Identification of the GST Gene Family and Functional Analysis of RcGSTF2 Related to Anthocyanin in Rosa chinensis ‘Old Blush’
Abstract
1. Introduction
2. Results
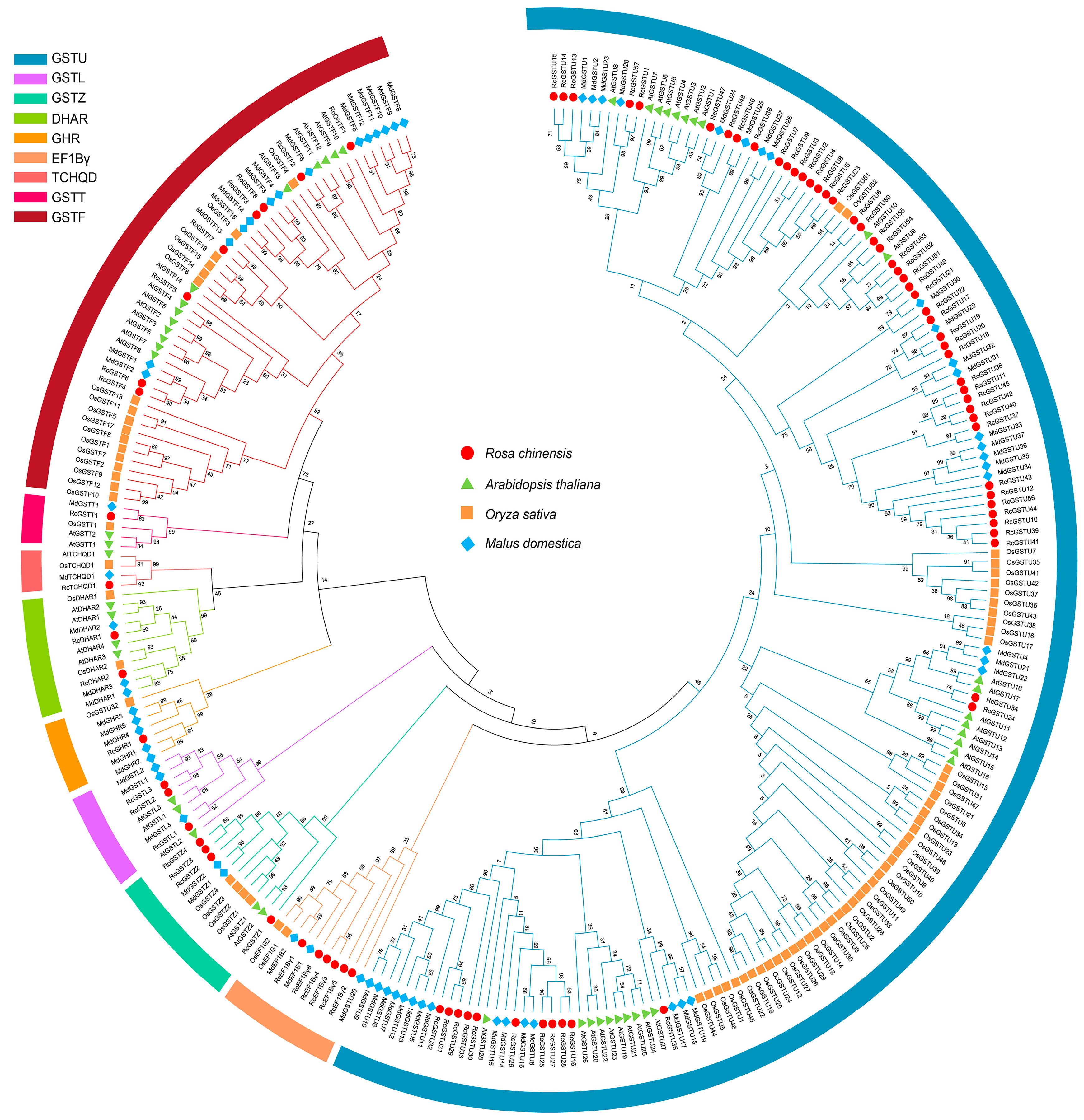
2.1. Identification and Characterization of GST Gene Family Members in Roses
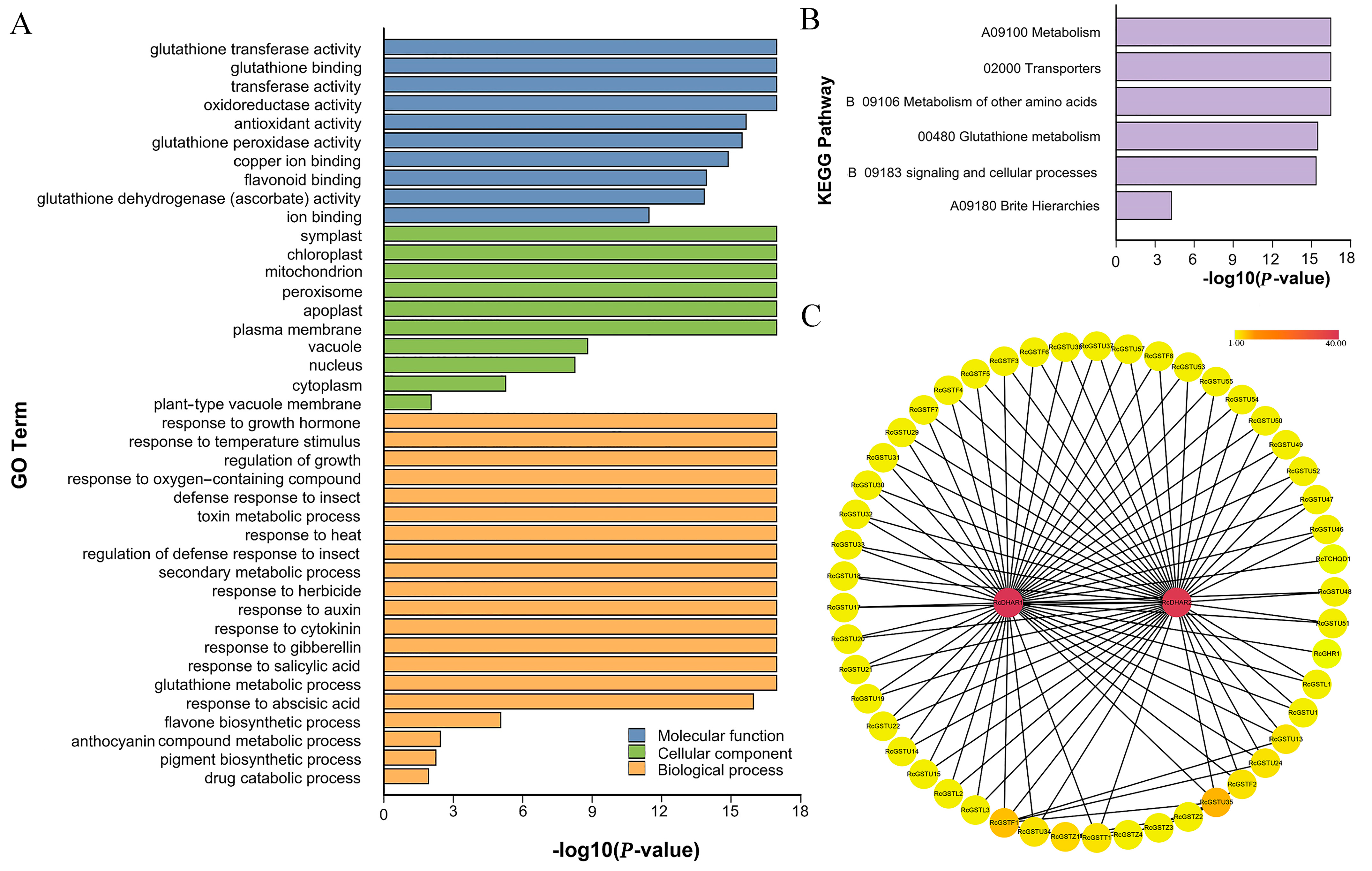
2.2. Functional Annotation of RcGSTs
2.3. Expression Analysis of RcGST Transcripts in Different Tissues
2.4. Characterization of RcGSTF2 Gene Related to Anthocyanin Accumulation in Rose
2.5. Regulatory Role of RcGSTF2 in Anthocyanin Accumulation
3. Discussion
4. Materials and Methods
4.1. Data Source and RcGST Gene Identification
4.2. Phylogenetic Analysis and Subfamily Categorization
4.3. Chromosomal Locations and Collinearity Analysis
4.4. Gene Structure and Protein Motif Analysis
4.5. Functional Analysis
4.6. RNA-Seq Data Analysis
4.7. Plant Material
4.8. Measurement of Anthocyanin Content
4.9. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.10. Molecular Docking
4.11. Genetic Transformation of RcGSTF2 in Rose Plants
4.12. Subcellular Localization Analysis of RcGSTF2
4.13. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Ogata, J.; Kanno, Y.; Itoh, Y.; Tsugawa, H.; Suzuki, M. Plant biochemistry: Anthocyanin biosynthesis in roses. Nature 2005, 435, 757–758. [Google Scholar] [CrossRef] [PubMed]
- Clifford, M.N. Anthocyanins—Nature, occurrence, and dietary burden. J. Sci. Food Agric. 2000, 80, 1063–1072. [Google Scholar] [CrossRef]
- Marrs, K.A. The functions and regulation of glutathione s-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef]
- Aloke, C.; Onisuru, O.O.; Achilonu, I. Glutathione S-transferase: A versatile and dynamic enzyme. Biochem. Biophys. Res. Commun. 2024, 734, 150774. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front Plant Sci. 2018, 9, 01836. [Google Scholar] [CrossRef]
- Gao, H.; Yu, C.; Liu, R.; Li, X.; Huang, H.; Wang, X.; Zhang, C.; Jiang, N.; Li, X.; Cheng, S.; et al. The Glutathione S-transferase PtGSTF1 improves biomass production and salt tolerance through regulating xylem cell proliferation, ion homeostasis and reactive oxygen species scavenging in poplar. Int. J. Mol. Sci. 2022, 23, 11288. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yu, K.; Zhang, S.; Li, Y.; Xu, C.; Qian, H.; Cui, Y.; Guo, Y.; Zhang, X.; Li, R.; et al. Poplar glutathione S-transferase PtrGSTF8 contributes to reactive oxygen species scavenging and salt tolerance. Plant Physiol. Biochem. 2024, 212, 108766. [Google Scholar] [CrossRef]
