1. Introduction
Rice is a staple food in China, which exhibits a long history of cultivation and widespread popularity among its population. Currently, over 6% of the global land area is increasingly affected by salt accumulation, which affects plant growth, crop production, and ecosystem balance [
1]. Soil salinization is a major abiotic stressor affecting agriculture worldwide. Sodium ions affect the structure and characteristics of soil, indirectly causing damage to plant roots. Under high-salt stress, plant cell walls undergo changes in composition and structure, and enhance polysaccharide synthesis and the expression of related modification genes to maintain cell integrity and improve adaptability to salt stress [
2].
Numerous studies have demonstrated the crucial role of NAC transcription factors in plant responses to abiotic stress. The Arabidopsis transcription factor
NAC016 inhibits the transcription of AREB1 through the tricarboxylic acid regulatory loop involving NAP, thereby enhancing the plant’s response to drought stress [
3]. The rice NAC transcription factor
ONAC066 is drought-responsive, and various stress treatments can significantly induce its expression of
ONAC066. Overexpression of
ONAC066 enhances drought tolerance in transgenic rice [
4].
VvNAC17 is a novel grape stress-responsive transcription factor that enhances the sensitivity of transgenic Arabidopsis to abscisic acid (ABA) and improves its salinity, freezing, and drought resistance [
5]. The NAC domain transcription factor
GmNAC06 enhances soybean salt tolerance. Combining overexpression technology with the CRISPR-Cas9 system revealed that
GmNAC06 facilitated the accumulation of proline (Pro) and betaine, thereby alleviating or preventing the negative effects of reactive oxygen species (ROS) [
6]. The wheat NAC transcription factor
TANAC29 improved the salt and drought tolerance of transgenic
Arabidopsis thaliana, and overexpressed plants
TANAC29 improved their tolerance to high salt and dehydration, and showed high sensitivity to ABA [
7]. Yeast one-hybrid experiments demonstrated that the protein encoded by
PsnNAC090 can bind to ABA-responsive elements (ABRE), and its ectopic expression enhances the salt tolerance and permeability of transgenic tobacco [
8]. IbNIEL-mediated degradation of
IbNAC087 regulates jasmonic acid-dependent salt and sweet potato drought resistance [
9].
A total of 158 NAC transcription factors have been identified in rice; however, only a limited number have been investigated or demonstrated to have functions [
10]. Therefore, investigating additional NAC transcription factors is necessary. Effective strategies for molecular breeding crops involve analyzing the tissue-specific and stress-induced expression characteristics of
OsNAC113 combined with CRISPR/Cas9 technology, site-directed mutagenesis of target genes, construction of a rice NAC transcription factor mutant library, and screening NAC transcription factors with notable research potential for further investigation. This study aimed to analyze the rice transcription factor
OsNAC113, elucidate its tissue-specific characteristics and stress response regulatory mechanisms, and provide comprehensive foundational data and a theoretical framework for further exploration of
OsNAC113. The acquisition of rice stress-responsive
OsNAC113 functional deletion mutants provides important experimental material for subsequent research. Through a joint omics analysis of the transcriptome and metabolome,
OsNAC113 was systematically analyzed at the gene regulation and cellular metabolic levels.
3. Discussion
CRISPR/Cas gene editing technology has recently enabled the rapid and accurate production of new germplasms within a short time period, which is of considerable importance for investigating rice genome function, expression regulation, and genetic improvement of germplasm resources. Hui et al. (2022) created a new allele of betaine aldehyde dehydrogenase 2 (
OsBADH2) and cultivated a three-line hybrid rice variety with enhanced grain aroma [
11], while Zhu et al. developed an efficient multi-gene vector system, Trans Gene Stacking II (TGSII), using CRISPR/Cas technology, which enables targeted synthesis of anthocyanins in the endosperm. Eight key genes related to anthocyanin synthesis have been transferred to rice receptors, creating a new germplasm, “Purple Crystal Rice,” rich in anthocyanins [
12]. Zhao et al. knocked out the rice histone demethylase
JMJ710 and observed that the JMJ710 gene, which regulates its synthesis, was a negative regulator of drought stress response genes, leading to strong drought resistance in rice mutants [
13]. Zu et al. isolated the inhibitor sop10 of the pseudouridine synthase gene
OsPUS1-1 in indica rice, and observed that mutants without sop10 function exhibited reduced levels of ROS, increased tolerance to low temperature stress, and maintained yield [
14].
NAC TFs are involved in the regulation of plant responses to various stressors, including salinity, cold, and drought [
15,
16]. The effects of salt stress on plants are mainly manifested in three aspects: (1) high salt content results in osmotic stress, thereby preventing plants from absorbing water; (2) plants absorb toxic Na
+ and Cl
− ions, disrupting the body’s ion balance; and (3) salt stress can inhibit plant growth and promote aging by disrupting redox homeostasis [
17]. The results of this study indicated that the
osnac113 mutant tends to be induced by salt stress.
