NAC Transcription Factor GmNAC035 Exerts a Positive Regulatory Role in Enhancing Salt Stress Tolerance in Plants
Abstract
1. Introduction
2. Results
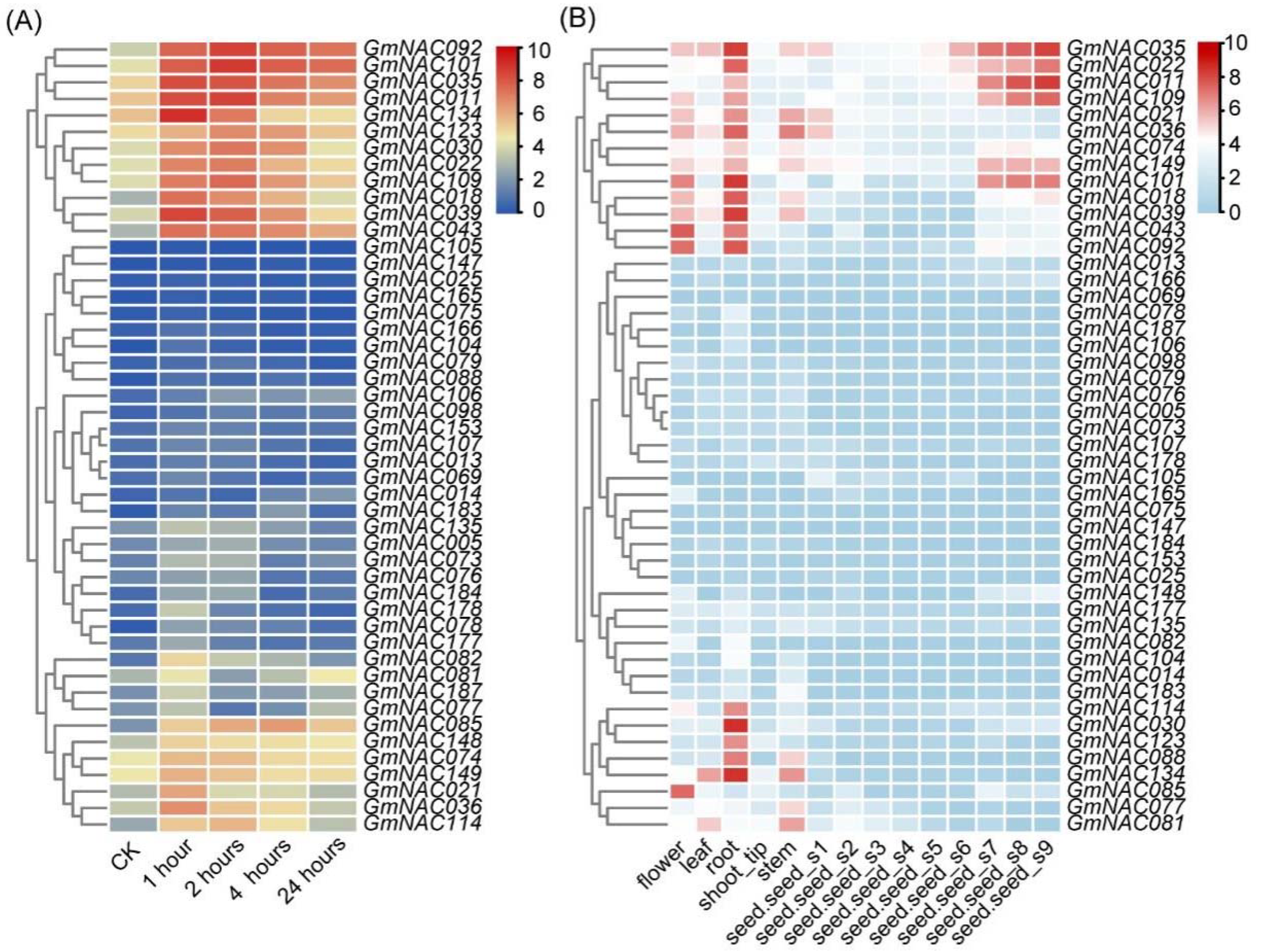
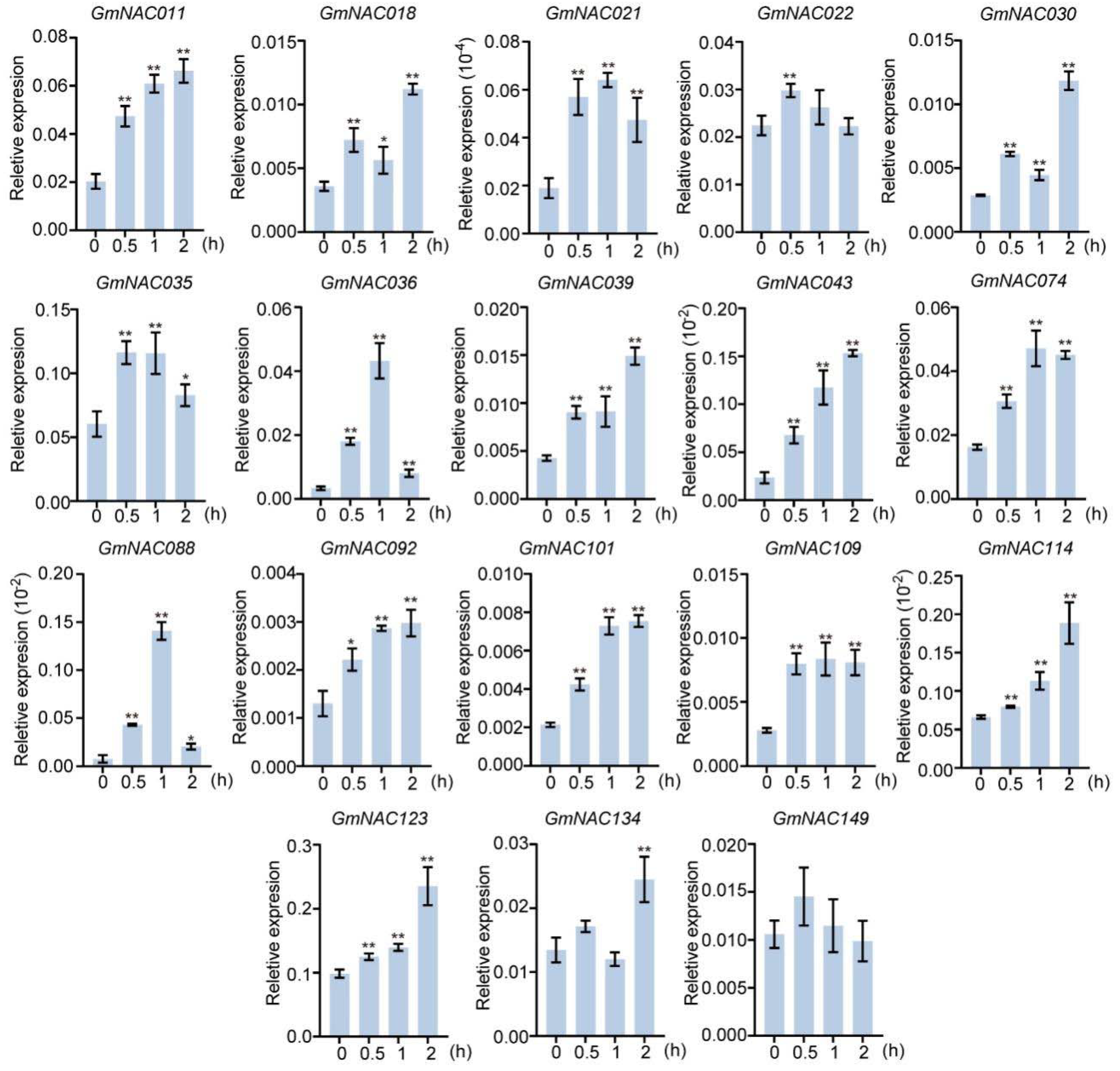
2.1. Expression Patterns of Predicted GmNAC Genes in Roots Under Salt Stress Conditions
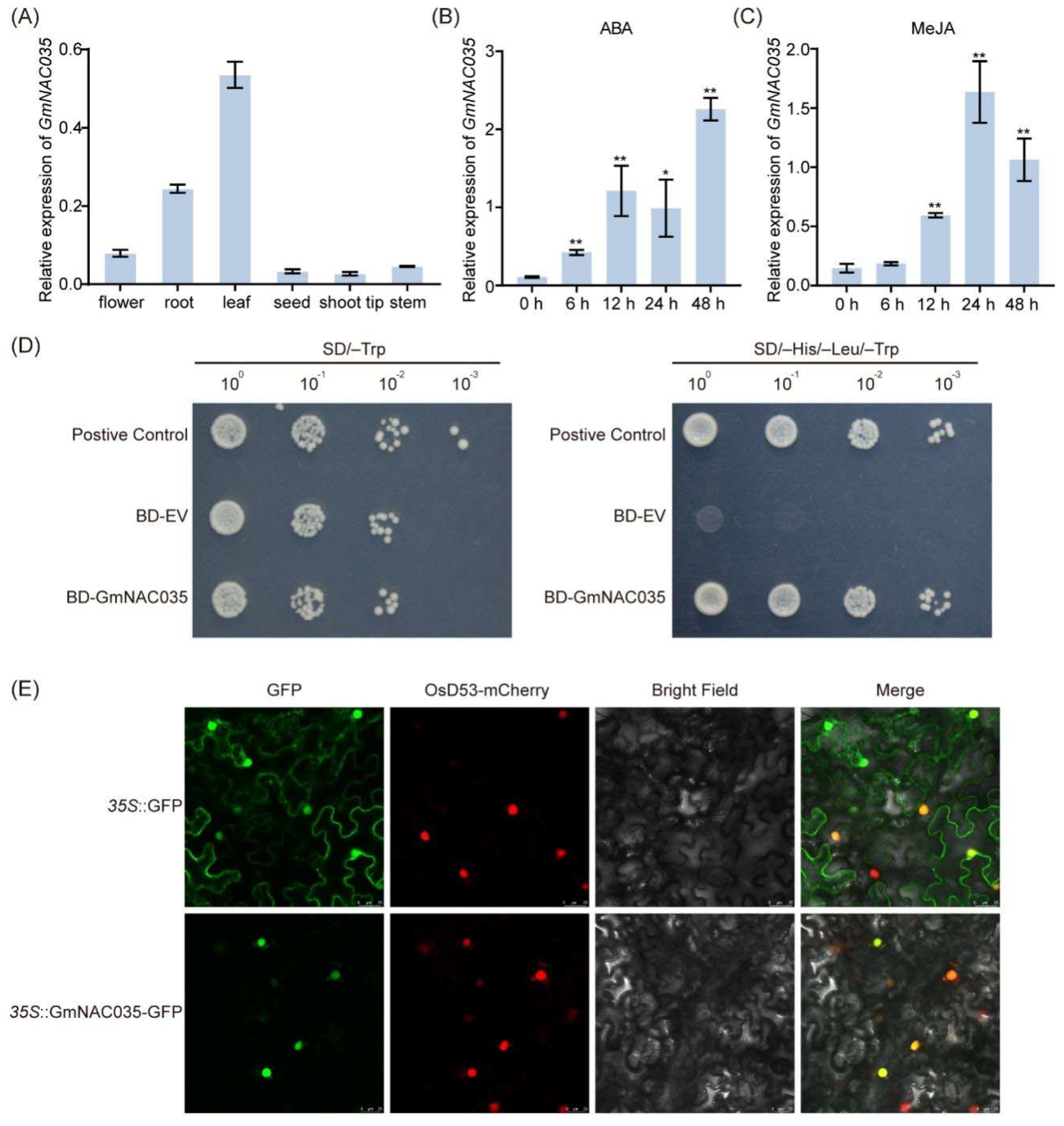
2.2. The Expression of GmNAC035 Is Induced by Hormonal Signals, and It Encodes a Nuclear-Localized Protein with Self-Transactivation Activity
2.3. The Overexpression of GmNAC035 Improves Salt Stresses Tolerance of Arabidopsis
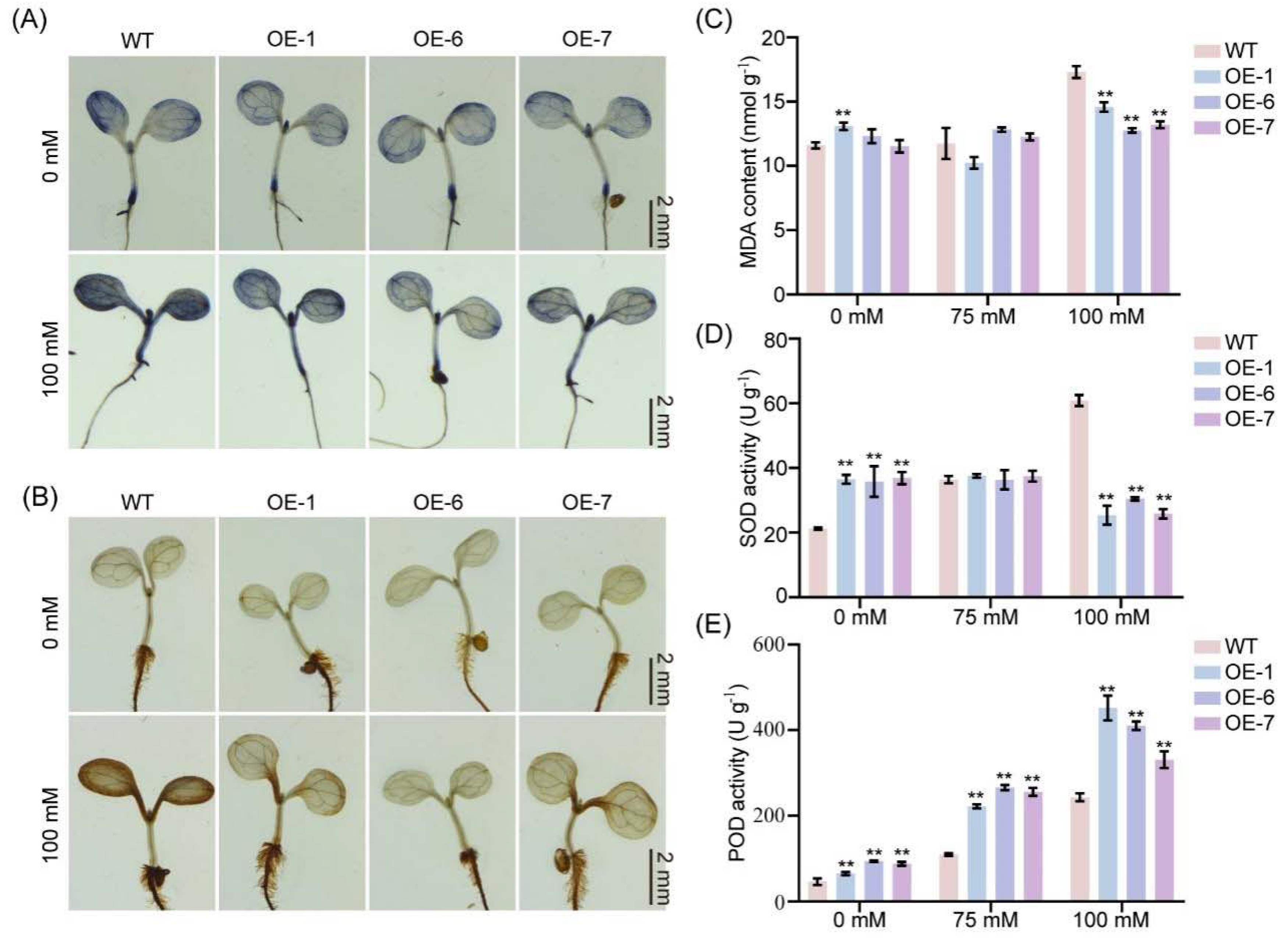
2.4. GmNAC035 Contributed to the Elimination of ROS
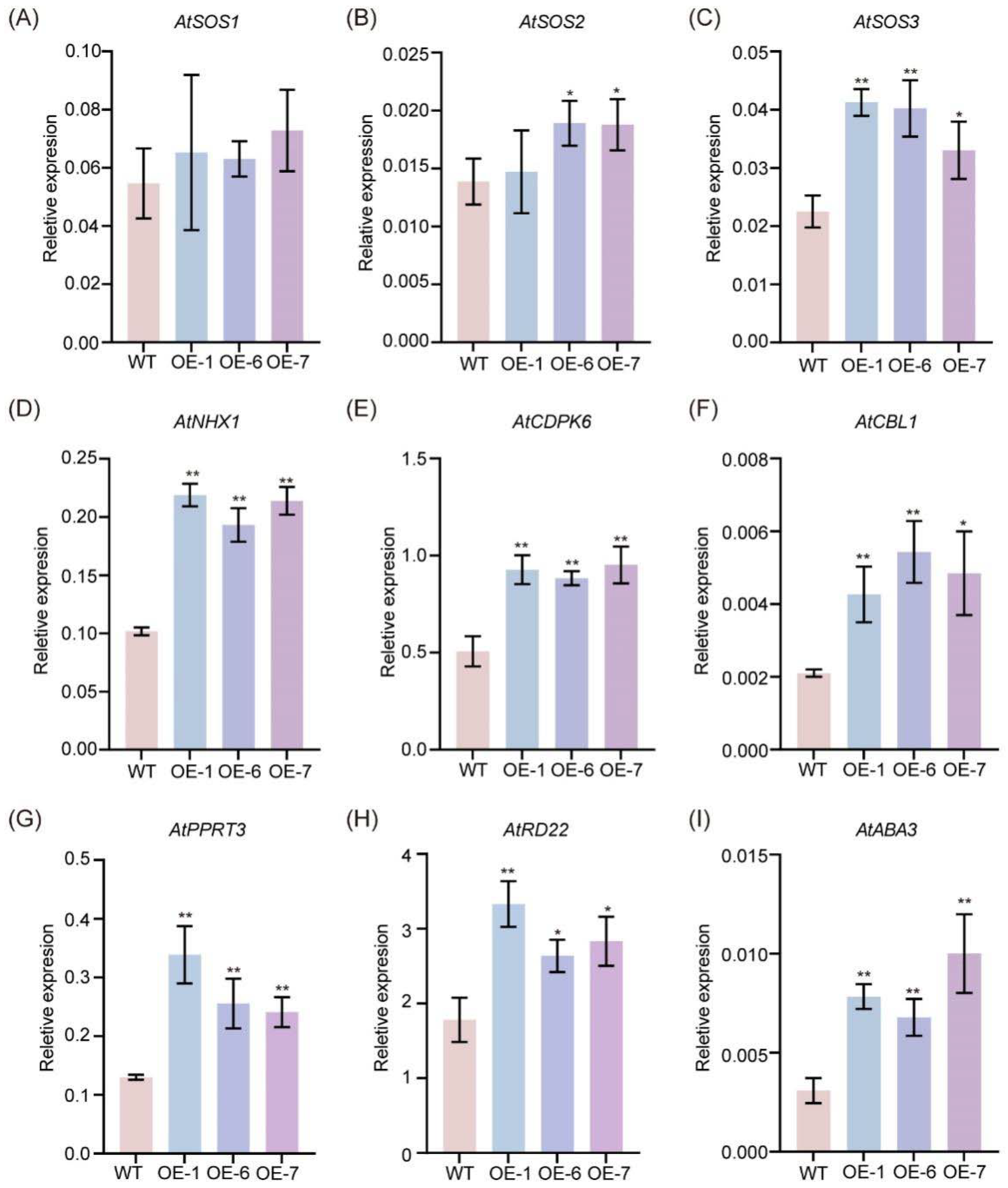
2.5. Overexpression of GmNAC035 Increases Expression of Genes Related to Abiotic Stress Resistance
3. Discussion
4. Materials and Methods
4.1. Identification of NAC Genes in the Soybean Genome and Analysis of Tissue-Specific Expression
4.2. Plant Materials, Growth Conditions, and Stress Treatments
4.3. RNA Extraction and RT-qPCR Analysis
4.4. Subcellular Localization of GmNAC035
4.5. Transcriptional Self-Activation Activity Analysis
4.6. Plant Transformation and Screening of Transgenic Plants
4.7. Salt Tolerance Analysis of GmNAC035 Overexpression Lines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Staniak, M.; Szpunar-Krok, E.; Kocira, A. Responses of soybean to selected abiotic stresses-photoperiod, temperature and water. Agriculture 2023, 13, 146. [Google Scholar] [CrossRef]