- Fang, X.; An, Y.; Zheng, J.; Shangguan, L.; Wang, L. Genome-wide identification and comparative analysis of GST gene family in apple (Malus domestica) and their expressions under ALA treatment. 3 Biotech. 2020, 10, 307. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, C.; Huang, X.; Hu, D. Genome-wide analysis of the glutathione S-transferase (GST) genes and functional identification of MdGSTU12 reveals the involvement in the regulation of anthocyanin accumulation in apple. Genes 2021, 12, 1733. [Google Scholar] [CrossRef]
- Duan, X.; Yu, X.; Wang, Y.; Fu, W.; Cao, R.; Yang, L.; Ye, X. Genome-wide identification and expression analysis of glutathione S-transferase gene family to reveal their role in cold stress response in cucumber. Front. Genet. 2022, 13, 1009883. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Wang, A.; Zhang, X.; Wu, Y.; Wang, R.; Cui, H.; Huang, R.L.; Luo, Y.H. Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses. BMC Plant Biol. 2017, 17, 225. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fu, H.; Zhao, J.; Wang, J.; Dong, S.; Yuan, X.; Li, X.; Chen, M. Genome-wide identification and expression profiling of glutathione s-transferase gene family in foxtail millet (Setaria italica L.). Plants 2023, 12, 1138. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhu, Y.; Liu, R.; Zhang, A.; Zhu, M.; Xu, W.; Lin, A.; Lu, K.; Li, J.N. Genome wide identification and comparative analysis of glutathione transferases (GST) family genes in Brassica napus. Sci. Rep. 2019, 9, 9196. [Google Scholar] [CrossRef]
- Csiszár, J.; Horváth, E.; Váry, Z.; Gallé, Á.; Bela, K.; Brunner, S.; Tari, I. Glutathione transferase supergene family in tomato: Salt stress-regulated expression of representative genes from distinct GST classes in plants primed with salicylic acid. Plant Physiol. Biochem. 2014, 78, 15–26. [Google Scholar] [CrossRef]
- Wang, X.; Dong, J.; Hu, Y.; Huang, Q.; Lu, X.; Huang, Y.; Sheng, M.; Cao, L.; Xu, B.; Li, Y.; et al. Identification and characterization of the glutathione S-transferase gene family in blueberry (Vaccinium corymbosum) and their potential roles in anthocyanin intracellular transportation. Plants 2024, 13, 1316. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhao, J.; Lai, B.; Qin, Y.; Wang, H.; Hu, G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016, 35, 831–843. [Google Scholar] [CrossRef]
