Isolation of Native Arbuscular Mycorrhizal Fungi within Young Thalli of the Liverwort Marchantia paleacea
Abstract
1. Introduction
2. Results and Discussion
2.1. Growth Condition of M. paleacea Young Thalli
2.2. Young Thalli (<2 mm) Colonized by Arbuscular Mycorrhizal Fungi
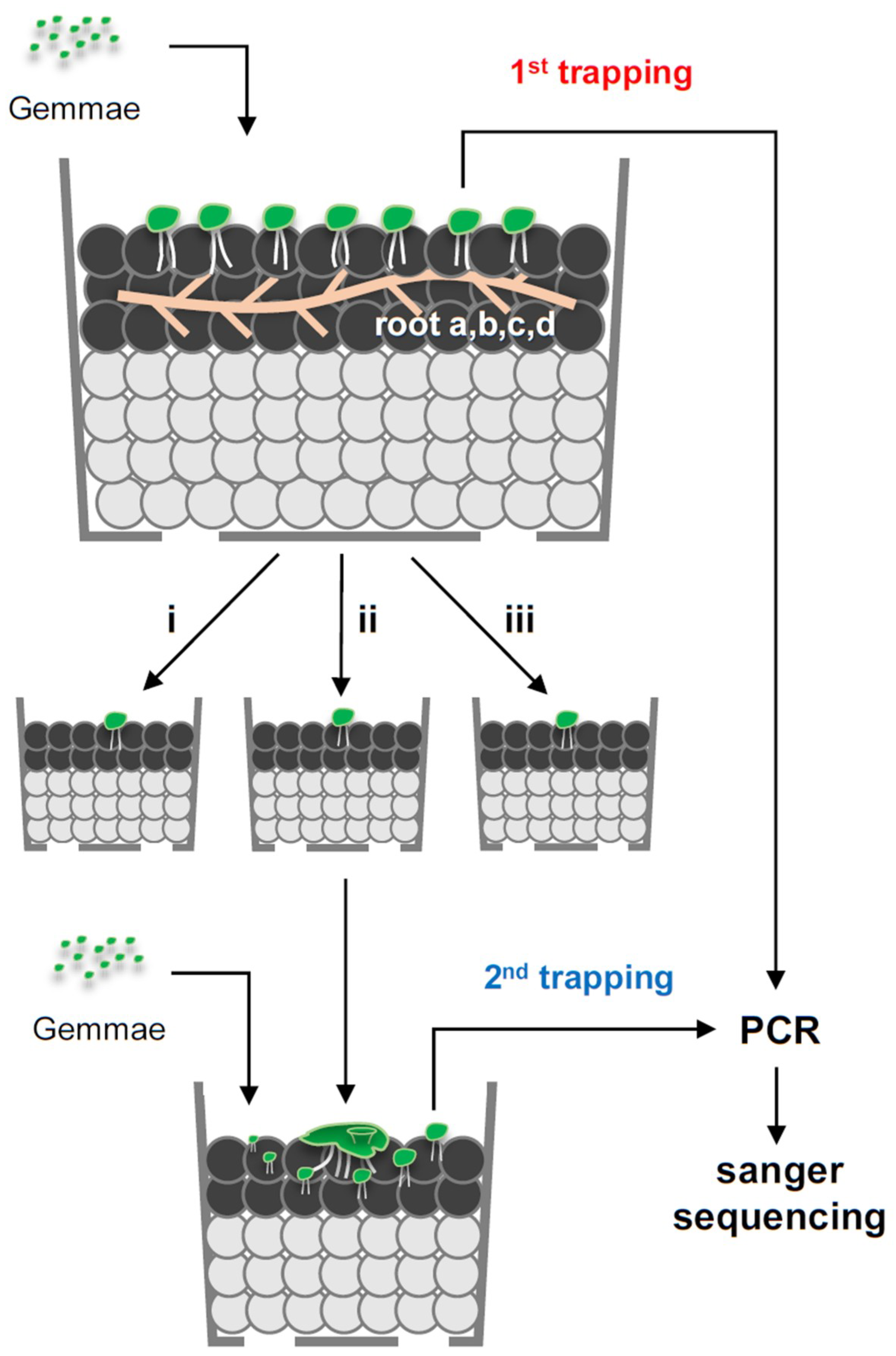
2.3. First Trapping of Arbuscular Mycorrhizal Fungi and Sequencing
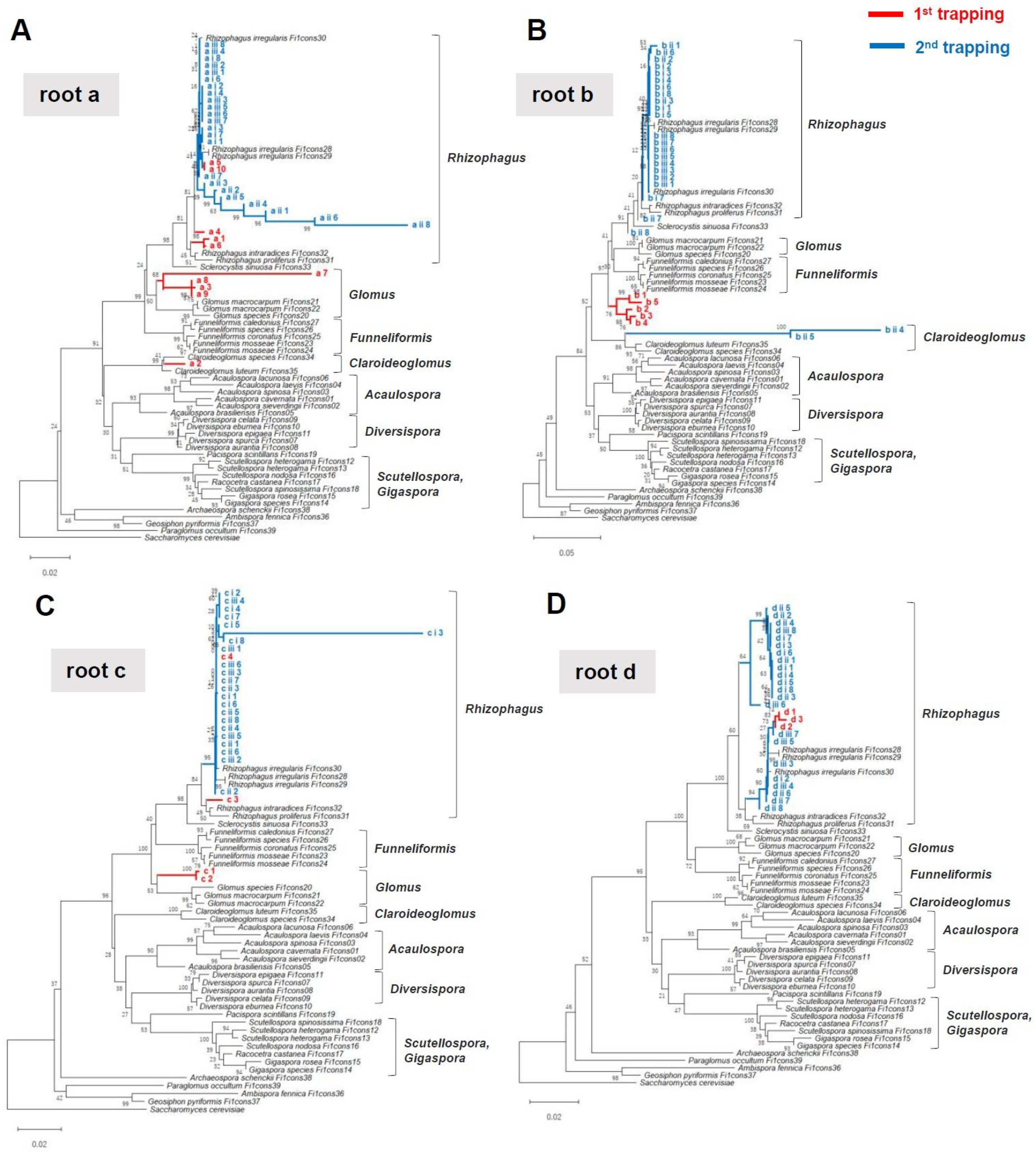
2.4. “Meta”-Sequences of Arbuscular Mycorrhizal Fungi rDNA in Young Thalli
2.5. Second Trapping Isolated Phylogenetically Uniform Arbuscular Mycorrhizal Fungi Species
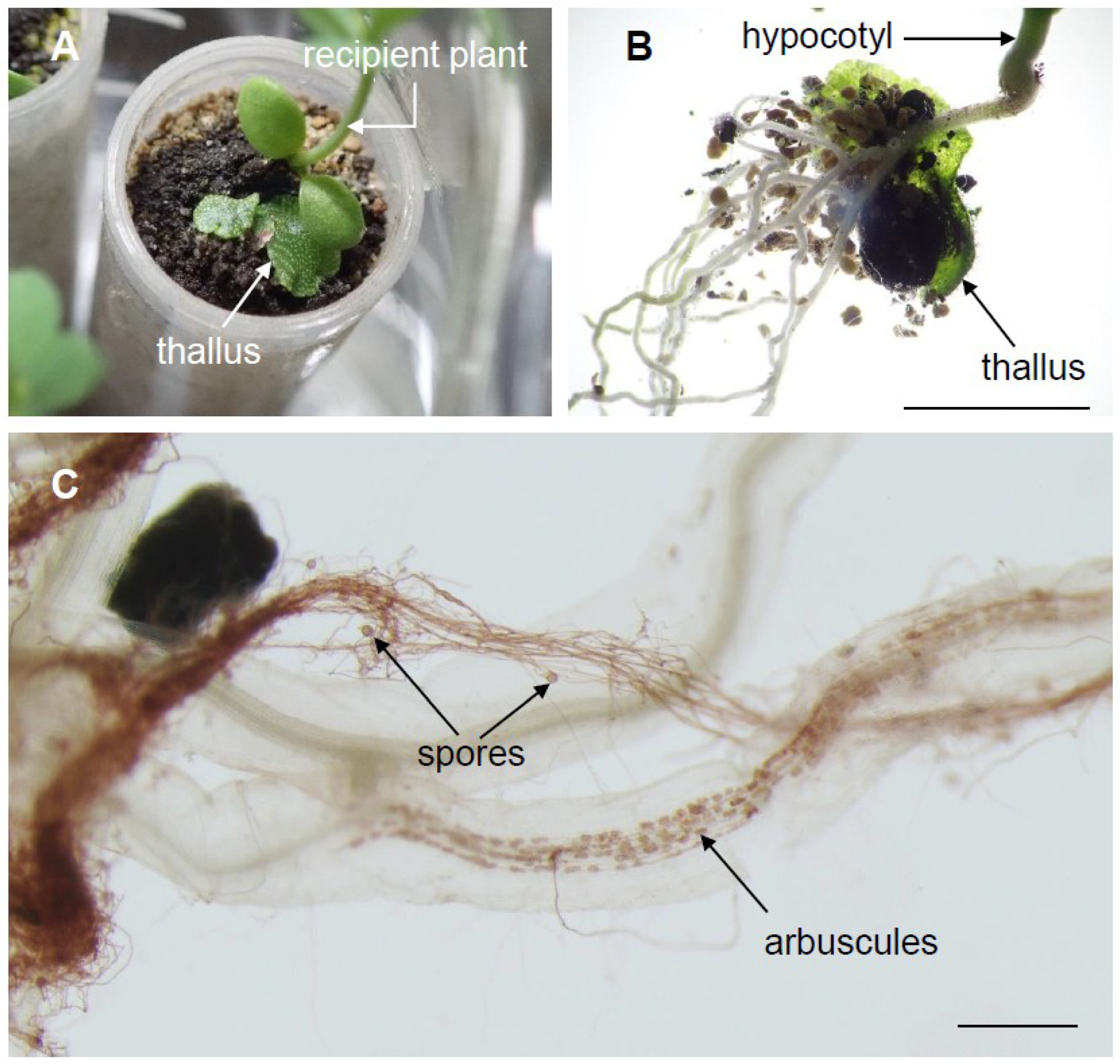
2.6. Mycothalli Can Be Used as an Arbuscular Mycorrhizal Fungi Inoculum in Pot Experiments
3. Materials and Methods
3.1. M. paleacea Thalli Growth Conditions
3.2. Inoculation Test
3.3. Arbuscular Mycorrhizal Fungi Cell Wall Staining
3.4. Trap Culture
3.5. PCR and Sequencing
3.6. Sequence Analysis and Classification
3.7. Mycothalli Inoculation Test
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nehls, U. Mastering ectomycorrhizal symbiosis: The impact of carbohydrates. J. Exp. Bot. 2008, 59, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Rillig, M.C.; Aguilar-Trigueros, C.A.; Camenzind, T.; Cavagnaro, T.R.; Degrune, F.; Hohmann, P.; Lammel, D.R.; Mansour, I.; Roy, J.; van der Heijden, M.G.A.; et al. Why farmers should manage the arbuscular mycorrhizal symbiosis. New Phytol. 2019, 222, 1171–1175. [Google Scholar] [CrossRef]
- Zhang, S.; Lehmann, A.; Zheng, W.; You, Z.; Rillig, M.C. Arbuscular mycorrhizal fungi increase grain yields: A meta-analysis. New Phytol. 2019, 222, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Qiu, Y. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 2006, 16, 299–363. [Google Scholar] [CrossRef] [PubMed]
