Genome-Wide Analysis of the Dof Gene Family in Soybean and Functional Identification of GmDof63 in Response to Phytophthora sojae Infection
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of the Dof Genes in Soybean
2.2. Chromosome Localization of GmDofs
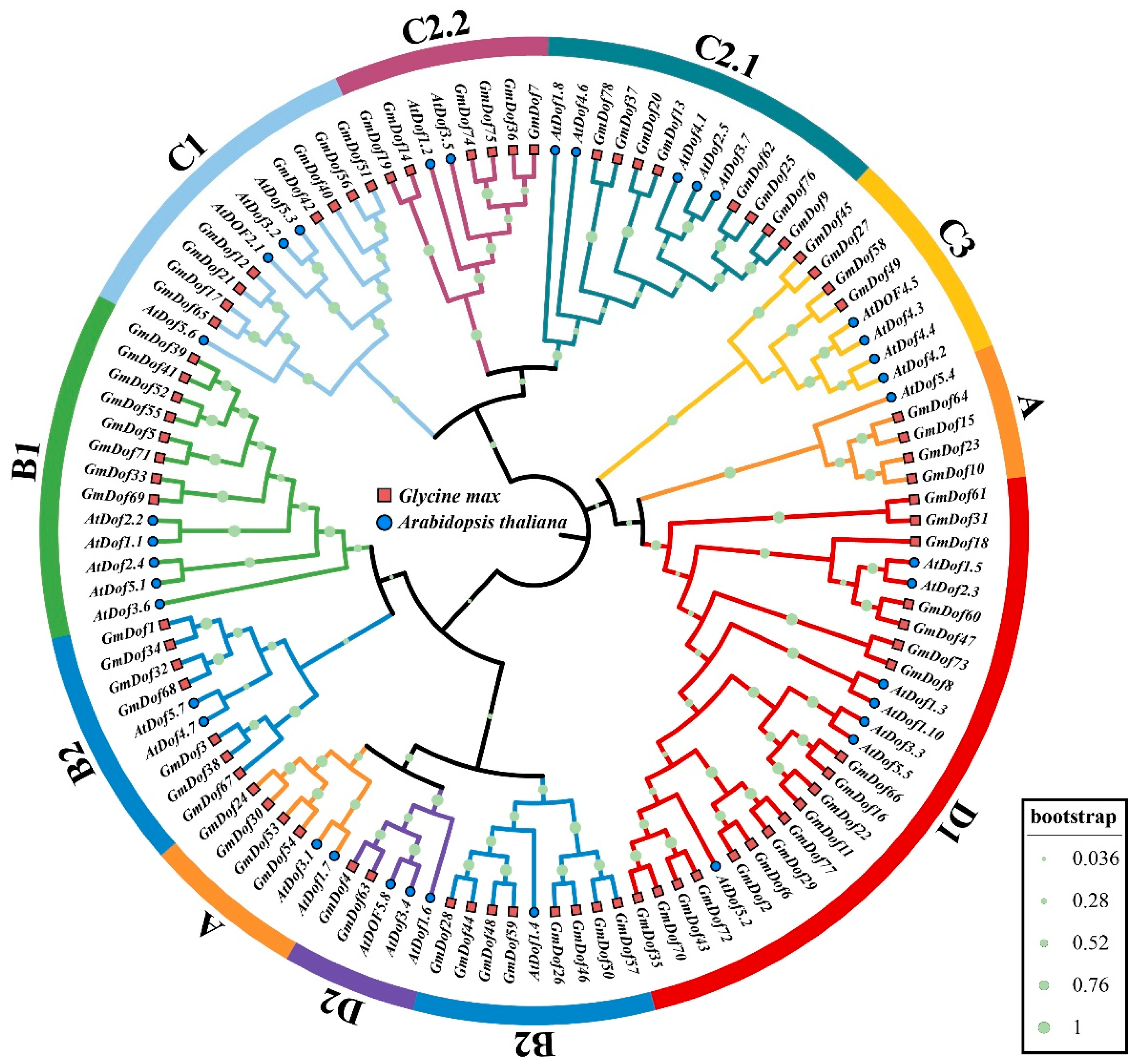
2.3. Phylogenetic Relationships of GmDofs
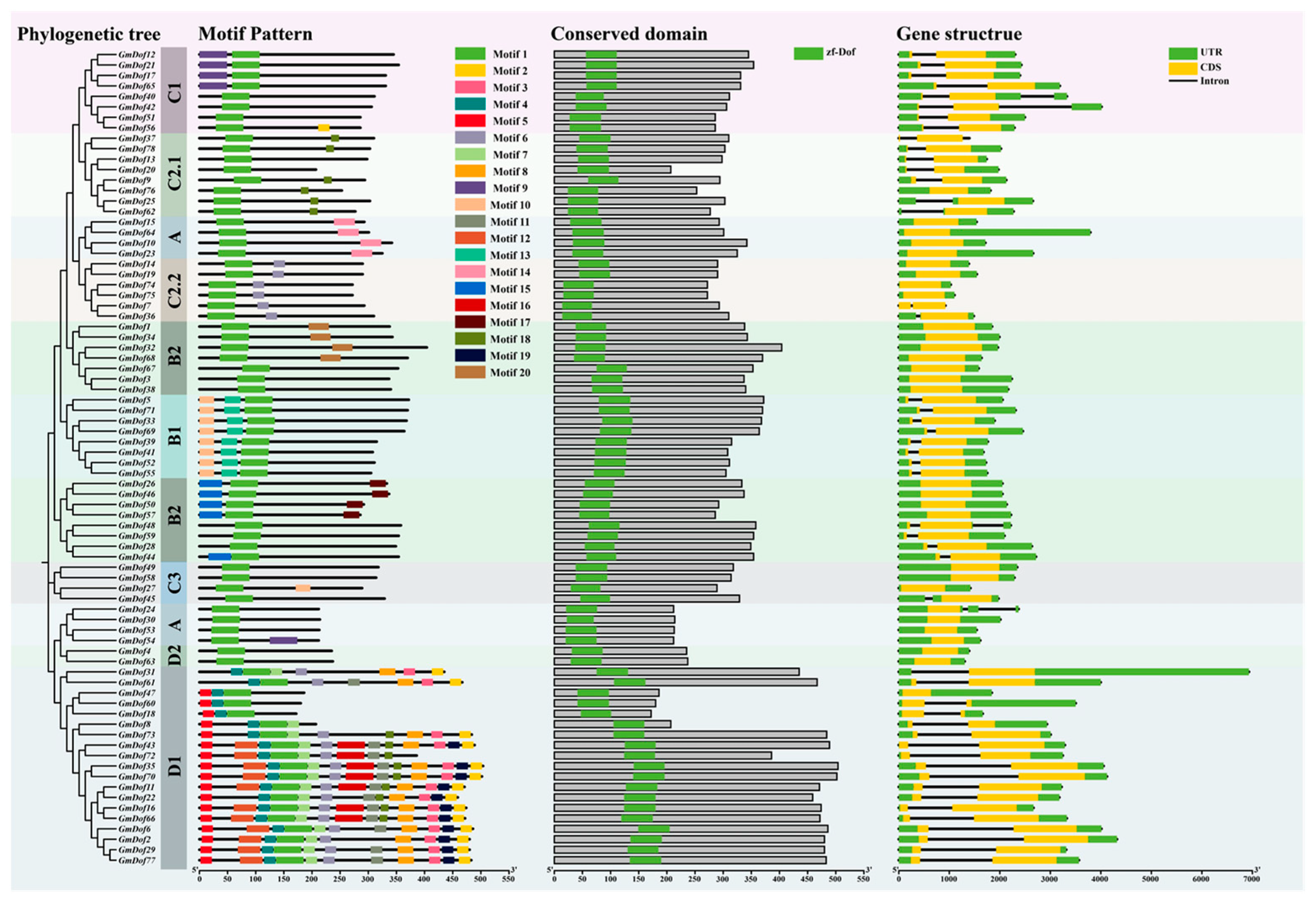
2.4. Gene Structures and Motif Distribution of GmDofs
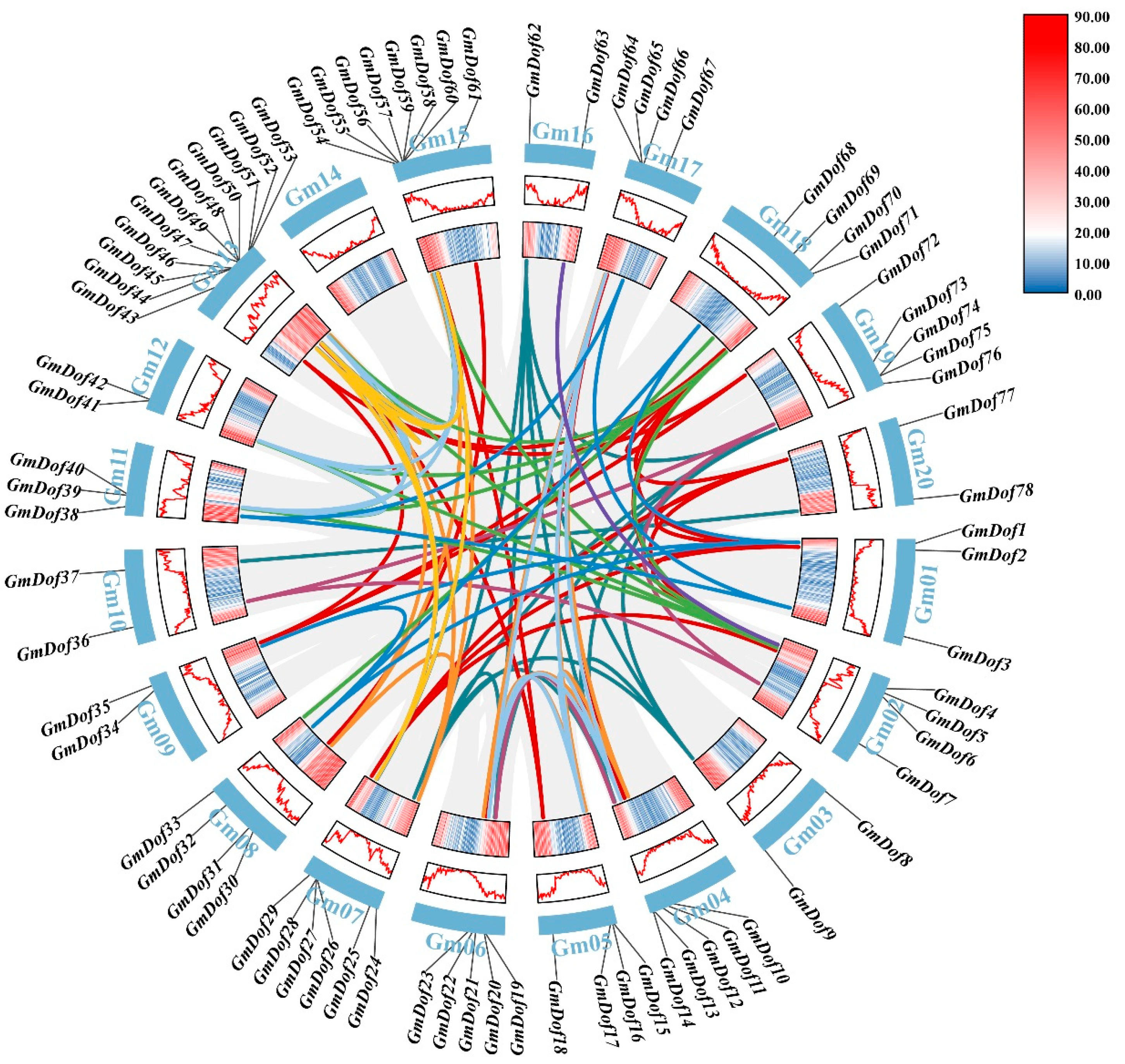
2.5. Collinearity of GmDofs
2.6. Interspecies Collinearity of GmDofs
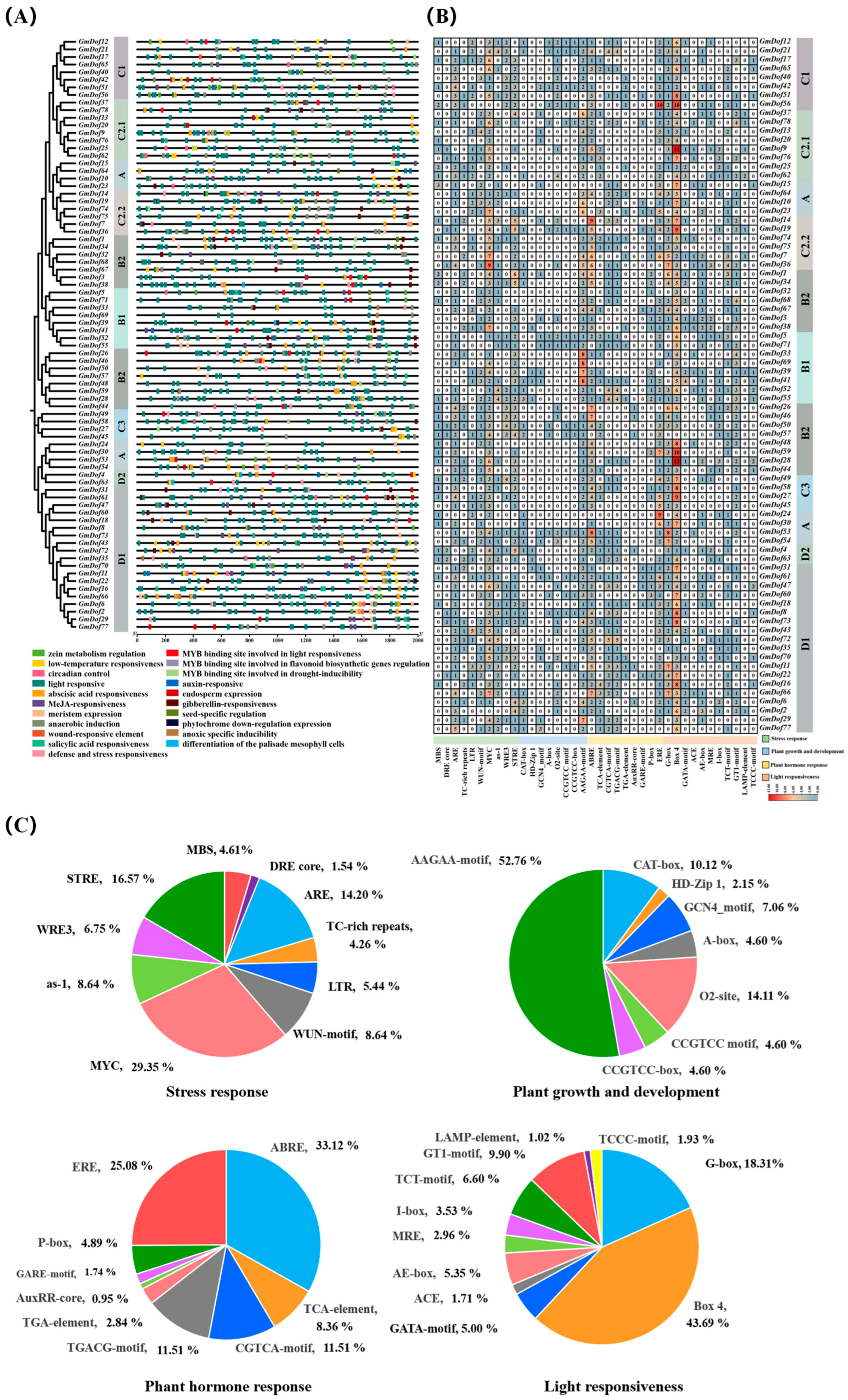
2.7. Predicted Cis-Acting Elements in the Promoters of GmDofs
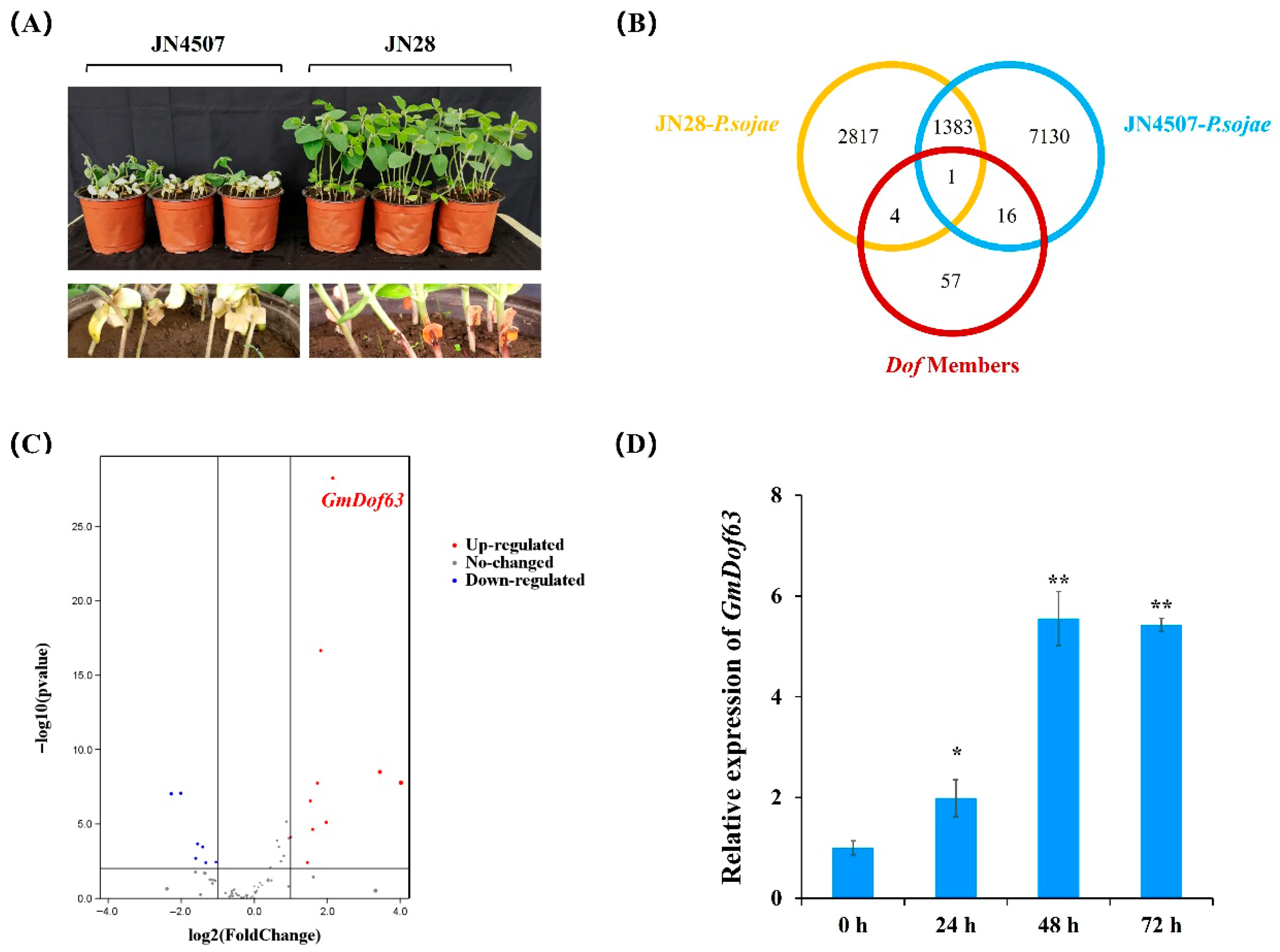
2.8. Screening of Dof Genes in Soybean After P. sojae Infection
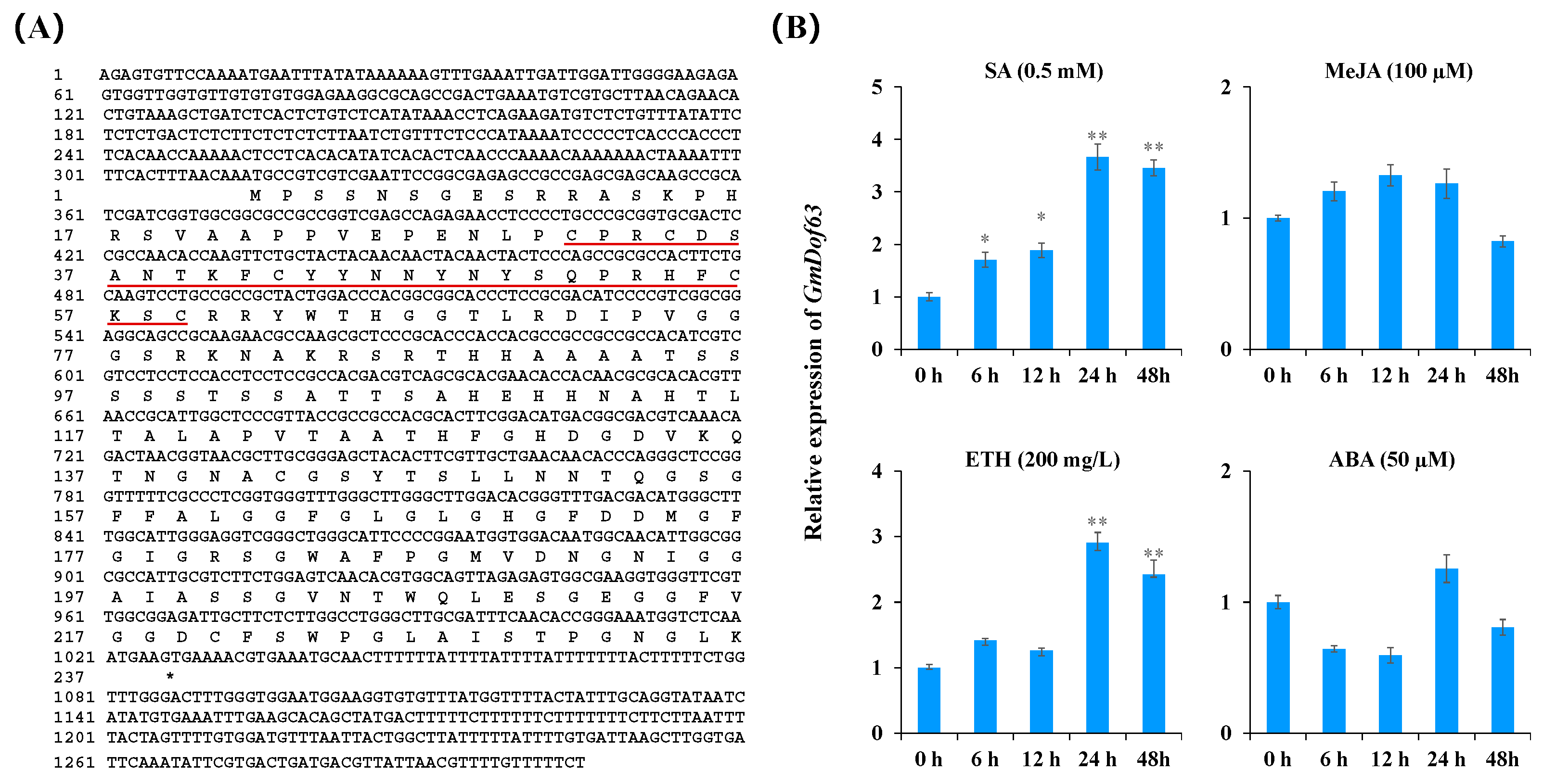
2.9. Sequence Characteristics and Expression Pattern of GmDof63
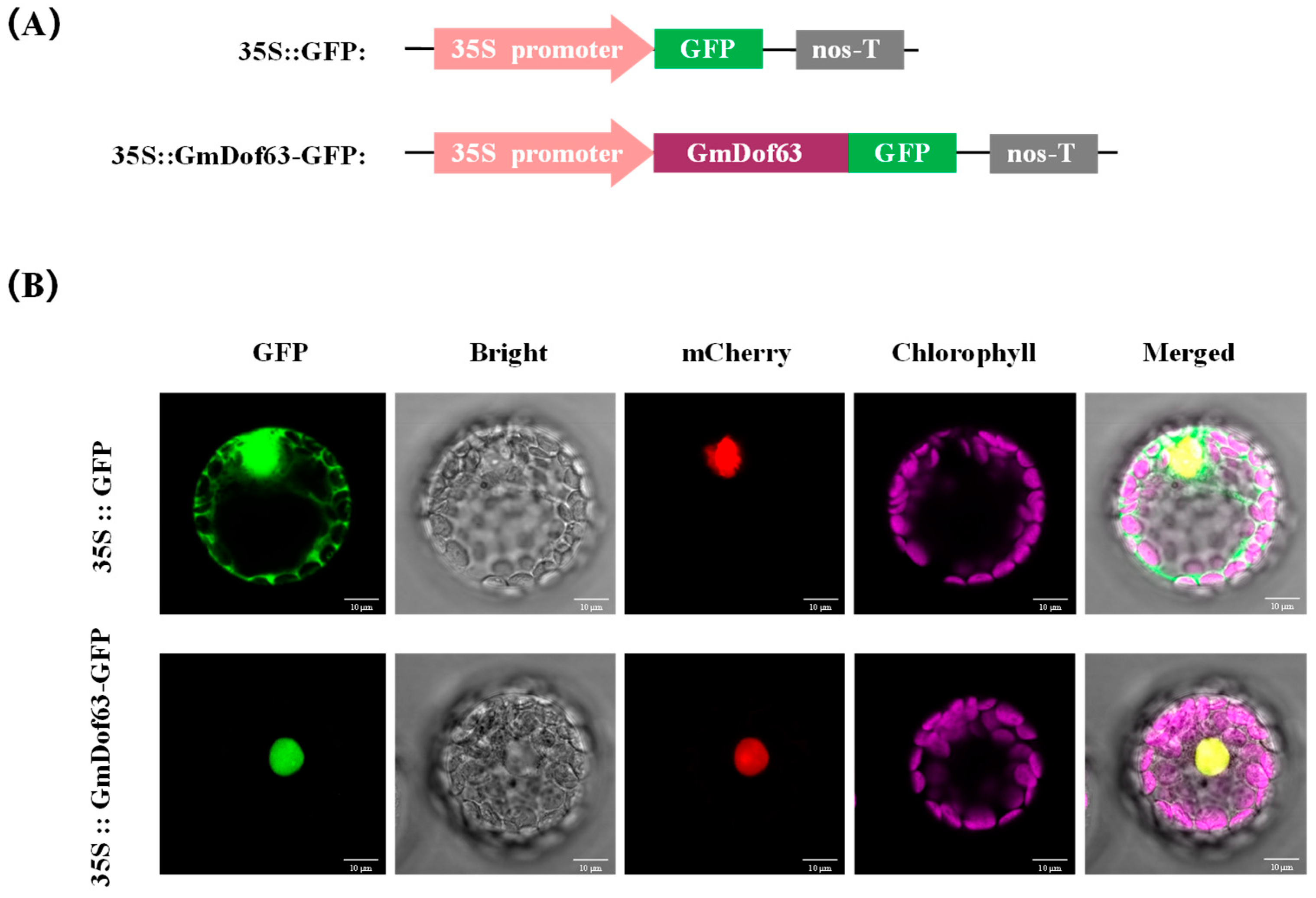
2.10. Subcellular Localization of GmDof63
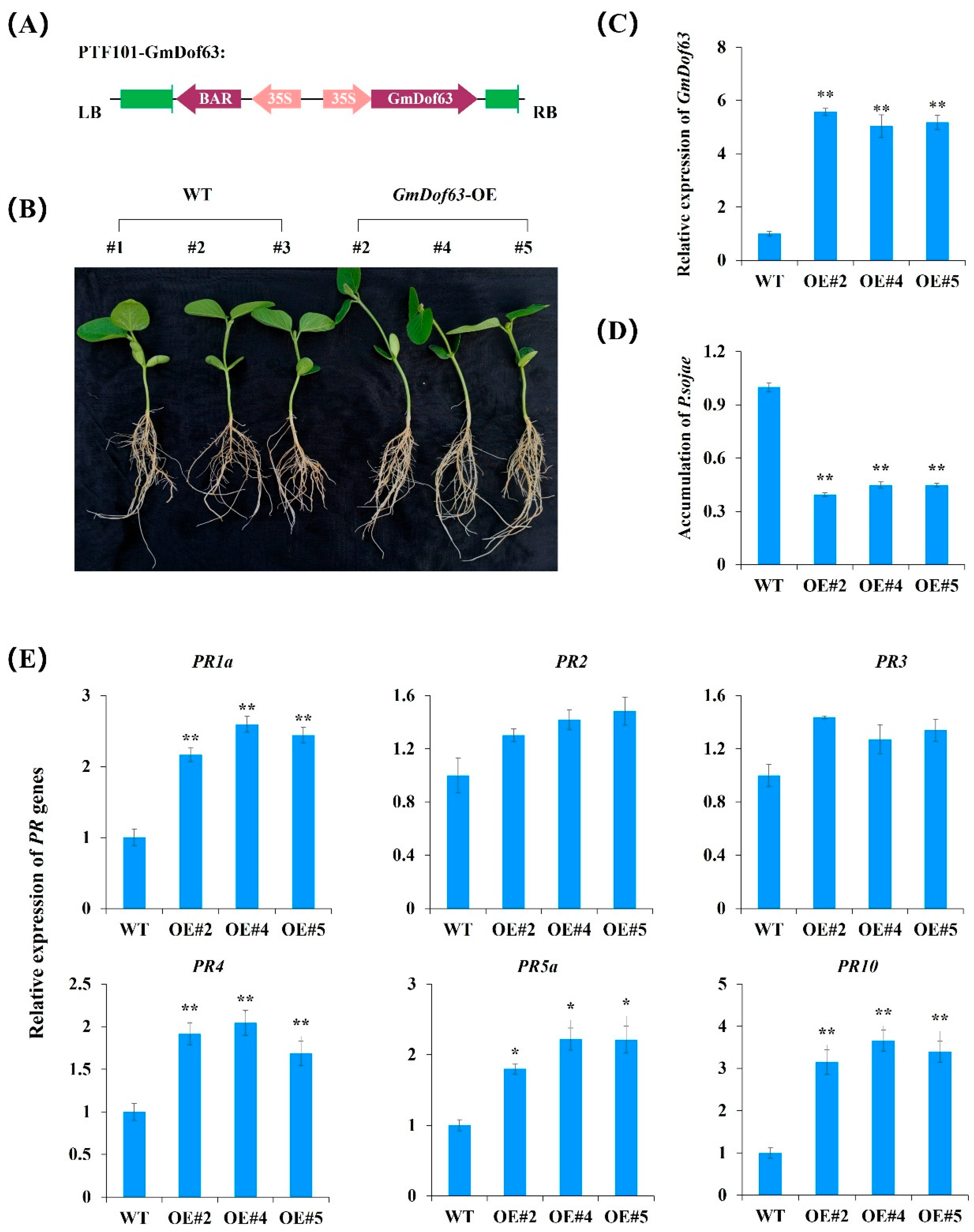
2.11. GmDof63 Enhances Resistance of Transgenic Soybean Seedlings to P. sojae
3. Discussion
4. Materials and Methods
4.1. Identification of Dof Members in Soybean Genome
4.2. Physicochemical Characterization of Soybean Dof Members
4.3. Chromosomal Location Analysis of Soybean Dof Members
4.4. Phylogenetic Analysis of Soybean Dof Members
4.5. Gene Structure and Motif Analysis of Soybean Dof Members
4.6. Analysis of Collinearity and KaKs of Soybean Dof Members
4.7. Prediction of Cis-Acting Elements of Soybean Dof Members
4.8. Plant Materials and Pathogen Strain
4.9. Resistance Identification, RNA Extraction, and Transcriptome Sequencing
4.10. Quantitative RT-PCR
4.11. Primer Sequences Used in the Present Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmitthenner, A.F. Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 1985, 69, 362–368. [Google Scholar] [CrossRef]
- Anderson, T.R.; Buzzell, R.I. Diversity and frequency of races of Phytophthora megasperma f. sp. glycinea in soybean fields in Essex County, Ontario, 1980–1989. Plant Dis. 1992, 76, 587–589. [Google Scholar] [CrossRef]
- Tyler, B.M. Phytophthora sojae: Root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 2007, 8, 1–8. [Google Scholar] [CrossRef]
- Chang, K.F.; Hwang, S.F.; Ahmed, H.U.; Zhou, Q.; Strelkov, S.E.; Conner, R.L.; McLaren, D.L.; Henriquez, M.A.; Harding, M.W.; Turnbull, G.D. First report of Phytophthora sojae causing root rot in soybean [Glycine max (L.) Merr.] in Alberta, Canada. Crop Prot. 2017, 91, 49–56. [Google Scholar] [CrossRef]