- Li, L.; Huang, Z.; Zhang, Y.; Mu, Y.; Li, Y.; Nie, L. Regulation of 2-acetyl-1-pyrroline (2-AP) biosynthesis and grain quality in fragrant rice under salt stress. Field Crop Res. 2025, 322, 109747. [Google Scholar] [CrossRef]
- Shahid, I.; Batool, S.; Hassan, M.; Ismail, H.; Mehnaz, S.; Deeba, F.; Anwar, M.; Zulfiqar, F.; Iqbal, R.; Ali, H.M. A decade of progress in rhizoengineering to exploit plant microbiome for salt stress amelioration. Plant Stress 2024, 11, 100325. [Google Scholar] [CrossRef]
- Singh, V.; Singh, J.; Singh, A. Salinity tolerance in soybeans: Physiological, molecular, and genetic perspectives. In Soybean Improvement: Physiological, Molecular and Genetic Perspectives; Springer: Berlin/Heidelberg, Germany, 2022; pp. 99–108. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Liang, X.; Li, J.; Yang, Y.; Jiang, C.; Guo, Y. Designing salt stress-resilient crops: Current progress and future challenges. J. Integr. Plant Biol. 2024, 66, 303–329. [Google Scholar] [CrossRef]
- Leung, H.S.; Chan, L.Y.; Law, C.H.; Li, M.W.; Lam, H.M. Twenty years of mining salt tolerance genes in soybean. Mol. Breed. 2023, 43, 45. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Zhao, Y.; Sun, Y.; Li, Y. NACs, generalist in plant life. Plant Biotechnol. J. 2023, 21, 2433–2457. [Google Scholar] [CrossRef]
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Li, Y.; Han, S.; Sun, X.; Khan, N.U.; Zhong, Q.; Zhang, Z.; Zhang, H.; Ming, F.; Li, Z.; Li, J. Variations in OsSPL10 confer drought tolerance by directly regulating OsNAC2 expression and ROS production in rice. J. Integr. Plant Biol. 2023, 65, 918–933. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Ding, J.; Zhang, B.; Xi, D.; Ming, F. OsNAC2 positively affects salt-induced cell death and binds to the OsAP37 and OsCOX11 promoters. Plant J. 2018, 94, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 2010, 284, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Chuong, N.N.; Hoa, T.T.K.; Doan, H.; Van, P.H.P.; Trang, L.D.M.; Huyen, P.N.T.; Le, D.T.; Tran, L.-S.P.; Thao, N.P. The drought-mediated soybean GmNAC085 functions as a positive regulator of plant response to salinity. Int. J. Mol. Sci. 2021, 22, 8986. [Google Scholar] [CrossRef]
- Yang, X.; Kim, M.Y.; Ha, J.; Lee, S.H. Overexpression of the soybean NAC gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic Arabidopsis plants. Front. Plant Sci. 2019, 10, 1036. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, X.; Tang, M.; Wang, Y.; Zhang, Q.; Li, H.; Zhou, Y.; Sun, F.; Cui, X. Molecular Characterization and Drought Resistance of GmNAC3 Transcription Factor in Glycine max (L.) Merr. Int. J. Mol. Sci. 2022, 23, 12378. [Google Scholar] [CrossRef]
- Yang, C.; Huang, Y.; Lv, W.; Zhang, Y.; Bhat, J.A.; Kong, J.; Xing, H.; Zhao, J.; Zhao, T. GmNAC8 acts as a positive regulator in soybean drought stress. Plant Sci. 2020, 293, 110442. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Huang, Y.; Lv, P.; Antwi-Boasiako, A.; Begum, N.; Zhao, T.; Zhao, J. NAC transcription factor GmNAC12 improved drought stress tolerance in soybean. Int. J. Mol. Sci. 2022, 23, 12029. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Tang, M.; Li, L.; Chang, J.; Yang, X.; Chang, H.; Zhou, J.; Liu, M.; Wang, Y.; Zhou, Y. Expression patterns and molecular mechanisms regulating drought tolerance of soybean [Glycine max (L.) Merr.] conferred by transcription factor gene GmNAC19. Int. J. Mol. Sci. 2024, 25, 2396. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Melo, B.P.; Fraga, O.T.; Silva, J.C.F.; Ferreira, D.O.; Brustolini, O.J.; Carpinetti, P.A.; Machado, J.P.B.; Reis, P.A.; Fontes, E.P. Revisiting the soybean GmNAC superfamily. Front. Plant Sci. 2018, 9, 1864. [Google Scholar] [CrossRef]
- Amin, N.; Du, Y.; Lu, L.; Khalifa, M.A.; Ahmad, N.; Ahmad, S.; Wang, P. GmNAC3 acts as a key regulator in soybean against drought stress. Curr. Plant Biol. 2024, 38, 100346. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Van Breusegem, F.; Van Montagu, M.; Inzé, D. Engineering stress tolerance in maize. Outlook Agric. 1998, 27, 115–124. [Google Scholar] [CrossRef]
- Bowler, C.; Montagu, M.v.; Inze, D. Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.K.; Shabala, S. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Lu, K.K.; Song, R.F.; Guo, J.X.; Zhang, Y.; Zuo, J.X.; Chen, H.H.; Liao, C.Y.; Hu, X.Y.; Ren, F.; Lu, Y.T. CycC1; 1–WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis. Plant Cell 2023, 35, 2570–2591. [Google Scholar] [CrossRef] [PubMed]
