A Comprehensive Review: Genetic Mapping of Genes Associated with Green Leaf Color Variations in Main Vegetable Crops
Abstract
1. Introduction
2. Source of Leaf Color Mutation in Vegetables
2.1. Spontaneous Mutation
2.2. Artificial Mutagenesis
3. Localization of Leaf Color Mutant Genes in Vegetables
3.1. Map-Based Cloning
3.2. BSA-Seq
3.2.1. QTL-Seq
3.2.2. Mut-Map
3.3. Transcriptome Sequencing
4. Molecular Mechanism of Leaf Color Mutation in Vegetables
4.1. Chloroplast Development Pathway
4.1.1. Photomorphogenesis
4.1.2. Hormone Signaling
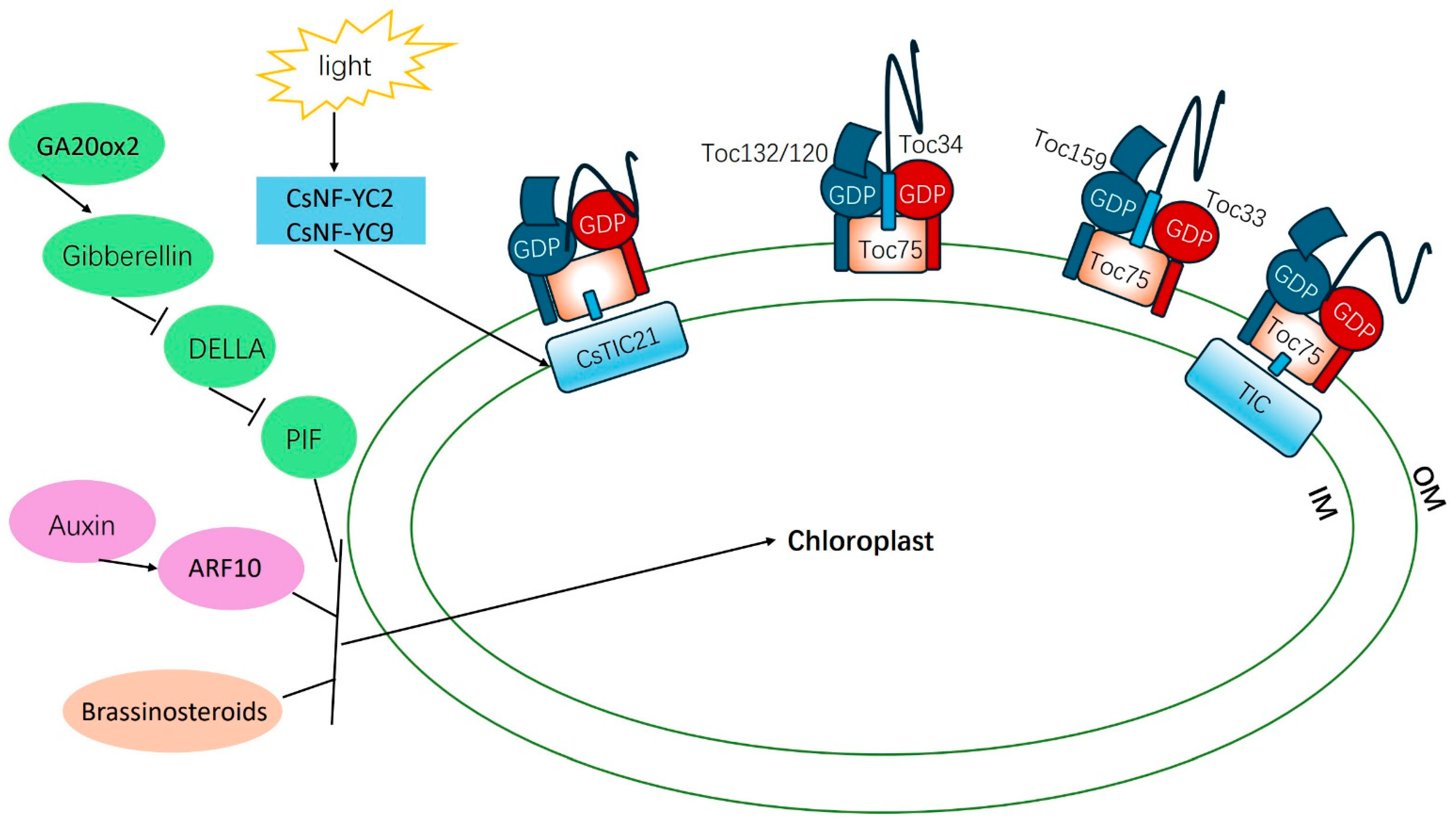
4.1.3. Nuclear Protein Transport and Chloroplast Protein
4.1.4. Chloroplast Division
4.2. Chlorophyll Biosynthesis Pathway
4.3. Genes Related to the Heme–Phytochrome Metabolic Pathway
4.4. Chlorophyll Degradation Pathway
5. Interaction Mechanisms Between Environmental and Genetic Factors in Regulating Leaf Color Development in Vegetables
6. Breeding Applications of Leaf Color Mutants
6.1. Enhancing Photosynthetic Efficiency
6.2. Developing Stress-Resistant Varieties
6.3. Improving Nutritional Quality
6.4. Ornamental and Specialty Varieties
6.5. Marker-Assisted Selection
6.6. Case Studies
7. Prospects
| Reference | Species | Source | Localization | The Localization on Chromosomes | Gene Name | Gene Number |
|---|---|---|---|---|---|---|
| [5] | Cucumis sativus L. | EMS | BSA-Seq Fine mapping | Within approximately 45.3 kb of the region defined by the two markers CAPS777-1 and Indel777-3 on chromosome 3 | CsHD | Csa3G836480 |
| [18] | Cucumis sativus L. | EMS | BSA-Seq Fine mapping | Within the 86.3 kb region between the molecular markers UW804200 and SSR05515 on chromosome 4 | CsVYL | Csa4G637110 |
| [20] | Cucumis sativus L. | EMS | BSA-Seq Fine mapping | Within the 73kb region between the molecular markers NSN and SNP16 on chromosome 3 | v-2 | Csa3G890020 |
| [30] | Cucumis sativus L. | EMS | BSA-Seq | Locus SNP12112564 on chromosome 3 | CsSE59 | CsaV3_3G016210 |
| [6] | Cucumis sativus L. | - | Fine mapping | The 50.4 kb region between the molecular markers v1SSR8 and CAPs15 on chromosome 6 | v-1 | CsaCNGCs |
| [7] | Cucumis sativus L. | Spontaneous | Whole-genome resequencing | The interval defined by the molecular markers SNP11124523-SNP11216771 on chromosome 4 | ygl1 | Csa4M286960 Csa4M287550 Csa4M288070 Csa4M288080 |
| [21] | Cucumis sativus L. | EMS | BSA-Seq Fine mapping | Within the 167kb region defined by the molecular markers Indel22 and SNP81 on chromosome 2 | CsYL2.1 | Csa2G263900 |
| [25] | Cucumis sativus L. | EMS | BSA-Seq Fine mapping | Within the 100 Kb region defined by the molecular markers AInd3-16 and AInd3-24 on chromosome 3 | Cscpftsy | CsaV3_3G009150 |
| [26] | Cucumis sativus L. | Spontaneous | Fine mapping whole-genome sequencing | The interval defined by the molecular markers InDel8 and SSR20583 on chromosome 7 | CsSRP43 | CsGy7G001220 |