- Demirbas, A.; Rector, B.G.; Lohnes, D.G.; Fioritto, R.J.; Graef, G.L.; Cregan, P.B.; Shoemarker, R.C.; Specht, J.E. Simple sequence repeat markers linked to the soybean Rps genes for Phytophthora resistance. Crop Sci. 2001, 41, 1220–1227. [Google Scholar] [CrossRef]
- Burnham, K.D.; Dorrance, A.E.; Francis, D.M. Rps8, a new locus in soybean for resistance to Phytophthora sojae. Crop Sci. 2003, 43, 101–110. [Google Scholar] [CrossRef]
- Ping, J.; Fitzgerald, J.C.; Zhang, C.; Lin, F.; Bai, Y.; Wang, D.; Aggarwal, R.; Rehman, M.; Crasta, O.; Ma, J. Identification and molecular mapping of Rps11, a novel gene conferring resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 2016, 129, 445–451. [Google Scholar] [CrossRef]
- Zou, X.; Sun, H. DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 2023, 14, 1044918. [Google Scholar] [CrossRef]
- Liu, J.; Jin, Y.; Wu, Y.S.; Liu, Y.; Wang, W.b.; Ren, S.S.; Diao, S.F.; Chen, Y.L. Advances on the structural characteristics and function of Dof gene in plant. Biotechnol. Bull. 2020, 36, 180–219. [Google Scholar] [CrossRef]
- Wang, T.; Yue, J.J.; Wang, X.J.; Xu, L.; Li, L.B.; Gu, X.P. Genome-wide identification and characterization of the Dof gene family in moso bamboo (Phyllostachys heterocycla var. Pubescens). Genes Genom. 2016, 38, 733–745. [Google Scholar] [CrossRef]
- Lijavetzky, D.; Carbonero, P.; Vicente-Carbajosa, J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003, 3, 17. [Google Scholar] [CrossRef]
- Gou, C.; Zhang, G.; Deng, Z.; Lin, C.; Li, H.; Liu, H.; Fang, X. Genome-wide analysis of the DNA-binding with one finger gene family reveals soybean expression pattern and functional analysis. Int. J. Mol. Sci. 2025, 26, 6192. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Izui, K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 1993, 268, 16028–16036. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Park, S.W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015, 94, 73–85. [Google Scholar] [CrossRef]
- Chen, M.; Liu, X.; Huan, L.; Sun, M.; Liu, L.; Chen, X.; Gao, D.; Li, L. Genome-wide analysis of Dof family genes and their expression during bud dormancy in peach (Prunus persica). Sci. Hortic. 2017, 214, 18–26. [Google Scholar] [CrossRef]