- Balestrini, R.; Bonfante, P. Cell wall remodeling in mycorrhizal symbiosis: A way towards biotrophism. Front Plant Sci. 2014, 5, 237. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef] [PubMed]
- Dickson, S.; Smith, F.A.; Smith, S.E. Structural differences in arbuscular mycorrhizal symbioses: More than 100 years after Gallaud, where next? Mycorrhiza 2007, 17, 375–393. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Smith, F.A. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis; Academic Press: Cambridge, UK, 2008. [Google Scholar]
- Javot, H.; Penmetsa, R.V.; Terzaghi, N.; Cook, D.R.; Harrison, M.J. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2007, 104, 1720–1725. [Google Scholar] [CrossRef]
- Yang, S.Y.; Grønlund, M.; Jakobsen, I.; Grotemeyer, M.S.; Rentsch, D.; Miyao, A.; Hirochika, H.; Kumar, C.S.; Sundaresan, V.; Salamin, N.; et al. Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell 2012, 24, 4236–4251. [Google Scholar] [CrossRef]
- Willmann, M.; Gerlach, N.; Buer, B.; Polatajko, A.; Nagy, R.; Koebke, E.; Jansa, J.; Flisch, R.; Bucher, M. Mycorrhizal phosphate uptake pathway in maize: Vital for growth and cob development on nutrient poor agricultural and greenhouse soils. Front. Plant Sci. 2013, 4, 533. [Google Scholar] [CrossRef]
- Strullu-Derrien, C.; Selosse, M.A.; Kenrick, P.; Martin, F.M. The origin and evolution of mycorrhizal symbioses: From palaeomycology to phylogenomics. New Phytol. 2018, 220, 1012–1030. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.M.; Uroz, S.; Barker, D.G. Ancestral alliances: Plant mutualistic symbioses with fungi and bacteria. Science 2017, 356, eaad4501. [Google Scholar] [CrossRef] [PubMed]
- Field, K.J.; Pressel, S. Unity in diversity: Structural and functional insights into the ancient partnerships between plants and fungi. New Phytol. 2018, 220, 996–1011. [Google Scholar] [CrossRef] [PubMed]