| [31] | Cucumis sativus L. | Spontaneous | BSA-Seq Fine mapping RNA-Seq | Within the region defined by the molecular markers UW084839 and SSR15124 on chromosome 3 | v-3 | Csa3G042730 |
| [38] | Cucumis sativus L. | EMS | Map-based cloning BSA-Seq | Within the 63.44Kb interval defined by the molecular markers SBP7349 and SNP0787 of chromosome 7 | CsTIC21 | Csa7G071680 |
| [35] | Cucumis sativus L. | EMS | BSA-Seq RNA-Seq | Chromosome 6 molecular marker SNP-18277305 | Cscs | Csa6G405290 |
| [36] | Brassica campestris L. | EMS | MutMap | Chromosome A03 | Brnym1 | BraA03g050600.3C |
| [12] | Brassica rapa L. | EMS | BSR-Seq Fine mapping whole-genome resequencing | Within the 64.25 kb region defined by the molecular markers INDEL-N14 and INDEL-I8 of chromosome A10 | Brpem1 Brpem2 | BraA10g021490.3C BraA10g021490.3C |
| [19] | Brassica napus | EMS | BSA-Seq Map-based cloning RNA-Seq | Within the 70kb interval defined by the molecular markers yvl-O10 and InDel-O6 of chromosome A03 | BnaA03.CHLH | BnaA03g04440D |
| [28] | Brassica napus | EMS | Fine mapping | BnCDE1 | BnaC08g34840D | |
| [22] | Solanum lycopersicum | EMS | whole-genome sequencing MutMap | Locus NC_015441.3 of chromosome 4 | FtsH-like protein precursor | LOC100037730 |
| [13] | Glycine max | Spontaneous | BSA-Seq Map-based cloning | YL1 is located at chromosome 11 (within the 270kb range defined by the markers BARCSOYSSR_11_0156 and BARCSOYSSR_11_0175); YL2 is located in chromosome 1 (within the 270kb interval defined by BARCSOYSSR_11_0164 and BARCSOYSSR_11_0169). | YL1 YL2 | glyma11g04660 glyma01g40650 |
| [14] | Capsicum annuum L. | 60Co γ-ray | Fine mapping | The 214 kb region defined by the molecular markers SNP5791587 to SNP6011215 on chromosome 9 | CaLY1 | Capana09g000166 |
| [48] | Brassica campestris L. | Spontaneous | BSA-Seq Fine mapping | Within the 81.01kb region defined by the molecular markers SSRWN27 and SSRWN30 of chromosome A03 | Brnye1 | Bra019346 |
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Awan, M.A.; Konzak, C.F.; Rutger, J.N.; Nilan, R.A. Mutagenic Effects of Sodium Azide in Rice1. Crop Sci. 1980, 20, 663–668. [Google Scholar] [CrossRef]
- Cheng, M.; Meng, F.; Mo, F.; Chen, X.; Zhang, H.; Wang, A. Insights into the molecular basis of a yellow leaf color mutant (ym) in tomato (Solanum lycopersicum). Sci. Hortic. 2022, 293, 110743. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, J.H.; Jang, Y.H.; Yu, J.; Bae, S.; Kim, M.S.; Cho, Y.G.; Jung, Y.J.; Kang, K.K. Transcriptome and Metabolite Profiling of Tomato SGR-Knockout Null Lines Using the CRISPR/Cas9 System. Int. J. Mol. Sci. 2022, 24, 109. [Google Scholar] [CrossRef] [PubMed]