- Apse, M.P.; Aharon, G.S.; Snedden, W.A.; Blumwald, E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, Y.; Zhong, M.; Hussian, J.; Tang, Y.; Liu, S.; Qi, G. Calcium signaling-mediated transcriptional reprogramming during abiotic stress response in plants. Theor. Appl. Genet. 2023, 136, 210. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tian, Y.S.; Peng, R.H.; Xiong, A.S.; Zhu, B.; Jin, X.F.; Gao, F.; Fu, X.Y.; Hou, X.L.; Yao, Q.H. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 2010, 231, 1251–1260. [Google Scholar] [CrossRef]
- Shi, S.; Chen, W.; Sun, W. Comparative proteomic analysis of the Arabidopsis cbl1 mutant in response to salt stress. Proteomics 2011, 11, 4712–4725. [Google Scholar] [CrossRef]
- Barrero, J.M.; Rodríguez, P.L.; Quesada, V.; Piqueras, P.; Ponce, M.R.; Micol, J.L. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 2006, 29, 2000–2008. [Google Scholar] [CrossRef]
- Yu, S.; Lan, X.; Zhou, J.; Gao, K.; Zhong, C.; Xie, J. Dioscorea composita WRKY3 positively regulates salt-stress tolerance in transgenic Arabidopsis thaliana. J. Plant Physiol. 2022, 269, 153592. [Google Scholar] [CrossRef]
- Chong, X.; Liu, Y.; Li, P.; Wang, Y.; Zhou, T.; Chen, H.; Wang, H. Heterologous expression of chrysanthemum TCP transcription factor CmTCP13 enhances salinity tolerance in Arabidopsis. Plants 2024, 13, 2118. [Google Scholar] [CrossRef]
- Liu, Y.; Peng, L.; Gao, X.; Liu, Y.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. AtPPRT3, a novel E3 ubiquitin ligase, plays a positive role in ABA signaling. Plant Cell Rep. 2020, 39, 1467–1478. [Google Scholar] [CrossRef]