- Clapp, J.P.; Fitter, A.H.; Young, J.P.W. Ribosomal small subunit sequence variation within spores of an arbuscular mycorrhizal fungus, Scutellospora sp. Mol. Ecol. 1999, 8, 915–921. [Google Scholar] [CrossRef] [PubMed]
- van Tuinen, D.; Jacquot, E.; Zhao, B.; Gollotte, A.; Gianinazzi-Pearson, V. Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol. Ecol. 1998, 7, 879–887. [Google Scholar] [CrossRef]
- Clapp, J.P.; Young, J.P.W.; Merryweather, J.W.; Fitter, A.H. Diversity of fungal symbionts in arbuscular mycorrhizas from a natural community. New Phytol. 1995, 130, 259–265. [Google Scholar] [CrossRef]
- Merryweather, J.; Fitter, A. The arbuscular mycorrhizal fungi of Hyacinthoides non-scripta I. Diversity of fungal taxa. New Phytol. 1998, 138, 117–129. [Google Scholar] [CrossRef]
- Ohsowski, B.M.; Zaitsoff, P.D.; Öpik, M.; Hart, M.M. Where the wild things are: Looking for uncultured Glomeromycota. New Phytol. 2014, 204, 171–179. [Google Scholar] [CrossRef]
- Declerck, S.; Strullu, D.G.; Plenchette, C. Monoxenic culture of the intraradical forms of Glomus sp. isolated from a tropical ecosystem: A proposed methodology for germplasm collection. Mycologia 1998, 90, 579–585. [Google Scholar] [CrossRef]
- Biermann, B.; Linderman, R.G. Use of vesicular-arbuscular mycorrhizal roots, intraradical vesicles and extraradical vesicles as inoculum. New Phytol. 1983, 95, 97–105. [Google Scholar] [CrossRef]
- Dalpé, Y.; Cranenbrouck, S.; Seguin, S.; Declerck, S. The monoxenic culture of arbuscular mycorrhizal fungi as a tool for systematics and biodiversity. In In Vitro Vulture of Mycorrhizas; Declerck, S., Fortin, A., Strullu, D.G., Eds.; Springer: Heidelberg, Germany, 2005; pp. 31–48. [Google Scholar]
- Boullard, B. Observations on the coevolution of fungi with hepatics. In Coevolution of Fungi with Plants and Animals; Pyrozynski, K.A., Hawksworth, D.L., Eds.; Academic Press: London, UK, 1988; pp. 107–124. [Google Scholar]