- Mueller, L.A.; Goodman, C.D.; Silady, R.A.; Walbot, V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000, 123, 1561–1570. [Google Scholar] [CrossRef]
- Sun, Y.; Li, H.; Huang, J.R. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 2012, 5, 387–400. [Google Scholar] [CrossRef]
- Luo, H.; Dai, C.; Li, Y.; Feng, J.; Liu, Z.; Kang, C. Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 2018, 69, 2595–2608. [Google Scholar] [CrossRef]
- Liu, Y.; Qi, Y.; Zhang, A.; Wu, H.; Liu, Z.; Ren, X. Molecular cloning and functional characterization of AcGST1, an anthocyanin-related glutathione S-transferase gene in kiwifruit (Actinidia chinensis). Plant Mol. Biol. 2019, 100, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Chen, M.; He, N.; Chen, X.; Wang, N.; Sun, Q.; Zhang, T.; Xu, H.; Fang, H.; Wang, Y.; et al. MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 2019, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, X.; Duan, R.; Han, C.; Yang, J.; Wang, L.; Wang, S.; Su, Y.; Wang, L.; Dong, Y.; et al. Genomic analysis of the glutathione S-transferase family in pear (Pyrus communis) and functional identification of PcGST57 in anthocyanin accumulation. Int. J. Mol. Sci. 2022, 23, 746. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, W.; Zhu, Y.; Allan, A.C.; Lin-Wang, K.; Xu, C. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 2020, 18, 1284–1295. [Google Scholar] [CrossRef]
- Shao, D.; Li, Y.; Zhu, Q.; Zhang, X.; Liu, F.; Xue, F.; Sun, J. GhGSTF12, a glutathione S-transferase gene, is essential for anthocyanin accumulation in cotton (Gossypium hirsutum L.). Plant Sci. 2021, 305, 110827. [Google Scholar] [CrossRef]
- Cao, Y.; Xu, L.; Xu, H.; Yang, P.; He, G.; Tang, Y.; Qi, X.; Song, M.; Ming, J. LhGST is an anthocyanin-related glutathione S-transferase gene in Asiatic hybrid lilies (Lilium spp.). Plant Cell Rep. 2021, 40, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhou, L.; Zou, H.; Yuan, M.; Wang, Y. PsGSTF3, an Anthocyanin-Related Glutathione S-Transferase Gene, Is Essential for Petal Coloration in Tree Peony. Int. J. Mol. Sci. 2022, 23, 1423. [Google Scholar] [CrossRef]
- Winkler, J.; Mylle, E.; De Meyer, A.; Pavie, B.; Merchie, J.; Grones, P.; Van Damme, D.L. Visualizing protein-protein interactions in plants by rapamycin-dependent delocalization. Plant Cell 2021, 33, 1101–1117. [Google Scholar] [CrossRef]