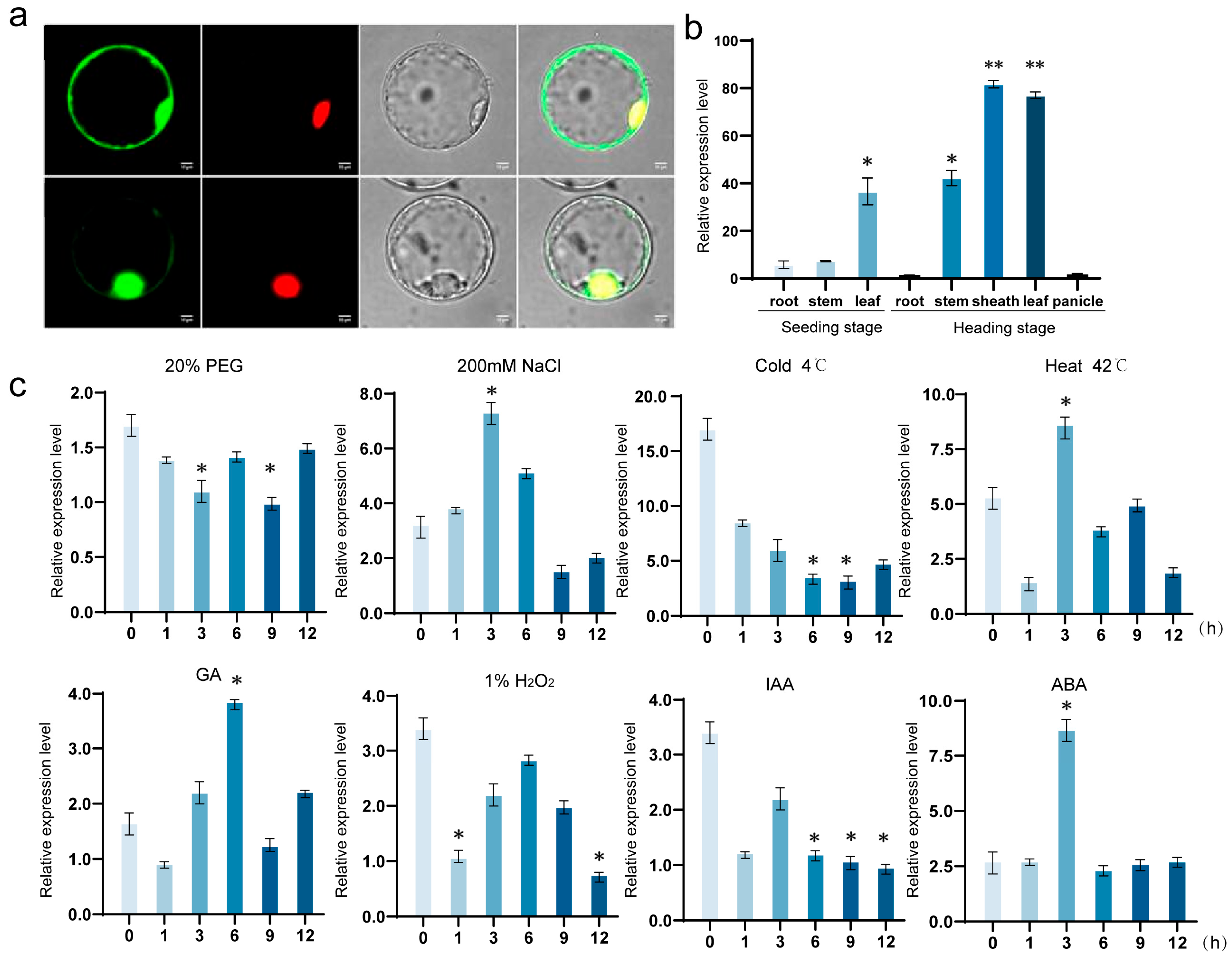
OsNAC113 was specifically induced under salt stress conditions, but showed no significant response to IAA and hydrogen peroxide stress. Under normal conditions,
OsNAC113 is mainly expressed in the leaves and leaf sheaths, with a relative expression level as high as 40–50 times that in the roots. In addition, we observed that the expression level of OsNAC113 increased within 1–6 h of salt treatment and reached its peak at 3 h after treatment. These results indicate that
OsNAC113 expression is tissue-specific and affects salt stress tolerance in rice.
In rice,
OsNCED3 promotes ABA synthesis and enhances abiotic stress tolerance [
18]. In higher plants, NCEDs are key enzymes that control ABA biosynthesis and belong to a differentially expressed gene family. The transcription factors DREBs/CBFs specifically interact with the dehydration response element/C-repeat (DRE/CRT) cis-acting element (nucleoid motif: G/ACCGAC). Overexpression of OsDREB1A in Arabidopsis can induce overexpression of the DREB1A gene, thereby enhancing Arabidopsis tolerance to various stresses, such as high salt [
19]. This indicates that OsDREB1A and DREB1A have similar functions; OsWRKY28 enhances salt tolerance in rice by directly binding to the OsDREB1B promoter and increasing its transcriptional activity, while negatively regulating ABA-mediated rice seedling establishment [
20]. RD20 is a stress-induced Arabidopsis gene belonging to the caleosin family. Compared to the wild type, rd20 knockout plants exhibited higher transpiration rates, which were related to increased stomatal opening and reduced drought tolerance. These results support the role of RD20 in drought tolerance under water-deficient conditions via stomatal control [
21]. The expression levels of
OsNCED3,
OsDREB1A,
OsDREB1B, and
OsRD20 significantly increased in the mutant, and the expression of these genes was closely related to the response of plants to abiotic stress. Therefore, the mutants may enhance their tolerance to salt stress by regulating the pathways in which they participate.