- Nanzyo, M. Unique properties of volcanic ash soils. Global Environ. Res. 2002, 6, 99–112. [Google Scholar]
- Breuillin, F.; Schramm, J.; Hajirezaei, M.; Ahkami, A.; Favre, P.; Druege, U.; Hause, B.; Bucher, M.; Kretzschmar, T.; Bossolini, E.; et al. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 2010, 64, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Balzergue, C.; Puech-Pagès, V.; Bécard, G.; Rochange, S.F. The regulation of arbuscular mycorrhizal symbiosis by phosphate in pea involves early and systemic signalling events. J. Exp. Bot. 2011, 62, 1049–1060. [Google Scholar] [CrossRef]
- Kobae, Y.; Ohmori, Y.; Saito, C.; Yano, K.; Ohtomo, R.; Fujiwara, T. Phosphate treatment strongly inhibits new arbuscule development but not the maintenance of arbuscule in mycorrhizal rice roots. Plant Physiol. 2016, 171, 566–579. [Google Scholar] [CrossRef]
- Hata, S.; Kobae, Y.; Banba, M. Interactions between plants and arbuscular mycorrhizal fungi. Int. Rev. Cell Mol. Biol. 2010, 281, 1–48. [Google Scholar]
- Humphreys, C.P.; Franks, P.J.; Rees, M.; Bidartondo, M.I.; Leake, J.R.; Beerling, D.J. Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat. Commun. 2010, 1, 103. [Google Scholar] [CrossRef]
- Kobae, Y.; Ohtomo, R.; Oka, N.; Morimoto, S. A simple model system for identifying arbuscular mycorrhizal fungal taxa that actively colonize rice roots grown in field soil. Soil. Sci. Plant Nutr. 2017, 63, 29–36. [Google Scholar] [CrossRef]
- Krüger, M.; Krüger, C.; Walker, C.; Stockinger, H.; Schussler, A. Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol. 2012, 193, 970–984. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Sanders, F. Ultrastructure of the host–fungus interface in a vesicular-arbuscular mycorrhiza. New Phytol. 1974, 73, 901–912. [Google Scholar] [CrossRef]