- Wu, Z.; Cheng, J.; Cui, J.; Xu, X.; Liang, G.; Luo, X.; Chen, X.; Tang, X.; Hu, K.; Qin, C. Genome-wide identification and expression profile of Dof transcription factor gene family in pepper (Capsicum annuum L.). Front. Plant Sci. 2016, 7, 574. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Malviya, N.; Kushwaha, H.; Nasim, J.; Bisht, N.C.; Singh, V.K.; Yadav, D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549–562. [Google Scholar] [CrossRef]
- Li, S.; Zhang, W.; Si, C.; Chen, J.; Huang, Y.; Li, M.; Liang, H.; Duan, J.; He, C. Genome-Wide identification and functional characterization of the Dof family in Dendrobium officinale. Int. J. Mol. Sci. 2025, 26, 2671. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Li, W.; Dai, G.; Chen, J. Genome-wide identification and expression analysis of the LbDof transcription factor family genes in Lycium barbarum. Plants 2025, 14, 1567. [Google Scholar] [CrossRef]
- Wang, P.; Wang, D.; Li, Y.; Li, J.; Liu, B.; Wang, Y.; Gao, C. The transcription factor ThDOF8 binds to a novel cis-element and mediates molecular responses to salt stress in Tamarix hispida. J. Exp. Bot. 2024, 75, 3171–3187. [Google Scholar] [CrossRef]
- Zhao, C.; Bai, H.; Li, C.; Pang, Z.; Xuan, L.; Lv, D.; Niu, S. Genome-wide identification of the Dof gene family in Kiwifruit (Actinidia chinensis) and functional validation of AcDOF22 in response to drought stress. Int. J. Mol. Sci. 2024, 25, 9103. [Google Scholar] [CrossRef]
- Dai, J.; Wu, Y.; Liu, J.; Wei, L.; Zeng, Y.; Liu, S.; Zhang, J.; Liu, G.; Huang, G. Genome-wide identification and expression analysis of Dof gene family members in mulberry trees (Morus notabilis L.) under drought stress. BMC Genomics 2025, 26, 744. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, N.; Deng, X.; Liu, D.; Li, M.; Cui, D.; Hu, Y.; Yan, Y. Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: Evolutionary characteristics and diverse abiotic stress responses. BMC Genom. 2020, 21, 276. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Parra, E.; Perianez-Rodriguez, J.; Navarro-Neila, S.; Gude, I.; Moreno-Risueno, M.A.; del Pozo, J.C. The transcription factor OBP4 controls root growth and promotes callus formation. New Phytol. 2017, 213, 1787–1801. [Google Scholar] [CrossRef]
- Li, T.; Wang, X.; Elango, D.; Zhang, W.; Li, M.; Zhang, F.; Pan, Q.; Wu, Y. Genome-wide identification, phylogenetic and expression pattern analysis of Dof transcription factors in blueberry (Vaccinium corymbosum L.). PeerJ 2022, 10, e14087. [Google Scholar] [CrossRef]