- Phang, T.H.; Shao, G.; Lam, H.M. Salt tolerance in soybean. J. Integr. Plant Biol. 2008, 50, 1196–1212. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Lv, R.; Li, J.; Lin, H.; Xi, D. Phytochrome A and B negatively regulate salt stress tolerance of Nicotiana tobacum via ABA–jasmonic acid synergistic cross-talk. Plant Cell Physiol. 2018, 59, 2381–2393. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Lu, M.; Riaz, M.; Tong, K.; Yu, H.; Gao, G.; Niu, Y. Exogenous proline enhances salt acclimation in soybean seedlings: Modifying physicochemical properties and controlling proline metabolism through the ornithine-glutamate dual pathway. Ecotoxicol. Environ. Saf. 2025, 294, 118012. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genomics 2014, 15, 950. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, R.; Zhao, X.; Xiong, W.; Tao, G.; Wu, S. Introduction, popularization and application of a new spring soybean variety “Tianlong No. 1” with high yield and good quality. Soybean Sci. Technol. 2014, 2, 49–52. [Google Scholar]
- Xu, C.; Shan, J.; Liu, T.; Wang, Q.; Ji, Y.; Zhang, Y.; Wang, M.; Xia, N.; Zhao, L. CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant Physiol. 2023, 191, 2427–2446. [Google Scholar] [CrossRef]
- Dong, L.; Hou, Z.; Li, H.; Li, Z.; Fang, C.; Kong, L.; Li, Y.; Du, H.; Li, T.; Wang, L. Agronomical selection on loss-of-function of GIGANTEA simultaneously facilitates soybean salt tolerance and early maturity. J. Integr. Plant Biol. 2022, 64, 1866–1882. [Google Scholar] [CrossRef]
