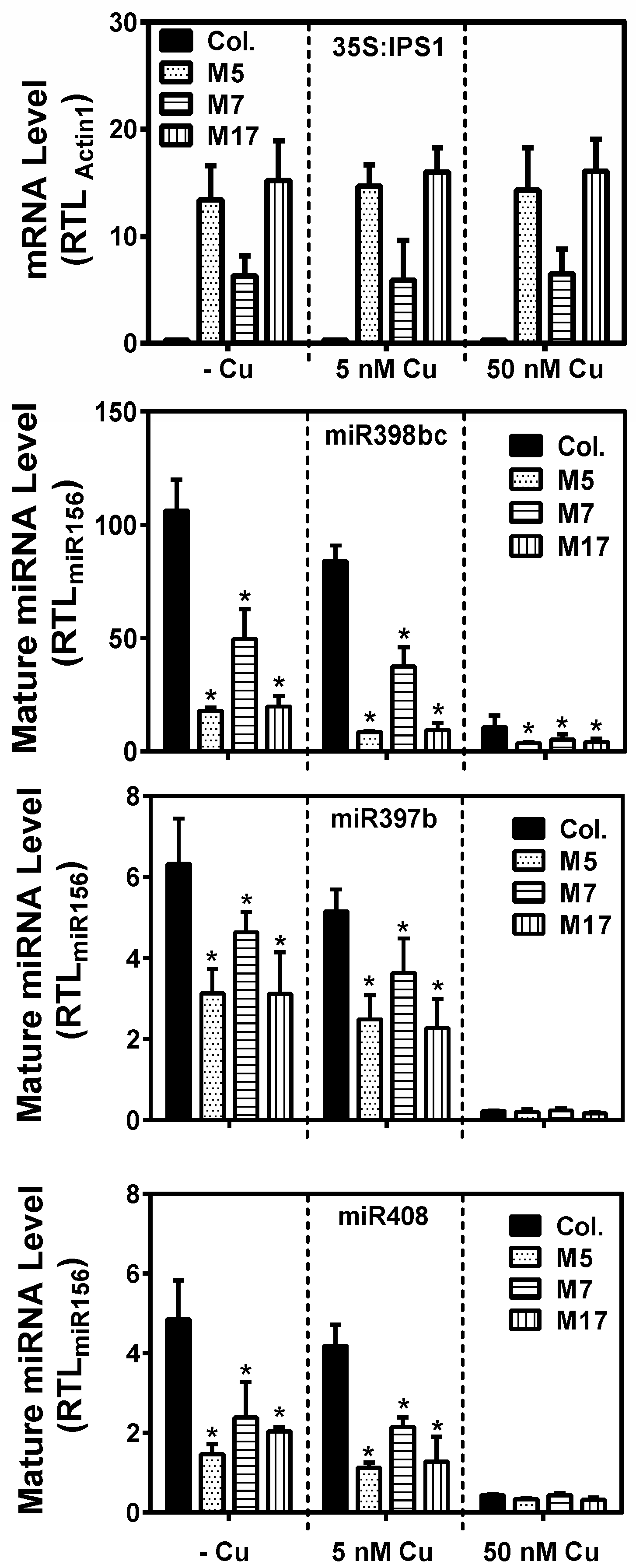
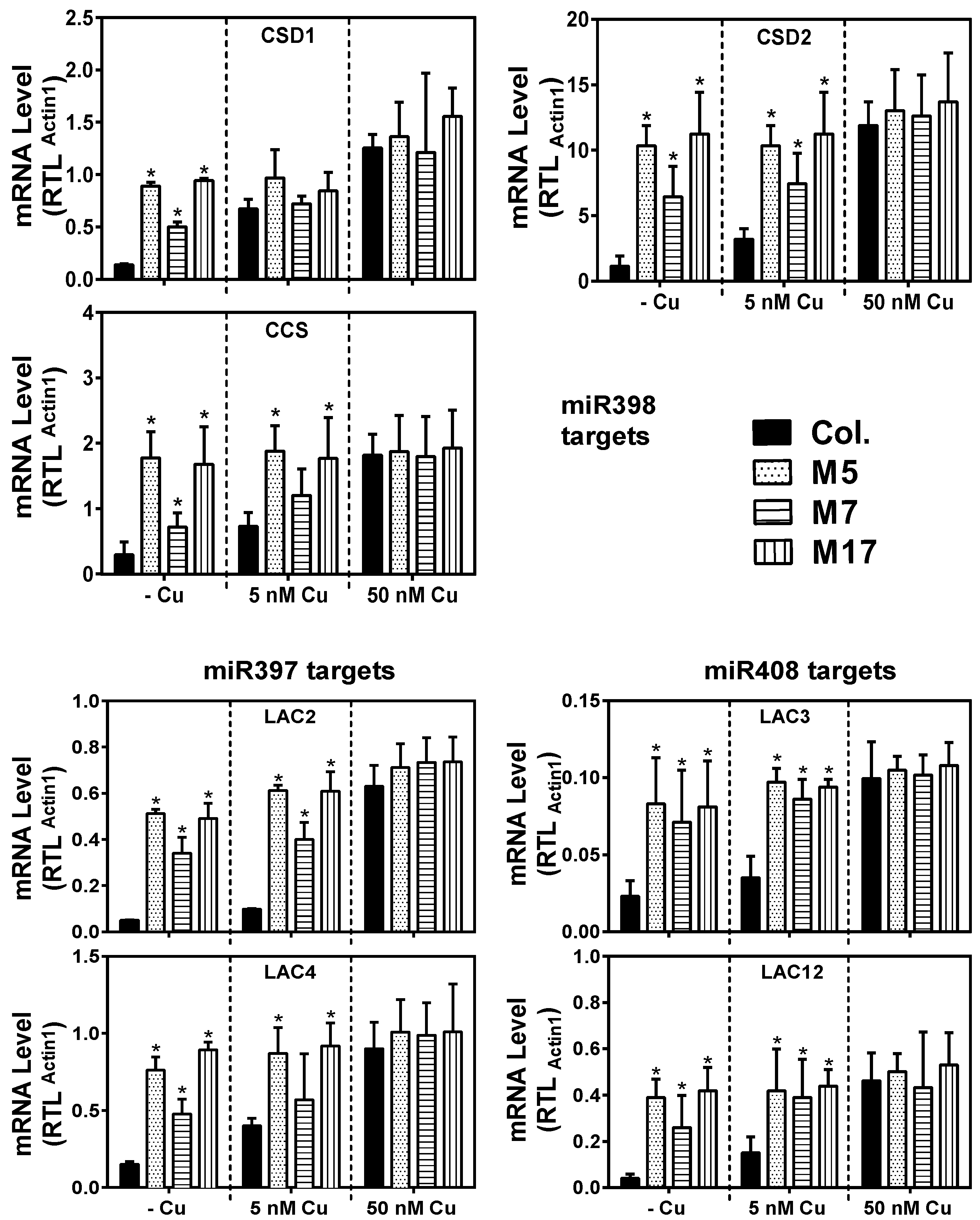
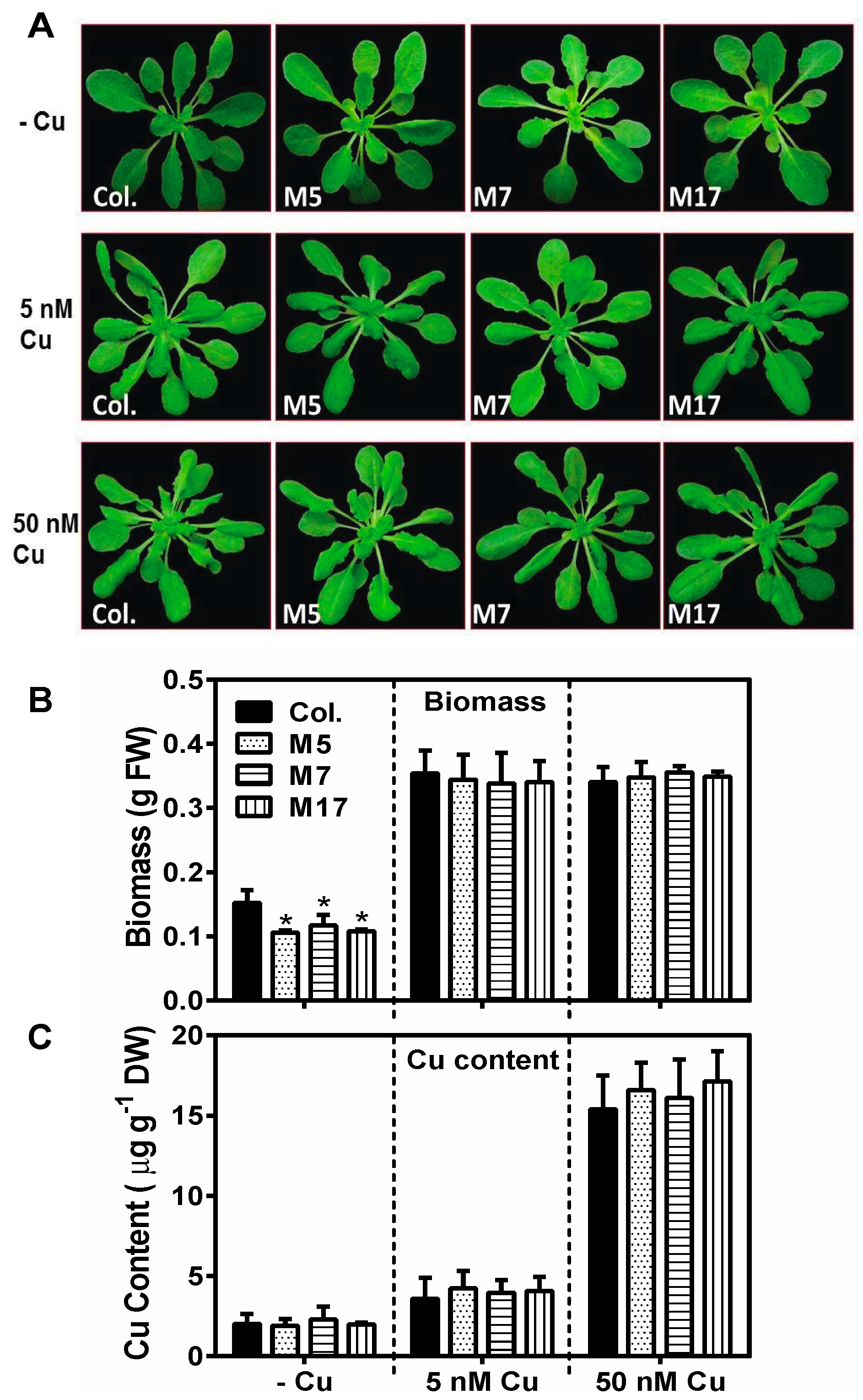
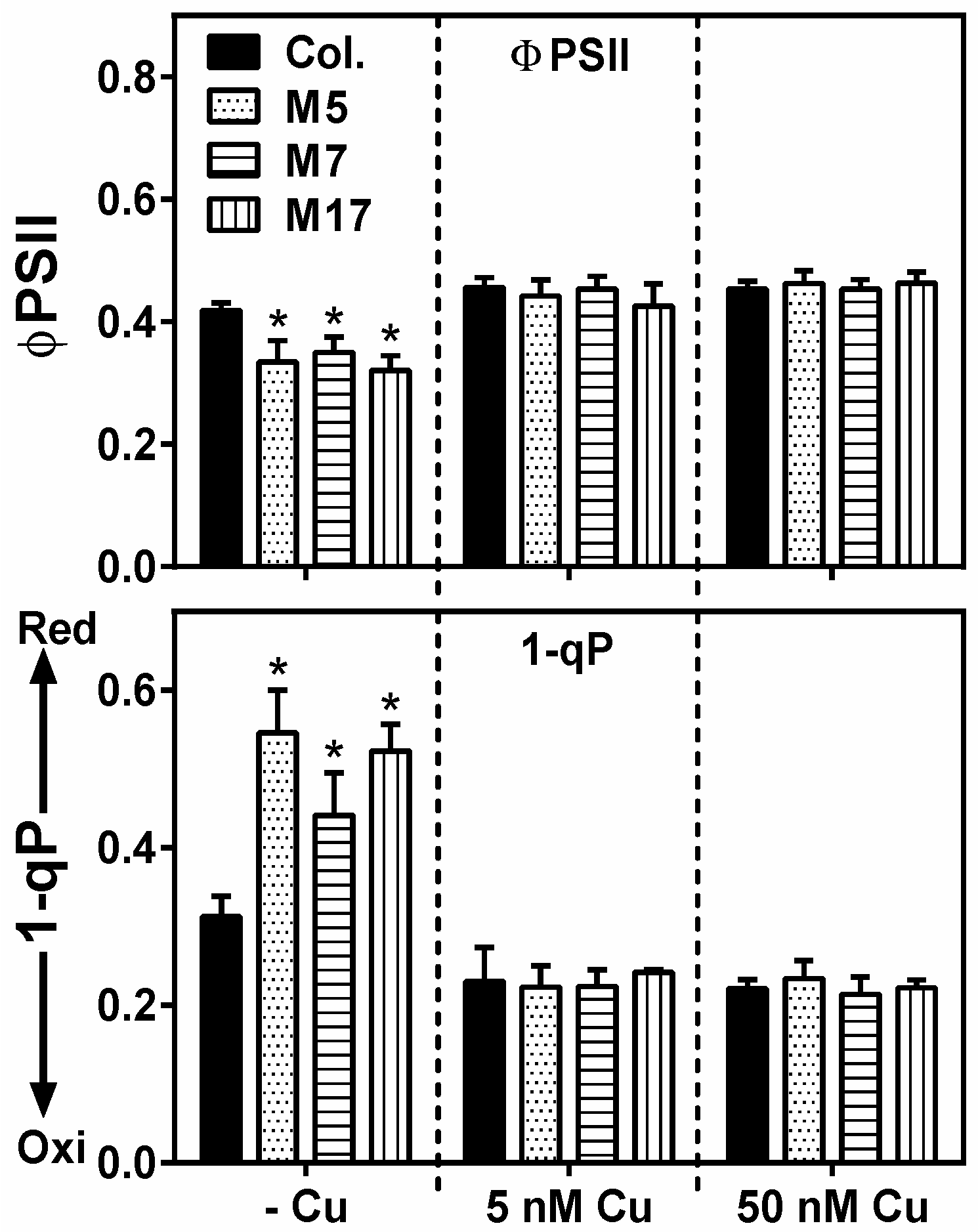
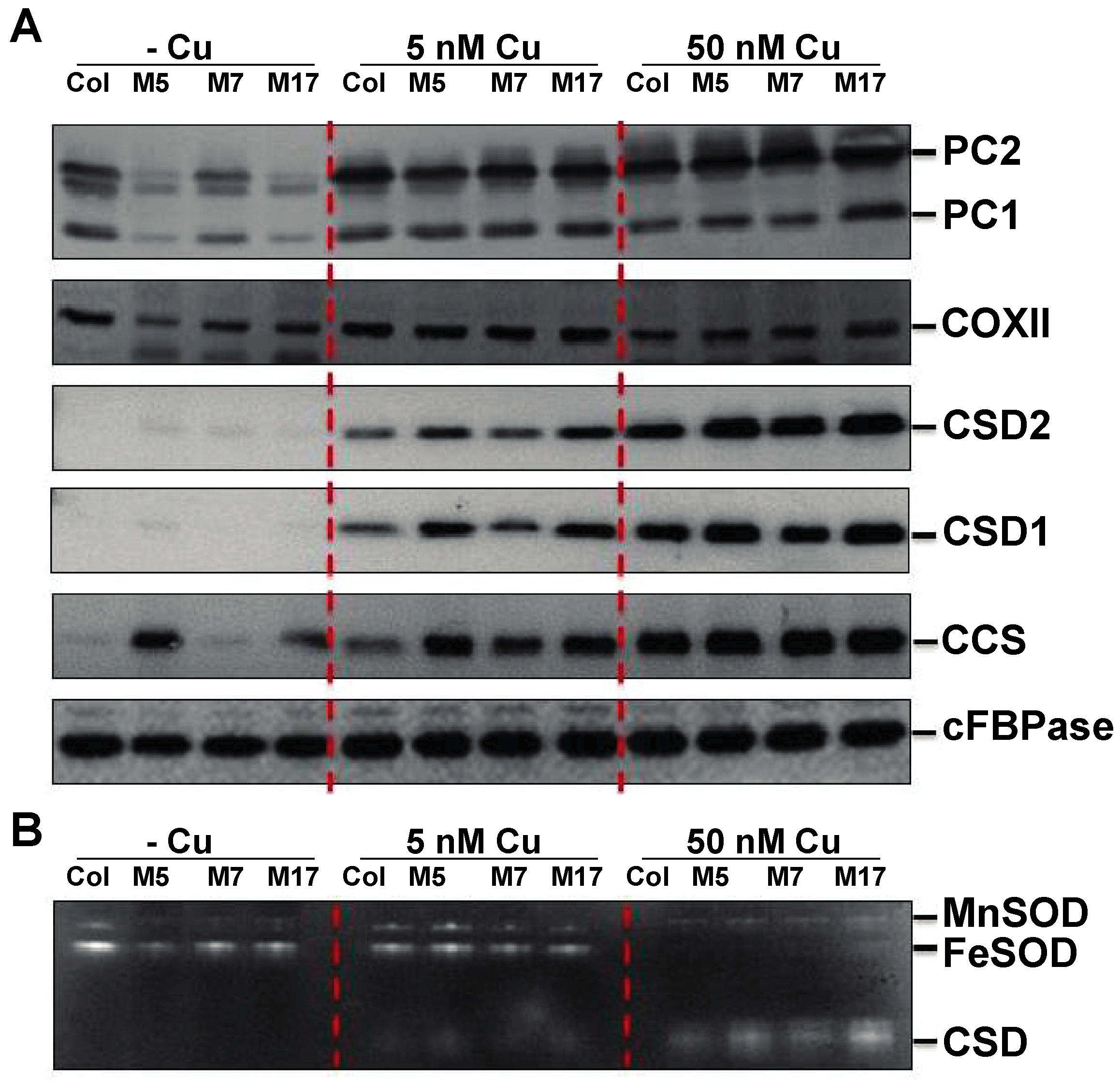
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Transgenic Lines
4.3. Elemental Analysis
4.4. Chlorophyll Fluorescence Measurements
4.5. Protein Accumulation
4.6. Quantitative Reverse Transcription-PCR
4.7. Mature miRNA Stem-Loop qRT-PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Marschner, H. Mineral Nutrition of Higher Plants; Academic Press: London. UK, 1995. [Google Scholar]
- Shikanai, T.; Muller-Moule, P.; Munekage, Y.; Niyogi, K.K.; Pilon, M. PAA1, a P-type ATPase of arabidopsis, functions in copper transport in chloroplasts. Plant Cell 2003, 15, 1333–1346. [Google Scholar] [CrossRef]
- Tapken, W.; Ravet, K.; Pilon, M. Plastocyanin controls the stabilization of the thylakoid Cu-transporting P-type ATPase PAA2/HMA8 in response to low copper in Arabidopsis. J. Biol. Chem. 2012, 287, 18544–18550. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A.; Evans, M.C.; Korb, R.E. The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosynth. Res. 1999, 60, 111–149. [Google Scholar] [CrossRef]
- Pesaresi, P.; Scharfenberg, M.; Weigel, M.; Granlund, I.; Schroeder, W.P.; Finazzi, G.; Rappaport, F.; Masiero, S.; Furini, A.; Jahns, P.; et al. Mutants, Overexpressors, and Interactors of Arabidopsis Plastocyanin Isoforms: Revised Roles of Plastocyanin in Photosynthetic Electron Flow and Thylakoid Redox State. Mol. Plant 2009, 2, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Ghany, S.E. Contribution of plastocyanin isoforms to photosynthesis and copper homeostasis in Arabidopsis thaliana grown at different copper regimes. Planta 2009, 229, 767–779. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Muller-Moule, P.; Niyogi, K.K.; Pilon, M.; Shikanai, T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 2005, 17, 1233–1251. [Google Scholar] [CrossRef]
- Bernal, M.; Casero, D.; Singh, V.; Wilson, G.T.; Grande, A.; Yang, H.; Dodani, S.C.; Pellegrini, M.; Huijser, P.; Connolly, E.L.; et al. Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 2012, 24, 738–761. [Google Scholar] [CrossRef] [PubMed]
- Molina-Heredia, F.P.; Wastl, J.; Navarro, J.A.; Bendall, D.S.; Hervás, M.; Howe, C.J.; De La Rosa, M.A. Photosynthesis: a new function for an old cytochrome? Nature 2003, 424, 33–34. [Google Scholar] [CrossRef]
- Weigel, M.; Varotto, C.; Pesaresi, P.; Finazzi, G.; Rappaport, F.; Salamini, F.; Leister, D. Plastocyanin Is Indispensable for Photosynthetic Electron Flow in Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 31286–31289. [Google Scholar] [CrossRef] [PubMed]
- Kliebenstein, D.J.; Monde, R.-A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclecticenzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef]