- Jáquez-Gutiérrez, M.; Atarés, A.; Pineda, B.; Angarita, P.; Ribelles, C.; García-Sogo, B.; Sánchez-López, J.; Capel, C.; Yuste-Lisbona, F.J.; Lozano, R.; et al. Phenotypic and genetic characterization of tomato mutants provides new insights into leaf development and its relationship to agronomic traits. BMC Plant Biol. 2019, 19, 141. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, H.; Xie, C.; Wang, J.; Zhang, J.; Wang, H.; Weng, Y.; Chen, P.; Li, Y. A mutation in CsHD encoding a histidine and aspartic acid domain-containing protein leads to yellow young leaf-1 (yyl-1) in cucumber (Cucumis sativus L.). Plant Sci. Int. J. Exp. Plant Biol. 2020, 293, 110407. [Google Scholar] [CrossRef]
- Miao, H.; Zhang, S.; Wang, M.; Wang, Y.; Weng, Y.; Gu, X. Fine Mapping of Virescent Leaf Gene v-1 in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2016, 17, 1602. [Google Scholar] [CrossRef]
- Ding, Y.; Yang, W.; Su, C.; Ma, H.; Pan, Y.; Zhang, X.; Li, J. Tandem 13-Lipoxygenase Genes in a Cluster Confers Yellow-Green Leaf in Cucumber. Int. J. Mol. Sci. 2019, 20, 3102. [Google Scholar] [CrossRef]
- Gebremeskel, H.; Umer, M.J.; Hongju, Z.; Li, B.; Shengjie, Z.; Yuan, P.; Xuqiang, L.; Nan, H.; Wenge, L. Genetic mapping and molecular characterization of the delayed green gene dg in watermelon (Citrullus lanatus). Front. Plant Sci. 2023, 14, 1152644. [Google Scholar] [CrossRef]
- Li, B.; Zhang, J.; Tian, P.; Gao, X.; Song, X.; Pan, X.; Wu, Y. Cytological, Physiological, and Transcriptomic Analyses of the Leaf Color Mutant Yellow Leaf 20 (yl20) in Eggplant (Solanum melongena L.). Plants 2024, 13, 855. [Google Scholar] [CrossRef]
- Zhao, X.; Hu, K.; Yan, M.; Yi, B.; Wen, J.; Ma, C.; Shen, J.; Fu, T.; Tu, J. Disruption of carotene biosynthesis leads to abnormal plastids and variegated leaves in Brassica napus. Mol. Genet. Genom. 2020, 295, 981–999. [Google Scholar] [CrossRef]
- Zhang, K.; Mu, Y.; Li, W.; Shan, X.; Wang, N.; Feng, H. Identification of two recessive etiolation genes (py1, py2) in pakchoi (Brassica rapa L. ssp. chinensis). BMC Plant Biol. 2020, 20, 68. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Huang, S.; Zhang, M.; Zhang, Y.; Feng, H. Mapping of a Pale Green Mutant Gene and Its Functional Verification by Allelic Mutations in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 2021, 12, 699308. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, Y.; Nie, Z.; Gai, J.; Bhat, J.A.; Kong, J.; Zhao, T. Double mutation of two homologous genes YL1 and YL2 results in a leaf yellowing phenotype in soybean [Glycine max (L.) Merr]. Plant Mol. Biol. 2020, 103, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, Z.; Chen, W.; Liang, C.; Li, X.; Liu, Z.; Cui, Q.; Ma, Y.; Zou, X. Fine-mapping and transcriptome analysis of the photosensitive leaf -yellowing gene CaLY1 in pepper (Capsicum annuum L.). Hortic. Plant J. 2023, 9, 122–132. [Google Scholar] [CrossRef]
- Arisha, M.H.; Shah, S.N.; Gong, Z.H.; Jing, H.; Li, C.; Zhang, H.X. Ethyl methane sulfonate induced mutations in M2 generation and physiological variations in M1 generation of peppers (Capsicum annuum L.). Front. Plant Sci. 2015, 6, 399. [Google Scholar] [CrossRef]