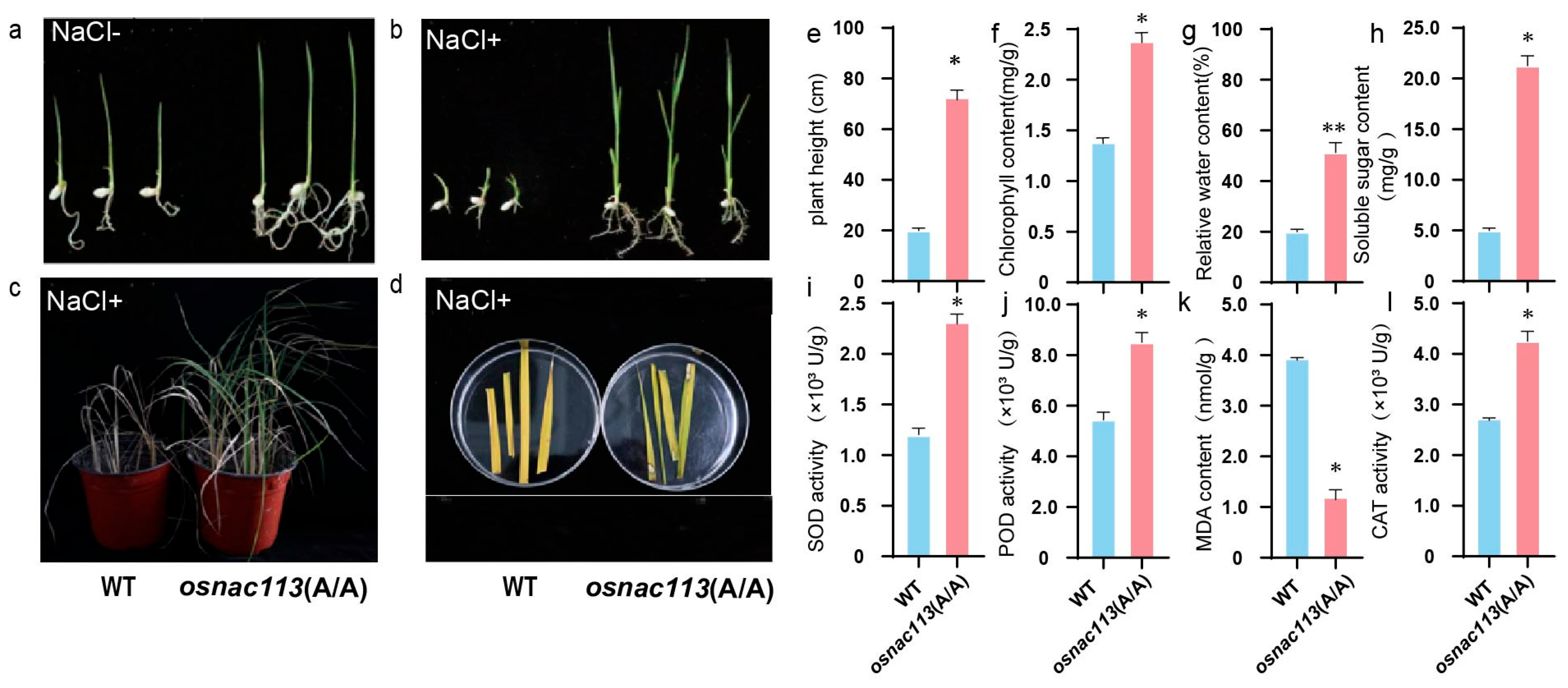
Excessive production of ROS is a fundamental plant response to salinity. Under salt stress, plant photosynthesis is inhibited, and the ROS produced during photosynthesis directly or indirectly affect the decomposition of chlorophyll and electron transfer. Plant chloroplasts and mitochondria are the prominent organelles producing ROS in cells and mediating the induction of pressure tolerance. The regulation of the antioxidant system includes both enzymatic and non-enzymatic components [
22]. In salt-tolerant rice lines, OsP5CS expression is upregulated, enhancing the accumulation of the key osmotic protective molecule pro. Plants can protect their cells from ROS damage and improve salt tolerance in rice [
23].
OsGPX2,
OsGPX4, and
OsGPX5 in the rice glutathione peroxidase (GPXs) gene family are associated with the AsA GSH pathway and contribute to improved salt tolerance in rice [
24]. Overexpression of the glutathione S-transferase gene
OsGST4 can clear ROS and increase salt tolerance [
25]. In this study, the
OsNAC113 mutant showed enhanced tolerance to salt stress. Under salt stress, the
OsNAC113 mutant showed an increase in chlorophyll content; enhanced activities of SOD, POD, and CAT; a decrease in MDA content; and an increase in soluble sugar content. The
OsNAC113 mutant exhibited increased resistance to salt stress, indicating that the
OsNAC113 transcription factor may regulate genes related to peroxidases in the antioxidant system.