- Su, Y.; Liang, W.; Liu, Z.; Wang, Y.; Zhao, Y.; Ijaz, B.; Hua, J. Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. J. Plant Physiol. 2017, 218, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis Dof transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Ma, C.N.; Du, L.D.; Xiang, Y.; Xiao, F.; Liu, Y.T.; Wang, C.K.; Li, W.K.; Zhao, T.T.; Hu, D.G. Two DNA-binding one zinc finger transcription factors, MdCDOF3 and MdDOF3.6, accelerate leaf senescence by activating cytokinin oxidase MdCKX7 in response to sorbitol signaling in apple. Hortic. Res. 2025, 12, uhaf120. [Google Scholar] [CrossRef]
- Ding, R.; Xiao, T.; Li, S.; Qiang, J.; Zhang, H.; Chang, H.; Yan, Y.; Li, X. Wheat endosperm-specific transcription factor TaDOF6 enhances grain development by regulating TaSWEET13h expression and facilitating sugar and gibberellin transport. Front. Plant Sci. 2025, 16, 1608090. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, C.; Shu, W.; Ye, Z.; Li, H.; Zhang, Y. The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem. Biophys. Res. Commun. 2016, 474, 736–741. [Google Scholar] [CrossRef]
- Wen, Y.; Tan, C.; Zhang, Y.; Wu, H.; Chen, D.; Yue, H.; Ding, Z.; Cao, S.; Zheng, K. Genome-wide characterization and functional analysis of CsDOF transcription factors in Camellia sinensis cv. Tieguanyin under combined heat-drought stress. Plants 2025, 14, 1829. [Google Scholar] [CrossRef]
- Song, F.; Goodman, R.M. Cloning and identification of the promoter of the tobacco Sar8.2b gene, a gene involved in systemic acquired resistance. Gene 2002, 290, 115–124. [Google Scholar] [CrossRef]
- Qiu, T.; Wei, S.; Fang, K.; Zhang, M.; Li, Y.; Feng, Y.; Cheng, Y.; Zhang, S.; Tian, J.; Gao, A.; et al. The atypical Dof transcriptional factor OsDes1 contributes to stay-green, grain yield, and disease resistance in rice. Sci. Adv. 2024, 10, eadp0345. [Google Scholar] [CrossRef] [PubMed]
- Trusov, Y.; Sewelam, N.; Rookes, J.E.; Kunkel, M.; Nowak, E.; Schenk, P.M.; Botella, J.R. Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant J. 2009, 58, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.L.; Kang, J.H.; Howe, G.A. Jasmonate-triggered plant immunity. J. Chem. Ecol. 2014, 40, 657–675. [Google Scholar] [CrossRef]
- Mersmann, S.; Bourdais, G.; Rietz, S.; Robatzek, S. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 2010, 154, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Charitha, P.A.J.; Emilee, R.M.S.; Jocelyn, A.O.; Stephen, E.S. The complex roles of plant hormones during clubroot disease development in the Brassicaceae. J. Plant Growth Regul. 2025, 44, 5692–5712. [Google Scholar] [CrossRef]