- Yan, J.; Liu, B.; Cao, Z.; Chen, L.; Liang, Z.; Wang, M.; Liu, W.; Lin, Y.; Jiang, B. Cytological, genetic and transcriptomic characterization of a cucumber albino mutant. Front. Plant Sci. 2022, 13, 1047090. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, P.; Fan, X.; Yang, H. Characterization and RNA-Seq Analysis of Yellow-Green Leaf Mutants in Tomato. Agronomy 2024, 14, 828. [Google Scholar] [CrossRef]
- Song, M.; Wei, Q.; Wang, J.; Fu, W.; Qin, X.; Lu, X.; Cheng, F.; Yang, K.; Zhang, L.; Yu, X.; et al. Fine Mapping of CsVYL, Conferring Virescent Leaf Through the Regulation of Chloroplast Development in Cucumber. Front. Plant Sci. 2018, 9, 432. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, L.; Safdar, L.B.; Xie, M.; Cheng, X.; Liu, Y.; Xiang, Y.; Tong, C.; Tu, J.; Huang, J.; et al. Characterization and Fine Mapping of a Yellow-Virescent Gene Regulating Chlorophyll Biosynthesis and Early Stage Chloroplast Development in Brassica napus. G3 Genes Genomes Genet. 2020, 10, 3201–3211. [Google Scholar] [CrossRef]
- Zhang, K.; Li, Y.; Zhu, W.; Wei, Y.; Njogu, M.K.; Lou, Q.; Li, J.; Chen, J. Fine Mapping and Transcriptome Analysis of Virescent Leaf Gene v-2 in Cucumber (Cucumis sativus L.). Front. Plant Sci. 2020, 11, 570817. [Google Scholar] [CrossRef]
- Xiong, L.; Du, H.; Zhang, K.; Lv, D.; He, H.; Pan, J.; Cai, R.; Wang, G. A Mutation in CsYL2.1 Encoding a Plastid Isoform of Triose Phosphate Isomerase Leads to Yellow Leaf 2.1 (yl2.1) in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2020, 22, 322. [Google Scholar] [CrossRef] [PubMed]
- Dechkrong, P.; Srima, S.; Sukkhaeng, S.; Utkhao, W.; Thanomchat, P.; de Jong, H.; Tongyoo, P. Mutation mapping of a variegated EMS tomato reveals an FtsH-like protein precursor potentially causing patches of four phenotype classes in the leaves with distinctive internal morphology. BMC Plant Biol. 2024, 24, 265. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liu, Z.; Li, D.; Yao, R.; Meng, Q.; Feng, H. Screening of Chinese cabbage mutants produced by 60Co γ-ray mutagenesis of isolated microspore cultures. Plant Breed 2014, 133, 480–488. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, X.; Yang, S. A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res. 2009, 19, 1205–1216. [Google Scholar] [CrossRef]
- Zha, G.; Yin, J.; Cheng, F.; Song, M.; Zhang, M.; Obel, H.O.; Wang, Y.; Chen, J.; Lou, Q. Fine mapping of CscpFtsY, a gene conferring the yellow leaf phenotype in cucumber (Cucumis sativus L.). BMC Plant Biol. 2022, 22, 570. [Google Scholar] [CrossRef]
- Zhang, T.; Dong, X.; Yuan, X.; Hong, Y.; Zhang, L.; Zhang, X.; Chen, S. Identification and characterization of CsSRP43, a major gene controlling leaf yellowing in cucumber. Hortic. Res. 2022, 9, uhac212. [Google Scholar] [CrossRef]