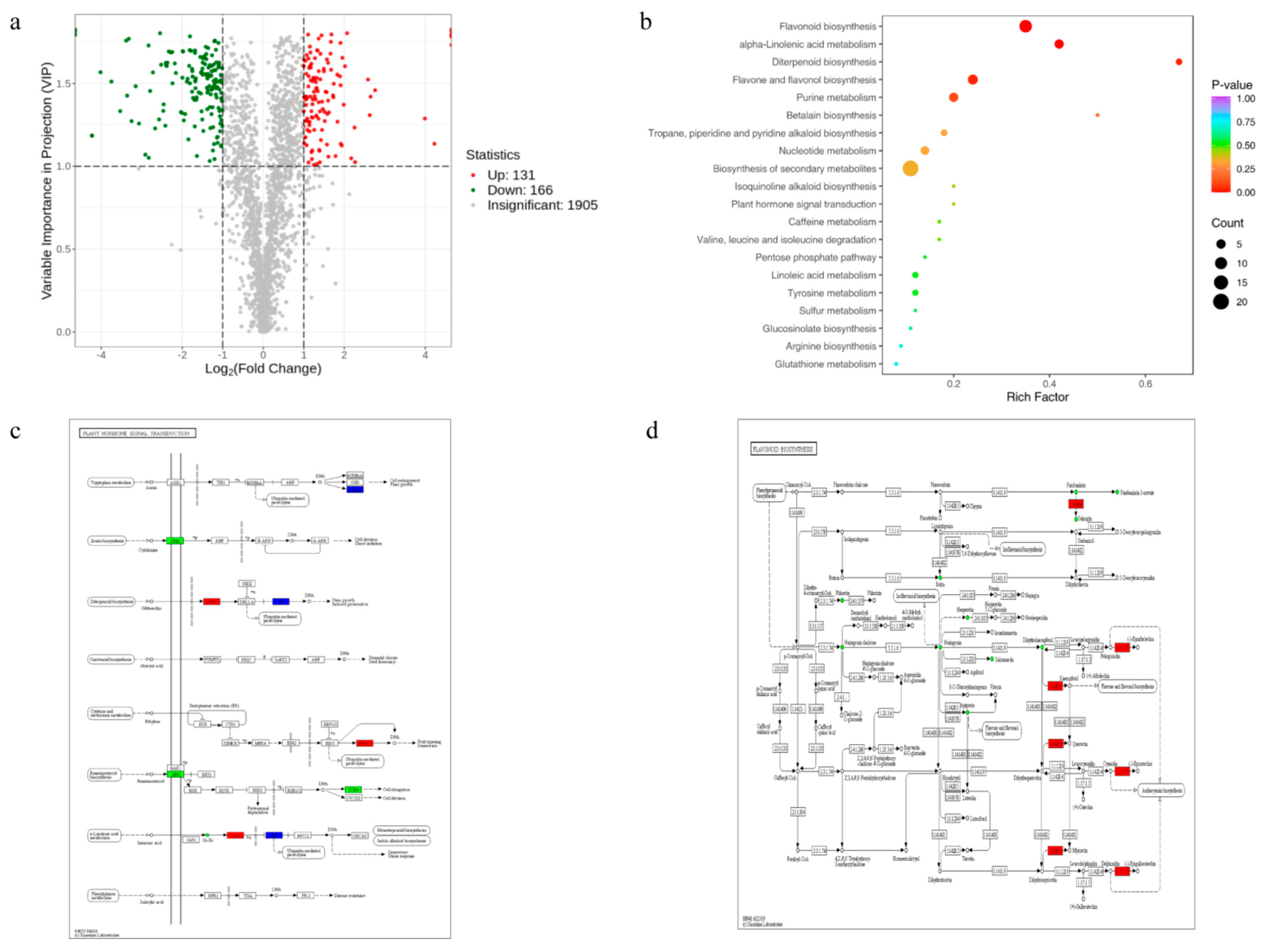
Plant hormones secreted by complex mechanisms are key factors affecting salt tolerance in rice and play a crucial role in plant adaptation to various stresses. Plant hormones are endogenous compounds that act as plant growth regulators at their synthesis sites, or when transported to other sites under different environmental and stress conditions. Abscisic acid is a stress-responsive plant hormone that facilitates the adaptation of plants to adverse environmental conditions by inhibiting seed germination and seedling growth. ABA plays an important regulatory role in salt stress processes. Jasmonic acid is involved in regulating plant life processes in response to various forms of abiotic stress, potentially contributing to enhanced resistance [
26]. The expression of the gene OsEIN2 in the ethylene signaling pathway stimulates salt tolerance in rice [
27]. In this study,
OsNAC113 responded to adversity stress, and ABA and GA
3 treatments induced the expression of
OsNAC113. During tryptophan metabolism, SAUR expression is upregulated, resulting in increased tryptophan production. Higher plants contain a substance that is structurally similar to IAA, and its precursor is tryptophan. The gene JAZ expression is upregulated, and JA is a lipid-derived natural plant hormone that can induce stomatal closure, which is beneficial for maintaining water in plants. The gene GID1 is upregulated during molecular biosynthesis. Gibberellin (GA) is a hormone that plays a crucial role in plant growth and regulates it under abiotic stress. In this study, we constructed a knockout mutant of
OsNAC113 and observed that
OsNAC113 was involved in regulating multiple regulatory pathways at the molecular metabolic level, thereby enhancing plant salt tolerance.
ABC transporters in plants are involved in various functions such as secondary metabolite transport [
28], heavy metal detoxification, antibiotic transport [
29], and plant hormone transport [
30]. Therefore, the functional annotation and analysis of differential metabolites observed in
OsNAC113 gene knockout plants indicate that changes in metabolites representing ABC transporters also affect the production of ABC transporters, thereby regulating the growth and development of rice. MAPKs are relatively conserved eukaryotic protein kinases, and they are involved in regulating signal transduction under stress and adversity [
31]. Salt stress leads to the activation of MPK6, which can phosphorylate the C-terminus of SOS1, thereby enhancing salt tolerance in Arabidopsis [
32]. MAPKs are involved in the response and adaptation of plants to stress, thereby regulating the homeostasis of intracellular ROS [
33]. Notable differences were observed in the gene expression levels of the ABC transporter and MAPK signaling pathways in the
OsNAC113 mutant, revealing the molecular mechanisms underlying the response regulation of
OsNAC113 under high-salt stress conditions.
Flavonoids play a crucial role in enabling plants to resist biotic or abiotic stresses and are beneficial for plants in resisting external stress [
34]. The Arabidopsis R2R3-MYB transcription factor PFG3 enhances plant tolerance to drought and osmotic stress by regulating flavonoid biosynthesis. In rice, the MYB-bHLH-WD40 protein complex synergistically regulates anthocyanin biosynthesis-related genes [
35]. The important structural genes OsPAL, OsC4H, and Os4CL, which are involved in anthocyanin biosynthesis, mainly play a role in lignin synthesis and stress resistance [
36]. In this study, flavonol synthase [EC: 1.14.20.6] was upregulated. Flavonoids are the most widely distributed class of compounds in plants. Flavonols protect plants from various environmental stimuli. The expression of anthocyanin reductase [EC: 1.3.1.77] was also upregulated. Anthocyanins play protective roles in plants. They can absorb ultraviolet radiation and act as antioxidants, preventing damage to photosynthetic pigments and other cellular structures and reducing the degree of damage. In summary,
OsNAC113 may affect the above pathways and respond to salt stress.