- Cai, M.; Lin, J.; Li, Z.; Lin, Z.; Ma, Y.; Wang, Y.; Ming, R. Allele specific expression of Dof genes responding to hormones and abiotic stresses in sugarcane. PLoS ONE 2020, 15, e0227716. [Google Scholar] [CrossRef]
- Virág, E.; Nagy, Á.; Tóth, B.B.; Kutasy, B.; Pallos, J.P.; Szigeti, Z.M.; Máthé, C.; Kardos, G.; Hegedűs, G. Master regulatory transcription factors in β-aminobutyric acid-induced resistance (BABA-IR): A perspective on phytohormone biosynthesis and signaling in Arabidopsis thaliana and Hordeum vulgare. Int. J. Mol. Sci. 2024, 25, 9179. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Xu, L.; Wang, C.; Luo, Y.; Feng, S.; Yuan, Y.; Yang, Q.; Feng, B. Genome-wide identification of DNA binding with one finger (dof) gene family in tartary buckwheat (Fagopyrum tataricum) and analysis of its expression pattern after exogenous hormone stimulation. Biology 2022, 11, 173. [Google Scholar] [CrossRef]
- Yu, Y.H.; Bian, L.; Wan, Y.T.; Jiao, Z.L.; Yu, K.K.; Zhang, G.H.; Guo, D.L. Grape (Vitis vinifera) VvDOF3 functions as a transcription activator and enhances powdery mildew resistance. Plant Physiol. Biochem. 2019, 143, 183–189. [Google Scholar] [CrossRef]
- Paz, M.M.; Shou, H.; Guo, Z.; Zhang, Z.; Banerjee, A.K.; Wang, K. Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explants. Euphytica 2004, 136, 167–179. [Google Scholar] [CrossRef]
- Li, S.; Cong, Y.; Liu, Y.; Wang, T.; Shuai, Q.; Chen, N.; Gai, J.; Li, Y. Optimization of Agrobacterium-mediated transformation in soybean. Front. Plant Sci. 2017, 8, 246. [Google Scholar] [CrossRef]
- Shrestha, S.D.; Chapman, P.; Zhang, Y.; Gijzen, M. Strain specific factors control effector gene silencing in Phytophthora sojae. PLoS ONE 2016, 11, e0150530. [Google Scholar] [CrossRef]
- Yang, X.; Jiang, X.; Yan, W.; Huang, Q.; Sun, H.; Zhang, X.; Zhang, Z.; Ye, W.; Wu, Y.; Govers, F.; et al. The mevalonate pathway is important for growth, spore production, and the virulence of Phytophthora sojae. Front. Microbiol. 2021, 12, 772994. [Google Scholar] [CrossRef]
- Alexander, D.; Goodman, R.M.; Gut-Rella, M.; Glascock, C.; Weymann, K.; Friedrich, L.; Maddox, D.; Ahl-Goy, P.; Luntz, T.; Ward, E. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. USA 1993, 90, 7327–7331. [Google Scholar] [CrossRef]