- Chu, C.C.; Lee, W.C.; Guo, W.Y.; Pan, S.M.; Chen, L.J.; Li, H.M.; Jinn, T.L. A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol. 2005, 139, 425–436. [Google Scholar] [CrossRef]
- Cohu, C.M.; Abdel-Ghany, S.E.; Gogolin Reynolds, K.A.; Onofrio, A.M.; Bodecker, J.R.; Kimbrel, J.A.; Niyogi, K.K.; Pilon, M. Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: regulation and unexpected phenotypes in an Arabidopsis mutant. Mol. Plant 2009, 2, 1336–1350. [Google Scholar] [CrossRef]
- Carr, H.S.; Winge, D.R. Assembly of Cytochrome c Oxidase within the Mitochondrion. Acc. Chem. Res. 2003, 36, 309–316. [Google Scholar] [CrossRef]
- Rodriguez, F.I.; Esch, J.J.; Hall, A.E.; Binder, B.M.; Schaller, G.E.; Bleecker, A.B. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 1999, 283, 996–998. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N. The Ethylene Signaling Pathway. Science 2004, 306, 1513–1515. [Google Scholar] [CrossRef]
- Dong, J.; Kim, S.T.; Lord, E.M. Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiol. 2005, 138, 778–789. [Google Scholar] [CrossRef]
- Pignocchi, C.; Fletcher, J.M.; Wilkinson, J.E.; Barnes, J.D.; Foyer, C.H. The function of ascorbate oxidase in tobacco. Plant Physiol. 2003, 132, 1631–1641. [Google Scholar] [CrossRef]
- Yamamoto, A.; Bhuiyan, M.N.; Waditee, R.; Tanaka, Y.; Esaka, M.; Oba, K.; Jagendorf, A.T.; Takabe, T. Suppressed expression of the apoplastic ascorbate oxidase gene increases salt tolerance in tobacco and Arabidopsis plants. J. Exp. Bot. 2005, 56, 1785–1796. [Google Scholar] [CrossRef]