4. Materials and Methods
4.1. Plant Materials and Cultivation Conditions
All rice materials used in this experiment, including wild-type Japanese clear rice (Nipponbare, Oryza sativa L. Japan) and osnac113 mutant plants, were grown in a light incubator at 28 °C for 16 h light (3000 Lux)/8 h dark conditions. The substrate used for tissue culture is MS medium. Wild-type and mutant plants with consistent growth are subjected to salt stress treatment simultaneously, and the above plant material was subjected to stress treatment with 200 mM saline solution one month after the growth of T1 seeds. The material growth environment is in a 3000 lux light incubator, and the photoperiod is 16 h light and 8 h darkness. After performing normal germination and growing in the soil for 4 weeks, different abiotic stresses were applied to rice to detect the response of OsNAC113 to different stimuli. Like low temperature stress of 4 °C, high temperature stress of 42 °C, simulated drought stress of PEG6000 (20%, w/v), high salt NaCl (200 mM), 1% H2O2 stress, 100 mM IAA hormone induction, 100 mM ABA hormone induction, and 100 mM GA3 induction. Plant RNA was extracted and transformed into cDNA for RT-qPCR analysis at 0, 1, 3, 6, 9, and 12 h. Each sample had three biological replicates.
4.2. Real-Time Quantitative PCR System and Conditions
The reaction solution was prepared using TB Green Premix Ex Taq II (R820A) from BioNTech (Mainz, Germany). The reaction mixture (20 μL) comprised the following components: total RNA reverse transcribed into cDNA (kit: PrimeScript™ RT reagent Kit, RR037Q), 1 μL cDNA, 0.5 μL upstream primer, 0.5 μL downstream primer, 10 μL 2× SYBR Green Mix, and 8 μL H2O. A qPCR reaction was performed on the My IQ quantitative PCR instrument of the BIO-RAD company (Hercules, CA, USA). The quantitative data were analyzed using BIO-RAD software (V2.0), and the relative expression values of genes were calculated using the ΔΔCt analysis method. The qPCR reaction is as follows: 94 °C pre-denaturation, 30 s, 1 cycle; denaturation at 94 °C, 5 s, 40 cycles; annealing at 58 °C. 15 s, 40 cycles; and extend at 72 °C for 40 cycles. The verification of the specificity of amplification was performed by melting curves (65 °C to 95 °C, increments of 0.5 °C). The ΔΔCt analysis method is used to calculate the relative expression values of genes for data analysis and calculation.
4.3. Subcellular Localization Requires the Construction of a Target Protein Expression Vector
In order to confirm the location of OsNAC113, the pBWA (V) HS-OsNAC113-Glosgfp10509 vector was constructed and incorporated into rice protoplasts. The plasmid encodes OsNAC113 fused to green fluorescent protein (GFP), and the empty GFP vector NLS::eGFP served as a control. Seedlings of rice were grown in the dark at 25 °C in a growth room for 1–2 weeks before isolating protoplasts. Healthy fresh rice seedlings were cut into fine segments and digested with an enzyme solution (1.5% cellulase R10, 0.75% Macerozyme R10, 0.6 M mannitol, 10 mM MES pH 5.7, 10 Mm CaCl2, and 0.1% BSA). After 3 h digestion with gentle shaking (20–30 rpm), protoplasts were isolated by filtration through 40 µm nylon meshes into round-bottom tubes. Pellets were collected at 80 g for 3 min and washed with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 Mm KCl, and 2 mM MES pH 5.7), and the protoplasts were kept on ice for 30 min. They were then resuspended in MMG solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES pH 5.7). Protoplast transformation was carried out in PEG solution (40% (w/v) PEG 4000, 2 M mannitol, and 0.1 CaCl2. Transformation mixtures (10 µg of target plasmid or, additionally, add 10 ug of marker plasmid for cotransformation with 200 µL protoplasts in 250 µL of PEG solution) were agitated gently. After 30 min at room temperature in the dark, protoplasts were harvested and washed with 800 ul of W5 solution. They were then centrifuged and resuspended in 2 mL W5 solution and cultured in the dark at room temperature, generally for 16–24 h. Then, the plasmid was viewed under a Laser scanning confocal microscope. The promoter used for the carrier is the 35S promoter. The eukaryotic resistance of the carrier is hygromycin, and the prokaryotic resistance is kanamycin. The microscope used was a Nikon C2-ER Laser Confocal Microscope, and the manufacturer is Nikon. After 24 h of cultivation, the proteins were separated and purified, and protein gel electrophoresis was performed to observe whether the green fluorescent protein was fused with osnac113. Finally, the positional distribution of green fluorescent protein was detected using a fluorescence microscope.