- Sessa, G.; Yang, X.Q.; Raz, V.; Eyal, Y.; Fluhr, R. Dark induction and subcellular localization of the pathogenesis-related PRB-1b protein. Plant Mol. Biol. 1995, 28, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.F.; Jiang, L.Y.; Wu, J.J.; Li, W.B.; Fan, S.J.; Zhang, S.Z. Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Mol. Biol. Rep. 2014, 41, 4899–4909. [Google Scholar] [CrossRef] [PubMed]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]
- Umemura, Y.; Ishiduka, T.; Yamamoto, R.; Esaka, M. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J. 2004, 37, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Chen, P.; Yan, M.; Li, L.; He, J.; Zhou, S.; Li, Z.; Niu, C.; Bao, C.; Zhi, F.; Ma, F.; et al. The apple DNA-binding one zinc-finger protein MdDof54 promotes drought resistance. Hortic. Res. 2020, 7, 195. [Google Scholar] [CrossRef]
- Khan, I.; Khan, S.; Zhang, Y.; Zhou, J.P. Genome-wide analysis and functional characterization of the Dof transcription factor family in rice (Oryza sativa L.). Planta 2021, 253, 101. [Google Scholar] [CrossRef]
- Yang, C.; Lai, Y.M.; Yao, N. Plant sphingolipids: Subcellular distributions and functions. Curr. Opin. Plant Biol. 2025, 85, 102704. [Google Scholar] [CrossRef]
- Marques, A.C.; Vinckenbosch, N.; Brawand, D.; Kaessmann, H. Functional diversification of duplicate genes through subcellular adaptation of encoded proteins. Genome Biol. 2008, 9, R54. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, L.; Chen, M.; Zhou, W.; Dong, Q.; Jiang, H.; Cheng, B. The Dof-domain transcription factor ZmDOF36 positively regulates starch synthesis in transgenic maize. Front. Plant Sci. 2019, 10, 465. [Google Scholar] [CrossRef]
- Luo, T.; Song, Y.; Gao, H.; Wang, M.; Cui, H.; Ji, C.; Wang, J.; Yuan, L.; Li, R. Genome-wide identification and functional analysis of Dof transcription factor family in Camelina sativa. BMC Genom. 2022, 23, 812. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Kondrashov, F.A. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 2012, 279, 5048–5057. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, X.; Yin, D.D.; Yuan, H.; Xie, Q.; Zhao, X.F.; Li, X.B.; Zhu, L.H.; Li, S.G.; Li, D.Y. Constitutive expression of OsDof4, encoding a C2-C2 zinc finger transcription factor, confesses its distinct flowering effects under long- and short-day photoperiods in rice (Oryza sativa L.). BMC Plant Biol. 2017, 17, 166. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.F.; Lee, S.K.; Yoo, Y.; Wei, J.; Kwon, S.Y.; Lee, S.W.; Jeon, J.S.; An, G. Rice transcription factor OsDOF11 modulates sugar transport by promoting expression of sucrose transporter and SWEET genes. Mol. Plant 2018, 11, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Wang, J.; Chen, X.B.; Wang, F.F.; Peng, P.; Zhou, Y.; Miao, Y.C.; Zhang, Y.O.; Gao, Y.D.; Qi, Y.D.; et al. Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 2019, 223, 798–813. [Google Scholar] [CrossRef] [PubMed]
- Shim, Y.; Kang, K.; An, G.; Paek, N.C. Rice DNA-binding one zinc finger 24 (OsDOF24) delays leaf senescence in a jasmonate-mediated pathway. Plant Cell Physiol. 2019, 60, 2065–2076. [Google Scholar] [CrossRef]
- Zhang, Y.; Verhoeff, N.I.; Chen, Z.; Chen, S.; Wang, M.; Zhu, Z.; Ouwerkerk, P.B.F. Functions of OsDof25 in regulation of OsC4PPDK. Plant Mol. Biol. 2015, 89, 229–242. [Google Scholar] [CrossRef]
- Zuo, Z.F.; Kang, H.G.; Park, M.Y.; Jeong, H.; Sun, H.J.; Song, P.S.; Lee, H.Y. Zoysia japonica MYC type transcription factor ZjICE1 regulates cold tolerance in transgenic Arabidopsis. Plant Sci. 2019, 289, 110254. [Google Scholar] [CrossRef]
- Mayordomo, I.; Estruch, F.; Sanz, P. Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (Stress Response Element)-regulated genes. J. Biol. Chem. 2002, 277, 35650–35656. [Google Scholar] [CrossRef]
- Lohani, N.; Babaei, S.; Singh, M.B.; Bhalla, P.L. Genome-wide in silico identification and comparative analysis of Dof gene family in Brassica napus. Plants 2021, 10, 709. [Google Scholar] [CrossRef]
- Liu, J.; Meng, Q.L.; Xiang, H.T.; Shi, F.M.; Ma, L.G.; Li, Y.C.; Liu, C.L.; Liu, Y.; Su, B.H. Genome-wide analysis of Dof transcription factors and their response to cold stress in rice (Oryza sativa L.). BMC Genom. 2021, 22, 800. [Google Scholar] [CrossRef]
- Wong, J.; Gao, L.; Yang, Y.; Zhai, J.X.; Arikit, S.; Yu, Y.; Duan, S.Y.; Chan, V.; Xiong, Q.; Yan, J.; et al. Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 2014, 79, 928–940. [Google Scholar] [CrossRef]
- Cheng, Q.; Dong, L.D.; Gao, T.J.; Liu, T.F.; Li, N.H.; Wang, L.; Chang, X.; Wu, J.J.; Xu, P.F.; Zhang, S.Z. The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 2018, 69, 2527–2541. [Google Scholar] [CrossRef]
- Li, W.; Zheng, X.; Cheng, R.; Zhong, C.; Zhao, J.; Liu, T.H.; Yi, T.; Zhu, Z.; Xu, J.; Meksem, K.; et al. Soybean ZINC FINGER PROTEIN03 targets two SUPEROXIDE DISMUTASE1s and confers resistance to Phytophthora sojae. Plant Physiol. 2023, 69, 2527–2541. [Google Scholar] [CrossRef]