- Frébort, I.; Sebela, M.; Svendsen, I.; Hirota, S.; Endo, M.; Yamauchi, O.; Bellelli, A.; Lemr, K.; Pec, P. Molecular mode of interaction of plant amine oxidase with the mechanism-based inhibitor 2-butyne-1,4-diamine. Eur. J. Biochem. 2000, 267, 1423–1433. [Google Scholar]
- Groß, F.; Rudolf, E.E.; Thiele, B.; Durner, J.; Astier, J. Copper amine oxidase 8 regulates arginine-dependent nitric oxide production in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2149–2162. [Google Scholar] [CrossRef]
- McCaig, B.C.; Meagher, R.B.; Dean, J.F. Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 2005, 221, 619–636. [Google Scholar] [CrossRef]
- Turlapati, P.V.; Kim, K.W.; Davin, L.B.; Lewis, N.G. The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 2001, 233, 439–470. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Davis, E.J.; Ballif, J.; Liang, M.; Bushman, E.; Haroldsen, V.; Torabinejad, J.; Wu, Y. Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 2006, 57, 2563–2569. [Google Scholar] [CrossRef] [PubMed]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cézard, L.; Le Bris, P.; Borrega, N.; Hervé, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 Results in Tissue-Specific Alterations to Lignification of Arabidopsis thaliana Stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef]