4.4. Construction of Rice sgRNA
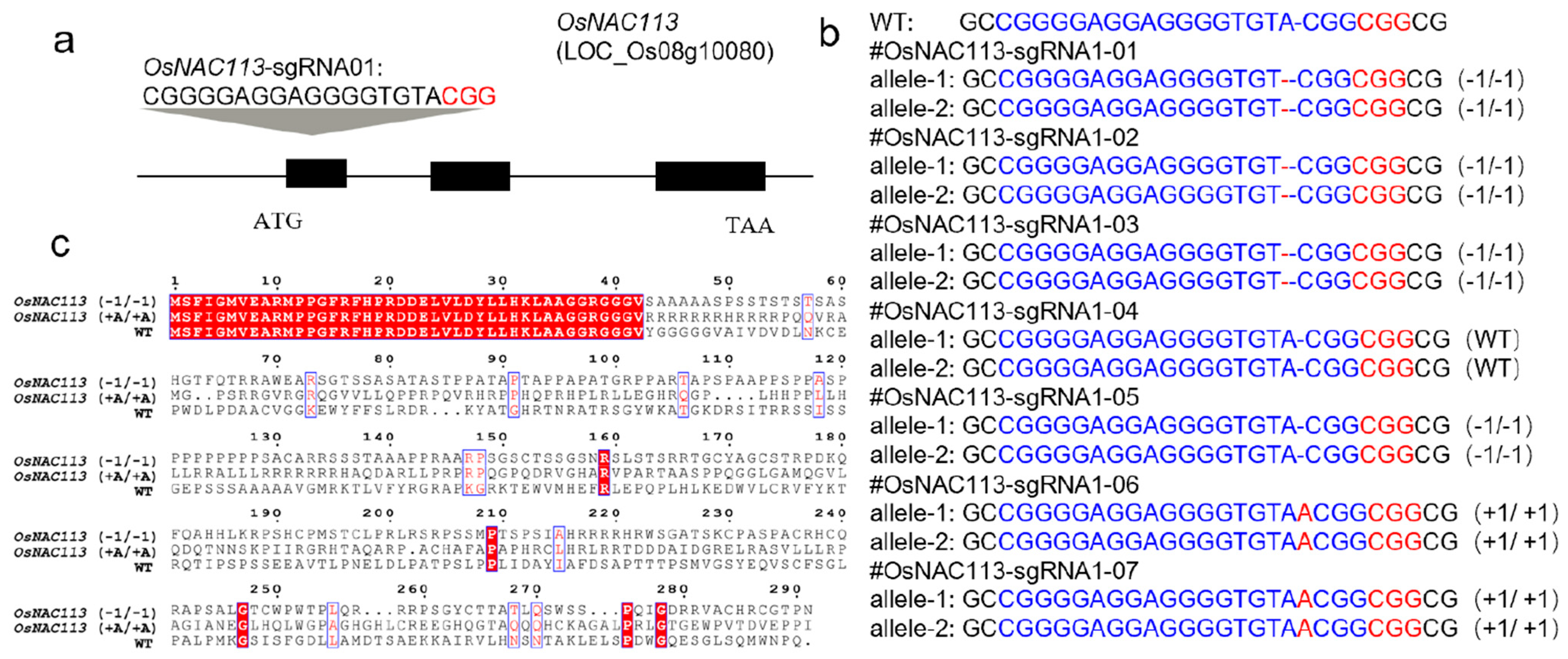
Based on the recognition and cleavage rules of CRISPR/Cas9 for target sites, the design of sgRNA in the
OsNAC113 (
https://planttfdb.gao-lab.org/tf.php?sp=Osj&did=LOC_Os08g10080.1, accessed on 11 June 2023) gene coding region was conducted using the database
http://CRISPR.hzau.edu.cn/CRISPR (accessed on 11 June 2023). The upstream and downstream primers of each target site were diluted to a concentration of 100 μmol. Additionally, 10 μL of each was collected, denatured for 5 min at 98 °C, cooled naturally, and the annealing product was diluted 20 times for later use. BsaI was used to perform single-enzyme digestion on pZHY988 [
37], and the restriction endonuclease instructions of New England Biolabs (NEB) were used to establish the digestion system and digestion conditions. Subsequently, NEB’s T4 ligase was used for ligation. The knockout site of the gene was selected at the position of the second exon (
Figure 2a), and the corresponding sgRNA was designed at this position.
4.5. Rice Knockout Vector Transformation
Agrobacterium-mediated transformation was used to infect and transform rice callus tissues. After infection, the callus was co-cultured in the dark for three days and then washed five times with sterile water. The callus was washed once with sterile water containing 500 mg/L carbenicillin (Carb) and transferred to a screened solid culture containing 500 mg/L Carb + 50 mg/L Hyg medium for 2 months. In the later stage, regenerated seedlings were induced to root in regeneration medium [
38].
4.6. Identification of OsNAC113 Knockout Mutant in Rice and Physiological Measurements
After 3 months of transformation, DNA was extracted from regenerated plants, and PCR product sequencing was used to identify the regenerated seedlings. After obtaining regenerated seedlings, individual plant genomes were extracted, and Sanger sequencing (Genewiz, Suzhou, China) was used to identify the genotype after gene knockout. The sequencing results were compared with the wild type to obtain the knockout genotype. Sequencing results were analyzed to identify the different knockout types of OsNAC113. Relative water content (RWC) testing: take a portion of the leaves and weigh the weight as fresh weight (FW); soak the leaf culture dish in water overnight; wipe off the moisture on the surface of the leaves and call the mass swelling (TW); 45 °C, 24 h, fully dry the leaves, and then weigh the mass as dry weight (DW). The formula for calculating relative moisture content is RWC = (FW-DW)/(TW-DW).
The method for determining chlorophyll content is as follows: Take 1 g of fresh leaves and grind them into powder with liquid nitrogen. Transfer all the powder to a 15 mL centrifuge tube, place it on ice, add 5 mL of 80% acetone for 30 min, and shake thoroughly three times during this period. At 4 °C, 10,000 rpm, centrifuge for 10 min, take the supernatant, and dilute it to 5 mL with 80% acetone. Use 80% acetone as a blank control to determine the OD values of the samples (OD645 and OD663). The formula for calculating chlorophyll content is C (a + b) (mg/L) = OD663 × 8.02 + OD645 × 20.2.