- Wei, Z.; Pan, T.; Zhao, Y.; Su, B.; Ren, Y.; Qiu, L. Rab5a and its GEFs are involved in post-Golgi trafficking of storage proteins in developing soybean cotyledon. J. Exp. Bot. 2020, 71, 808–822. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Lin, Q.; Ren, Y.; Lan, J.; Miao, R.; Feng, M.; Wang, X.; Liu, X.; Zhang, S.; Pan, T. A CYP78As-small grain4-coat protein complex Ⅱ pathway promotes grain size in rice. Plant Cell 2023, 35, 4325–4346. [Google Scholar] [CrossRef]
- Wang, F.; Cheng, Z.; Wang, J.; Zhang, F.; Zhang, B.; Luo, S.; Lei, C.; Pan, T.; Wang, Y.; Zhu, Y. Rice STOMATAL CYTOKINESIS DEFECTIVE2 regulates cell expansion by affecting vesicular trafficking in rice. Plant Physiol. 2022, 189, 567–584. [Google Scholar] [CrossRef]
- Höfgen, R.; Willmitzer, L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988, 16, 9877. [Google Scholar] [CrossRef]
- Pan, T.; Wang, Y.; Jing, R.; Wang, Y.; Wei, Z.; Zhang, B.; Lei, C.; Qi, Y.; Wang, F.; Bao, X. Post-Golgi trafficking of rice storage proteins requires the small GTPase Rab7 activation complex MON1-CCZ1. Plant Physiol. 2021, 187, 2174–2191. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L. D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, K.; Zhou, M.; Gao, Y.; Huang, H.; Liu, C.; Fan, Y.; Fan, Z.; Wang, Y.; Li, X. GmNAC181 promotes symbiotic nodulation and salt tolerance of nodulation by directly regulating GmNINa expression in soybean. New Phytol. 2022, 236, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Gao, S.; Xia, Y.; Hu, M.; Zheng, Y.; Ye, S.; Zhan, Y.; Yan, M.; Liu, H.; Gan, Y. GmGIF5 promotes cell expansion by negatively regulating cell wall modification. Int. J. Mol. Sci. 2025, 26, 492. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Li, W.; Zeng, Y.; Yin, F.; Wei, R.; Mao, X. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in sunflower during salt and drought stress. Sci. Rep. 2021, 11, 19865. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, W.; Ye, S.; Xin, Y.; Jin, H.; Hu, M.; Zheng, Y.; Zhan, Y.; Liu, H.; Gan, Y.; Zheng, Z.; et al. NAC Transcription Factor GmNAC035 Exerts a Positive Regulatory Role in Enhancing Salt Stress Tolerance in Plants. Plants 2025, 14, 1391. https://doi.org/10.3390/plants14091391
Shi W, Ye S, Xin Y, Jin H, Hu M, Zheng Y, Zhan Y, Liu H, Gan Y, Zheng Z, et al. NAC Transcription Factor GmNAC035 Exerts a Positive Regulatory Role in Enhancing Salt Stress Tolerance in Plants. Plants. 2025; 14(9):1391. https://doi.org/10.3390/plants14091391
Chicago/Turabian StyleShi, Wanting, Sixin Ye, Yiting Xin, Hongmiao Jin, Meiling Hu, Yueping Zheng, Yihua Zhan, Hongbo Liu, Yi Gan, Zhifu Zheng, and et al. 2025. "NAC Transcription Factor GmNAC035 Exerts a Positive Regulatory Role in Enhancing Salt Stress Tolerance in Plants" Plants 14, no. 9: 1391. https://doi.org/10.3390/plants14091391
APA StyleShi, W., Ye, S., Xin, Y., Jin, H., Hu, M., Zheng, Y., Zhan, Y., Liu, H., Gan, Y., Zheng, Z., & Pan, T. (2025). NAC Transcription Factor GmNAC035 Exerts a Positive Regulatory Role in Enhancing Salt Stress Tolerance in Plants. Plants, 14(9), 1391. https://doi.org/10.3390/plants14091391