- Walker, N.; Smith, S.E. The quantitative study of mycorrhizal infection, II. The relation of rate of infection and speed of fungal growth to propagule density, the mean length of the infection unit and the limiting value of the fraction of the root infected. New Phytol. 1984, 96, 55–69. [Google Scholar] [CrossRef]
- Ropars, J.; Toro, K.S.; Noel, J.; Pelin, A.; Charron, P.; Farinelli, L.; Marton, T.; Krüger, M.; Fuchs, J.; Brachmann, A.; et al. Evidence for the sexual origin of heterokaryosis in arbuscular mycorrhizal fungi. Nat. Microbiol. 2016, 1, 16033. [Google Scholar] [CrossRef]
- Chen, E.C.H.; Morin, E.; Beaudet, D.; Noel, J.; Yildirir, G.; Ndikumana, S.; Charron, P.; St-Onge, C.; Giorgi, J.; Krüger, M.; et al. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 2018. [Google Scholar] [CrossRef]
- Maeda, T.; Kobayashi, Y.; Kameoka, H.; Okuma, N.; Takeda, N.; Yamaguchi, K.; Bino, T.; Shigenobu, S.; Kawaguchi, M. Evidence of non-tandemly repeated rDNAs and their intragenomic heterogeneity in Rhizophagus irregularis. Commun. Biol. 2018, 1, 87. [Google Scholar] [CrossRef]
- Angelard, C.; Colard, A.; Niculita-Hirzel, H.; Croll, D.; Sanders, I.R. Segregation in a mycorrhizal fungus alters rice growth and symbiosis-specific gene transcription. Curr. Biol. 2010, 20, 1216–1221. [Google Scholar] [CrossRef]
- Ehinger, M.O.; Croll, D.; Koch, A.M.; Sanders, I.R. Significant genetic and phenotypic changes arising from clonal growth of a single spore of an arbuscular mycorrhizal fungus over multiple generations. New Phytol. 2012, 196, 853–861. [Google Scholar] [CrossRef]
- Angelard, C.; Tanner, C.J.; Fontanillas, P.; Niculita-Hirzel, H.; Masclaux, F.; Sanders, I.R. Rapid genotypic change and plasticity in arbuscular mycorrhizal fungi is caused by a host shift and enhanced by segregation. ISME J. 2014, 8, 284. [Google Scholar] [CrossRef]