The method for the determination of superoxide dismutase (SOD) activity is as follows: Take leaves from the same location of wild-type and mutant plants. If the treatment conditions require simultaneous processing, take samples and grind 5 mL of pre-cooled 0.05 mol/L PBS (pH 7.8) on ice until homogenized, then transfer to a centrifuge tube; centrifuge at 4 °C, 10,000 rpm for 20 min, then collect the supernatant and store at 4 °C. Prepare reaction mixture: 40.5 mL 14.5 mmol/L Met solution + 1.5 mL 3 mmol/L EDTA-Na2 solution + 1.5 mL 2.25 mmol/L NBT solution + 1.5 mL 60 µ mol/L riboflavin solution. Take 3 mL of the reaction mixture and 40 µL of the enzyme solution separately and place them in test tubes as sample tubes. Simultaneously perform two control tubes: control—3 mL reaction mixture + 40 µL PBS to measure the maximum photoreduction value, and initial control—set to zero when measuring 3 mL reaction mixture + 40 µL PBS (tin foil paper in the dark). The sample tube and control tube undergo a 4000 Lx light reaction at 25 °C for 20 min. The initial control was zeroed, and the absorbance value OD560 was measured sequentially for the samples. The calculation formula is SOD activity (U/g FW) = [(ACK-AE) × Vt]/(half ACK × W × VS), where ACK is the absorbance value of the control tube, AE is the absorbance value of the sample tube, Vt is the total volume of enzyme solution, W is the fresh weight of the sample, and vs. is the amount of enzyme solution used during measurement.
The method for the determination of peroxidase (POD) activity is as follows: weigh an appropriate amount of leaves from the same part of different strains (subjected to stress treatment for the same time) and place them in a mortar. Add 5 mL of pre-cooled 0.05 mol/L PBS (pH 7.8), grind them thoroughly on ice until homogenized, and then pour them into a 10 mL centrifuge tube; centrifuge at 4 °C and 12,000 rpm for 20 min, then collect the supernatant and store at 4 °C; preparation of reaction mixture: 50 mL of 0.1 mol/L PBS (pH 6.0) + 28 µL of guaiacol (2-methoxyphenol) are heated and stirred, cooled, and then mixed with 19 µL of 30% H2O2 and stored at 4 °C; take 3 mL of reaction mixture and 40 µL of enzyme solution separately and place them in test tubes as sample tubes. An amount of 3 mL reaction mixture + 40 µL PBS control zeroing; measure the change in OD470 at 40 s for 3 mL reaction mixture + 40 µL PBS. The formula for calculating POD activity is (U/g FW) = (△ OD470 × Vt)/(W × VS × 0.01 × T), where △ OD470 is the absorbance change within 40 s, Vt is the total volume of enzyme solution used, W is the fresh weight of the sample, VS is the amount of enzyme solution used during measurement, and T is the reaction time.
The method for catalase (CAT) activity assay is as follows: weigh leaves of different strains and parts of the same mass (under identical stress conditions and time), and place them in a mortar. Grind 5 mL of pre-cooled 0.05 mol/L PBS (pH 7.8) on ice until homogenized, then pour into a centrifuge tube for storage. At 4 °C, 10,000 rpm, centrifuge for 20 min and take the supernatant, which is the crude enzyme extract. Store the crude enzyme extract at 4 °C. Reaction mixture: Mix 100 mL of 0.1 mol/L PBS (pH 7.0) and 0.1546 mL of 30% H
2O
2 evenly. PBS was used as a control to zero, and the changes in OD240 of the 2.9 mL reaction mixture and the 0.1 mL enzyme solution were measured within 40 s. The calculation formula for CAT activity (U/g FW) is (△ OD240 × Vt)/(W × VS × 0.01 × T). Among them, △ OD240 is the absorbance change within the reaction time, Vt is the total volume of enzyme solution, W is the fresh weight of the sample, VS is the amount of enzyme solution used during the measurement, and T is the reaction time [
38].
Determination of soluble sugar content by anthrone colorimetric method: soluble sugars generate furfural under the action of sulfuric acid, which then undergoes dehydration and condensation reactions with anthrone to form a green complex. The color intensity and sugar content are linearly related. Measure the absorbance value of soluble sugars using a spectrophotometer with a wavelength of 625 nm. Based on the standard curve, the soluble sugar content in the sample can be calculated. It should be noted that the color presented by the reaction between anthrone reagent and sugar changes over time. In order to accurately measure, it is necessary to complete the measurement within the specified time and record the data.
For phenotypic analysis of seedlings, WT and
OsNAC113 mutant seeds were grown to the 4-week-old seedling stage in soil in pots, then subjected to salt stress. After 7 days of treatment, physiological measurements were carried out as described in our previous study [
39].
4.7. Off-Target Efficiency Detection
Using the Cas9 OFF-finder (
www.rgenome.net/cas-offinder/, accessed on 11 December 2024) website for prediction, we selected the 10 most likely off-target positions for detection, searched for their corresponding site sequences, designed primers based on the search sequences, and performed sequencing analysis of the amplified PCR products to verify whether there were off-target conditions.