- Han, Y.; Yu, J.; Zhao, T.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Dissecting the genome-wide evolution and function of R2R3-MYB transcription factor family in Rosa chinensis. Genes 2019, 10, 823. [Google Scholar] [CrossRef]
- Han, Y.; Wan, H.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis. Sci. Rep. 2017, 7, 43382. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Li, H.; Wang, W.; Zheng, H.; Tao, J. Genome-wide identification of glutathione S-transferase and expression analysis in response to anthocyanin transport in the flesh of the new teinturier grape germplasm ‘Zhongshan-HongYu’. Int. J. Mol. Sci. 2022, 23, 7717. [Google Scholar] [CrossRef] [PubMed]
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Millar, A.H.; Singh, K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009, 58, 53–68. [Google Scholar] [CrossRef]
- Dixon, D.P.; Lapthorn, A.; Edwards, R. Plant glutathione transferases. Genome Biol. 2002, 3, 3004. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Estévez, I.H.; Hernández, M.R. Plant glutathione S-transferases: An overview. Plant Gene 2020, 23, 100233. [Google Scholar] [CrossRef]
- Edwards, R.; Dixon, D.P.; Walbot, V. Plant glutathione S-transferases: Enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000, 5, 193–198. [Google Scholar] [CrossRef]
- Dixon, D.P.; Skipsey, M.; Edwards, R. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 2010, 71, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, 607–613. [Google Scholar] [CrossRef]
- Kou, M.; Liu, Y.; Li, Z.; Zhang, Y.; Tang, W.; Yan, H.; Wang, X.; Chen, X.; Su, Z.; Arisha, M.H.; et al. A novel glutathione S-transferase gene from sweetpotato, IbGSTF4, is involved in anthocyanin sequestration. Plant Physiol. Biochem. 2019, 135, 395–403. [Google Scholar] [CrossRef]
- Pérez-Díaz, R.; Madrid-Espinoza, J.; Salinas-Cornejo, J.; González-Villanueva, E.; Ruiz-Lara, S. Differential Roles for VviGST1, VviGST3, and VviGST4 in Proanthocyanidin and Anthocyanin Transport in Vitis vinifera. Front. Plant Sci. 2016, 7, 1166. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, X.; Wu, H.; Yao, Y. The nuclear and cytoplasmic colocalization of MdGST12 regulated by MdWRKY26 and MdHY5 promotes anthocyanin accumulation by forming homodimers and interacting with MdUFGT and MdDFR under light conditions in Malus. Int. J. Biol. Macromol. 