- Yang, M.; Wan, S.; Chen, J.; Chen, W.; Wang, Y.; Li, W.; Wang, M.; Guan, R. Mutation to a cytochrome P450 -like gene alters the leaf color by affecting the heme and chlorophyll biosynthesis pathways in Brassica napus. Plant J. Cell Mol. Biol. 2023, 116, 432–445. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef]
- Pan, J.; Song, J.; Sharif, R.; Xu, X.; Li, S.; Chen, X. A mutation in the promoter of the yellow stripe-like transporter gene in cucumber results in a yellow cotyledon phenotype. J. Integr. Agric. 2024, 23, 849–862. [Google Scholar] [CrossRef]
- Zhou, Y.; Liao, L.; Liu, L.; Xiao, L.; Zhou, Z.; Zhou, Y.; Hu, Z.; Liu, S. CsSE59 Encoding Invertase/Pectin Methyl Esterase Inhibitor Is a Candidate Gene Conferring the Virescent True Leaf Phenotype in Cucumber. Horticulturae 2023, 9, 951. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Xing, G.; Li, M.; Li, S. Integrating physiology, genetics, and transcriptome to decipher a new thermo-sensitive and light-sensitive virescent leaf gene mutant in cucumber. Front. Plant Sci. 2022, 13, 972620. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Gao, Y.; Zhou, Q.; Ping, X.; Li, J.; Liu, L.; Yin, J. Genetic mapping and physiological analysis of chlorophyll-deficient mutant in Brassica napus L. BMC Plant Biol. 2022, 22, 244. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.K.; Rubiales, D.; Wang, Y.; Fang, P.; Sun, T.; Liu, N.; Xu, P. Omics resources and omics-enabled approaches for achieving high productivity and improved quality in pea (Pisum sativum L.). TAG. Theoretical and applied genetics. Theor. Und Angew. Genet. 2021, 134, 755–776. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef]
- Cao, W.; Du, Y.; Wang, C.; Xu, L.; Wu, T. Cscs encoding chorismate synthase is a candidate gene for leaf variegation mutation in cucumber. Breed. Sci. 2018, 68, 571–581. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, Y.; Huang, S.; Liu, Z.; Li, C.; Feng, H. Defect in Brnym1, a magnesium-dechelatase protein, causes a stay-green phenotype in an EMS-mutagenized Chinese cabbage (Brassica campestris L. ssp. pekinensis) line. Hortic. Res. 2020, 7, 8. [Google Scholar] [CrossRef]
- Huo, J.; Zhang, N.; Gong, Y.; Bao, Y.; Li, Y.; Zhang, L.; Nie, S. Effects of different light intensity on leaf color changes in a Chinese cabbage yellow cotyledon mutant. Front. Plant Sci. 2024, 15, 1371451. [Google Scholar] [CrossRef]
- Ke, X.; Shen, J.; Niu, Y.; Zhao, H.; Guo, Y.; Sun, P.; Yang, T.; Jiang, Y.; Zhao, B.; Wang, Z.; et al. Cucumber NUCLEAR FACTOR-YC2/-YC9 target translocon component CsTIC21 in chloroplast photomorphogenesis. Plant Physiol. 2023, 192, 2822–2837. [Google Scholar] [CrossRef]
- Xiao, J.; Li, H.; Zhang, J.; Chen, R.; Zhang, Y.; Ouyang, B.; Wang, T.; Ye, Z. Dissection of GA 20-oxidase members affecting tomato morphology by RNAi-mediated silencing. Plant Growth Regul. 2006, 50, 179–189. [Google Scholar] [CrossRef]