4.8. Omics Sequencing and Data Analysis
RNA-seq and Metabolome sequencing were performed by Wuhan Maiweier Biotechnology Co., Ltd. (Wuhan, China) The transcriptome sequencing instrument system is Illumina NovaSeq 6000 (Thermo Fisher Scientific, Waltham, MA, USA). The liquid phase system used for data collection is Thermo’s ultra-high-pressure liquid phase system, UltiMate 3000 UPLC. The chromatographic column model and specification used is ACQUITY UPLC T3 (100 mm × 2.1 mm, 1.8 µm, Waters, Wilmslow, UK). The high-resolution mass spectrometer used for collection is a TripleTOF 6600 (SCIEX, Framingham, MA, USA) time-of-flight mass spectrometer. Wild-type and OsNAC113(+A/+A) with consistent growth were subjected to salt treatment (200 mM) for seven days before sampling. Transcriptome sequencing included total RNA extraction, mRNA enrichment, double-stranded cDNA synthesis, end-effector repair, fragment selection, PCR amplification, and library detection. The experimental process was as follows: after the library was constructed, the RNA sample concentration was ≥250 ng μL−1, pure OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, and the quality RIN > 7.0. By using fastp and Trimomatic to perform quality control on the raw data, high-quality quality-control data can be obtained. Using Hisat software (V1.0) to compare quality control data to the reference genome, assembling and estimating transcript abundance using StringTie software (V1.0), and predicting differential expression of genes and transcripts using DESeq2 software (V1.0). Perform GO and KEGG enrichment analysis of differentially expressed genes using the clusterProfiler package in R language.
Collect and process samples. Conventional samples (cells, etc.) are washed with PBS and centrifuged at 4 °C to obtain a precipitate. Precipitation of proteins and extraction of metabolites. QC (Quality Control) sample preparation: Take 10 µL of each sample and mix it to form a QC sample. Metabolomics studies were conducted using high-throughput mass spectrometers such as SCIEX 6500+.
4.9. Bioinformatics Analysis
The NCBI database was used to query the upstream 2 Kb sequence of the
OsNAC113 start codon (ATG) as the promoter sequence, and EXPASy, Netphos3.1, TMHMM2.0, ProtScale (V1.0), and pfam were used to predict the gene characteristics and secondary structure of
OsNAC113 (
Supplementary Table S2).
4.10. Statistical Analysis
Based on transcriptome data, RESM software was used to perform background correction and standardization on gene expression data. Filter genes with low expression levels and low coefficient of variation. Using R software (R version 4.1) and WGCNAWeighted. Gene Correlation Analysis (WGCNA) was performed using the R version 1.6.6 package and the Pearson correlation coefficient. Soft Threshold power β estimates e-value to fit the network into a scale-free topology, and the topology of each gene pair overlapping measurement (TOM) is used to construct dissimilarity matrices and to perform hierarchical clustering with arithmetic mean (UPGMA). Unweighted grouping method for classes: The best clustering scheme is implemented using the Dynamic Tree Cut R package to screen genes co-expressed with OsNAC113.
LC-MS/MS analyses were performed using a UHPLC system (Vanquish, Thermo Fisher Scientific) with a Waters ACQUITY UPLC BEH Amide (2.1 mm × 50 mm, 1.7 μm) coupled to an Orbitrap Exploris 120 mass spectrometer (Orbitrap MS, Thermo). The mobile phase consisted of 25 mmol/L ammonium acetate and 25 mmol/L ammonia hydroxide in water (pH = 9.75) (A) and acetonitrile (B). The auto-sampler temperature was 4 °C, and the injection volume was 2 μL. The Orbitrap Exploris 120 mass spectrometer was used for its ability to acquire MS/MS spectra on information-dependent acquisition (IDA) mode in the control of the acquisition software (Xcalibur, Thermo). In this mode, the acquisition software continuously evaluates the full scan MS spectrum. The ESI source conditions were set as follows: sheath gas flow rate as 50 Arb, Aux gas flow rate as 15 Arb, capillary temperature 320 °C, full MS resolution as 60,000, MS/MS resolution as 15,000, collision energy SNCE 20/30/40, and spray voltage as 3.8 kV (positive) or −3.4 kV (negative), respectively. The raw data were converted to the mzXML format using ProteoWizard and processed with an in-house program, which was developed using R and based on raw data, were converted to the mzXML format using ProteoWizard and processed with an in-house program, which was developed using R and based on XCMS, for peak detection, extraction, alignment, and integration. Then, an in-house MS2 database was applied in metabolite annotation. The cutoff for annotation was set at 0.3
5. Conclusions
The expression level of OsNAC113 is highest in the leaves during both the seedling and mature stages. Its expression changes indicate that it responds to various adverse biological stresses. Through genetic transformation of rice, knocked-out OsNAC113-regenerated plants were obtained. After Sanger sequencing identification and analysis, mutant OsNAC113 (−1/−1) and OsNAC113 (+A/+A) were obtained, providing a material basis for subsequent experiments. Under salt stress conditions during germination and seedling stages, knocking out OsNAC113 enhances plant salt tolerance. Physiological indicators testing showed that the relative water content, chlorophyll content, and oxidoreductase-related activity of the mutant were higher than those of the wild type, while the accumulation of MDA and other harmful substances was higher in the wild type. Analyze the whole genome expression changes in wild-type and mutant under stress based on RNA-seq analysis. After stress, significantly differentially expressed genes were screened through bioinformatics analysis. By annotating information, it was preliminarily revealed that OsNAC113 is involved in multiple biological processes, such as “plant hormone signaling pathway”, “MAPK signaling pathway”, “amino acid transport and metabolism”, “carbohydrate transport and metabolism”, and “lipid transport and metabolism and replication recombination and repair”, and responds to high-salt stress. Based on metabolomics sequencing results, knocking out OsNAC113 resulted in changes in various important plant biosynthetic pathways, including flavonoid biosynthesis, plant hormone signaling pathways, and ABC transporters, which responded to various abiotic stress conditions and improved the salt tolerance of OsNAC113.