- Zhao, Y.; Lin, S.; Qiu, Z.; Cao, D.; Wen, J.; Deng, X.; Wang, X.; Lin, J.; Li, X. MicroRNA857 is Involved in the Regulation of Secondary Growth of Vascular Tissues in Arabidopsis. Plant Physiol. 2015, 169, 2539–2552. [Google Scholar] [CrossRef] [PubMed]
- Pilon, M. The copper microRNAs. New Phytol. 2017, 213, 1030–1035. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Ann. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Yamasaki, H.; Abdel-Ghany, S.E.; Cohu, C.M.; Kobayashi, Y.; Shikanai, T.; Pilon, M. Regulation of Copper Homeostasis by Micro-RNA in Arabidopsis. J. Biol. Chem. 2007, 282, 16369–16378. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-mediated Systemic Down-regulation of Copper Protein Expression in Response to Low Copper Availability in Arabidopsis. J. Biol. Chem. 2008, 283, 15932–15945. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell. 2009, 21, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, L. SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J. 2013, 74, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, X.; Li, J.; Cai, H.; Deng, X.W.; Li, L. MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated respose to light and copper. Plant Cell 2014, 26, 4933–4953. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of MIRNA genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef]
- Ravet, K.; Danford, F.L.; Dihle, A.; Pittarello, M.; Pilon, M. Spatial-temporal analysis of Cu homeostasis in Populus trichocarpa reveals an integrated molecular remodeling for a preferential allocation of copper to plastocyanin in the chloroplasts of developing leaves. Plant Physiol. 2011, 157, 1300–1312. [Google Scholar] [CrossRef][Green Version]
- Shahbaz, M.; Ravet, K.; Peers, G.; Pilon, M. Prioritization of copper for the use in photosynthetic electron transport in developing leaves of hybrid poplar. Front. Plant Sci. 2015, 6, 407. [Google Scholar] [CrossRef]
- Yan, J.; Chia, J.C.; Sheng, H.; Jung, H.I.; Zavodna, T.O.; Zhang, L.; Huang, R.; Jiao, C.; Craft, E.J.; Fei, Z.; et al. Arabidopsis Pollen Fertility Requires the Transcription Factors CITF1 and SPL7 That Regulate Copper Delivery to Anthers and Jasmonic Acid Synthesis. Plant Cell 2017, 29, 3012–3029. [Google Scholar] [CrossRef]
- Dugas, D.V.; Bartel, B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol. Biol. 2008, 67, 403–417. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Zhang, J.P.; Yu, Y.; Feng, Y.Z.; Zhou, Y.F.; Zhang, F.; Yang, Y.W.; Lei, M.Q.; Zhang, Y.C.; Chen, Y.Q. MiR408 Regulates Grain Yield and Photosynthesis via a Phytocyanin Protein. Plant Physiol. 2017, 175, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Kropat, J.; Tottey, S.; Birkenbihl, R.P.; Depege, N.; Huijser, P.; Merchant, S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 2005, 102, 18730–18735. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Huang, D.; Guo, Z.; Kuang, Z.; Zhang, H.; Xie, X.; Ma, Z.; Gao, S.; Lerdau, M.T.; Chu, C.; et al. Overexpression of microRNA408 enhances photosynthesis, growth, and seed yield in diverse plants. J. Integr. Plant Biol. 2018, 60, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Zhang, L.; Wang, Y.; Li, H.; Li, S.; Zhao, H.; Zhang, H. Constitutive Expression of miR408 Improves Biomass and Seed Yield in Arabidopsis. Front. Plant Sci. 2018, 8, 2114. [Google Scholar] [CrossRef] [PubMed]
- Carrió-Seguí, À.; Ruiz-Rivero, O.; Villamayor-Belinchón, L.; Puig, S.; Perea-García, A.; Peñarrubia, L. The Altered Expression of microRNA408 Influences the Arabidopsis Response to Iron Deficiency. Front. Plant Sci. 2019, 10, 324. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated micro-RNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, J.; Song, A.; Chen, S.; Shan, H.; Luo, H.; Gu, C.; Sun, J.; Zhu, L.; Fang, W.; et al. Ambient temperature enhanced freezing tolerance of Chrysanthemum dichrum CdICE1 Arabidopsis via miR398. BMC Biol. 2013, 11, 121. [Google Scholar] [CrossRef]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef]
- Waters, B.M.; McInturf, S.A.; Stein, R.J. Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 5903–5918. [Google Scholar] [CrossRef]
- Robinson, N.J.; Procter, C.M.; Connolly, E.L.; Guerinot, M.L. A ferric-chelate reductase for iron uptake from soils. Nature 1999, 397, 694–697. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Epstein, E.; Bloom, A. Mineral Nutrition in Plants: Principles and Perspectives, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2005. [Google Scholar]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Pilon-Smits, E.A.; Hwang, S.; Lytle, C.M.; Zhu, Y.; Tai, J.C.; Bravo, R.C.; Chen, Y.; Leustek, T.; Terry, N. Overexpression of ATP sulfurylase in Indian mustard leads to increased selenite uptake, reduction and tolerance. Plant Physiol. 1999, 119, 123–132. [Google Scholar] [CrossRef]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—a practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Hellens, R.P. Quantitative stem-loop RT-PCR for detection of microRNAs. Methods Mol. Biol. 2007, 744, 145–157. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahbaz, M.; Pilon, M. Conserved Cu-MicroRNAs in Arabidopsis thaliana Function in Copper Economy under Deficiency. Plants 2019, 8, 141. https://doi.org/10.3390/plants8060141
Shahbaz M, Pilon M. Conserved Cu-MicroRNAs in Arabidopsis thaliana Function in Copper Economy under Deficiency. Plants. 2019; 8(6):141. https://doi.org/10.3390/plants8060141
Chicago/Turabian StyleShahbaz, Muhammad, and Marinus Pilon. 2019. "Conserved Cu-MicroRNAs in Arabidopsis thaliana Function in Copper Economy under Deficiency" Plants 8, no. 6: 141. https://doi.org/10.3390/plants8060141
APA StyleShahbaz, M., & Pilon, M. (2019). Conserved Cu-MicroRNAs in Arabidopsis thaliana Function in Copper Economy under Deficiency. Plants, 8(6), 141. https://doi.org/10.3390/plants8060141