- Sakurai, K.; Tomiyama, K.; Kawakami, Y.; Ochiai, N.; Yabe, S.; Nakagawa, T.; Asakawa, Y. Volatile components emitted from the liverwort Marchantia paleacea subsp. diptera. Nat. Prod. Commun. 2016, 11, 263–264. [Google Scholar] [CrossRef]
- Kobae, Y.; Ohtomo, R. An improved method for bright-field imaging of arbuscular mycorrhizal fungi in plant roots. Soil. Sci. Plant Nutr. 2016, 62, 27–30. [Google Scholar] [CrossRef]
- Trouvelot, S.; van Tuinen, D.; Hijri, M.; Gianinazzi-Pearson, V. Visualization of ribosomal DNA loci in spore interphasic nuclei of glomalean fungi by fluorescence in situ hybridization. Mycorrhiza 1999, 8, 203–206. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Young, J.P.W. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 2008, 65, 339–349. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobae, Y.; Ohtomo, R.; Morimoto, S.; Sato, D.; Nakagawa, T.; Oka, N.; Sato, S. Isolation of Native Arbuscular Mycorrhizal Fungi within Young Thalli of the Liverwort Marchantia paleacea. Plants 2019, 8, 142. https://doi.org/10.3390/plants8060142
Kobae Y, Ohtomo R, Morimoto S, Sato D, Nakagawa T, Oka N, Sato S. Isolation of Native Arbuscular Mycorrhizal Fungi within Young Thalli of the Liverwort Marchantia paleacea. Plants. 2019; 8(6):142. https://doi.org/10.3390/plants8060142
Chicago/Turabian StyleKobae, Yoshihiro, Ryo Ohtomo, Sho Morimoto, Daiki Sato, Tomomi Nakagawa, Norikuni Oka, and Shusei Sato. 2019. "Isolation of Native Arbuscular Mycorrhizal Fungi within Young Thalli of the Liverwort Marchantia paleacea" Plants 8, no. 6: 142. https://doi.org/10.3390/plants8060142
APA StyleKobae, Y., Ohtomo, R., Morimoto, S., Sato, D., Nakagawa, T., Oka, N., & Sato, S. (2019). Isolation of Native Arbuscular Mycorrhizal Fungi within Young Thalli of the Liverwort Marchantia paleacea. Plants, 8(6), 142. https://doi.org/10.3390/plants8060142