- Chory, J.; Nagpal, P.; Peto, C.A. Phenotypic and Genetic Analysis of det2, a New Mutant That Affects Light-Regulated Seedling Development in Arabidopsis. Plant Cell 1991, 3, 445–459. [Google Scholar] [CrossRef]
- Bauer, J.; Chen, K.; Hiltbunner, A.; Wehrli, E.; Eugster, M.; Schnell, D.; Kessler, F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 2000, 403, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gao, W.; Liao, X.; Xiong, C.; Yu, G.; Yang, Q.; Yang, C.; Ye, Z. The tomato WV gene encoding a thioredoxin protein is essential for chloroplast development at low temperature and high light intensity. BMC Plant Biol. 2019, 19, 265. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.; Kabeya, Y.; Suzuki, K.; Mori, T.; Ichikawa, T.; Matsui, M.; Nakanishi, H.; Miyagishima, S.Y. The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 2009, 21, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Fu, Y.; Hu, G.; Si, H.; Zhu, L.; Wu, C.; Sun, Z. Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.). Planta 2007, 226, 785–795. [Google Scholar] [CrossRef]
- Gao, M.; Hu, L.; Li, Y.; Weng, Y. The chlorophyll-deficient golden leaf mutation in cucumber is due to a single nucleotide substitution in CsChlI for magnesium chelatase I subunit. TAG. Theoretical and applied genetics. Theor. Und Angew. Genet. 2016, 129, 1961–1973. [Google Scholar] [CrossRef]
- Terry, M.J.; Kendrick, R.E. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 1999, 119, 143–152. [Google Scholar] [CrossRef]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef]
- Wang, N.; Liu, Z.; Zhang, Y.; Li, C.; Feng, H. Identification and fine mapping of a stay-green gene (Brnye1) in pakchoi (Brassica campestris L. ssp. chinensis). TAG. Theoretical and applied genetics. Theor. Und Angew. Genet. 2018, 131, 673–684. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Wang, X.; Wang, Y.; Yang, X.; Li, X.; Chen, J.; Feng, S. A Comprehensive Review: Genetic Mapping of Genes Associated with Green Leaf Color Variations in Main Vegetable Crops. Plants 2025, 14, 1609. https://doi.org/10.3390/plants14111609
Wang M, Wang X, Wang Y, Yang X, Li X, Chen J, Feng S. A Comprehensive Review: Genetic Mapping of Genes Associated with Green Leaf Color Variations in Main Vegetable Crops. Plants. 2025; 14(11):1609. https://doi.org/10.3390/plants14111609
Chicago/Turabian StyleWang, Menghao, Xinyin Wang, Yue Wang, Xiyue Yang, Xiabing Li, Junrong Chen, and Shengjun Feng. 2025. "A Comprehensive Review: Genetic Mapping of Genes Associated with Green Leaf Color Variations in Main Vegetable Crops" Plants 14, no. 11: 1609. https://doi.org/10.3390/plants14111609
APA StyleWang, M., Wang, X., Wang, Y., Yang, X., Li, X., Chen, J., & Feng, S. (2025). A Comprehensive Review: Genetic Mapping of Genes Associated with Green Leaf Color Variations in Main Vegetable Crops. Plants, 14(11), 1609. https://doi.org/10.3390/plants14111609