2025, 289, 138666. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, 605–612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ubi, B.E.; Honda, C.; Bessho, H.; Kondo, S.; Wada, M.; Kobayashi, S.; Moriguchi, T. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 2006, 170, 571–578. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Židek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Sanner, M.F.; Olson, A.J.; Spehner, J.C. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers 1996, 38, 305–320. [Google Scholar] [CrossRef]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Tuo, D.; Shen, W.; Deng, H.; Zhou, P.; Gao, X. A Nimble Cloning-compatible vector system for high-throughput gene functional analysis in plants. Plant Commun. 2023, 4, 100471. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, J.; Wei, B.; Li, Y.; Fang, X.; Zhong, Y.; Wang, L. Transcription factor MdNAC33 is involved in ala-induced anthocyanin accumulation in apples. Plant Sci. 2024, 339, 111949. [Google Scholar] [CrossRef]
- Liu, Q.; Gao, G.; Shang, C.; Li, T.; Wang, Y.; Li, L.; Feng, X. Screening and verification of proteins that interact with the anthocyanin-related transcription factor PbrMYB114 in ‘Yuluxiang’ pear. PeerJ 2024, 12, e17540. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Wu, H.; Sun, Y.; Zhang, P.; Li, L.; Luo, D.; Wu, Z. Identification of the GST Gene Family and Functional Analysis of RcGSTF2 Related to Anthocyanin in Rosa chinensis ‘Old Blush’. Plants 2025, 14, 932. https://doi.org/10.3390/plants14060932
Zhang T, Wu H, Sun Y, Zhang P, Li L, Luo D, Wu Z. Identification of the GST Gene Family and Functional Analysis of RcGSTF2 Related to Anthocyanin in Rosa chinensis ‘Old Blush’. Plants. 2025; 14(6):932. https://doi.org/10.3390/plants14060932
Chicago/Turabian StyleZhang, Ting, Han Wu, Yujia Sun, Peiheng Zhang, Lixia Li, Dan Luo, and Zhe Wu. 2025. "Identification of the GST Gene Family and Functional Analysis of RcGSTF2 Related to Anthocyanin in Rosa chinensis ‘Old Blush’" Plants 14, no. 6: 932. https://doi.org/10.3390/plants14060932
APA StyleZhang, T., Wu, H., Sun, Y., Zhang, P., Li, L., Luo, D., & Wu, Z. (2025). Identification of the GST Gene Family and Functional Analysis of RcGSTF2 Related to Anthocyanin in Rosa chinensis ‘Old Blush’. Plants, 14(6), 932. https://doi.org/10.3390/plants14060932