- Zhu, X.; Guo, L.; Zhu, R.; Zhou, X.; Zhang, J.; Li, D.; He, S.; Qiao, Y. Phytophthora sojae effector PsAvh113 associates with the soybean transcription factor GmDPB to inhibit catalase-mediated immunity. Plant Biotechnol. J. 2023, 21, 1393–1407. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Singh, K.B. Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: Overexpression of OBP3 leads to growth defects. Plant J. 2000, 21, 329–339. [Google Scholar] [CrossRef]
- van Verk, M.C.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J.M. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008, 146, 1983–1995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, W.; Kinkema, M.; Li, X.; Dong, X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 1999, 96, 6523–6528. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gou, X.P.; Yuan, T.; Lin, H.H.; Asami, T.; Yoshida, S.; Russell, S.D.; Li, J. BAK1 and BKK1 regulate Brassinosteroid-dependent growth and Brassinosteroidindependent cell-death pathways. Curr. Biol. 2007, 17, 1109–1115. [Google Scholar] [CrossRef]
- Ward, E.R.; Uknes, S.J.; Williams, S.C.; Dincher, S.S.; Wiederhold, D.L.; Alexander, D.C.; Ahl-Goy, P.; Metraux, J.P.; Ryals, J.A. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 1991, 3, 1085–1094. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Chen, H.; Huo, Y.; Song, Y.; Wang, P.; Zhang, Z.; Jiang, L. Genome-Wide Analysis of the Dof Gene Family in Soybean and Functional Identification of GmDof63 in Response to Phytophthora sojae Infection. Plants 2025, 14, 3621. https://doi.org/10.3390/plants14233621
Fan S, Chen H, Huo Y, Song Y, Wang P, Zhang Z, Jiang L. Genome-Wide Analysis of the Dof Gene Family in Soybean and Functional Identification of GmDof63 in Response to Phytophthora sojae Infection. Plants. 2025; 14(23):3621. https://doi.org/10.3390/plants14233621
Chicago/Turabian StyleFan, Sujie, Haiyuan Chen, Yuhan Huo, Yang Song, Piwu Wang, Zhuo Zhang, and Liangyu Jiang. 2025. "Genome-Wide Analysis of the Dof Gene Family in Soybean and Functional Identification of GmDof63 in Response to Phytophthora sojae Infection" Plants 14, no. 23: 3621. https://doi.org/10.3390/plants14233621
APA StyleFan, S., Chen, H., Huo, Y., Song, Y., Wang, P., Zhang, Z., & Jiang, L. (2025). Genome-Wide Analysis of the Dof Gene Family in Soybean and Functional Identification of GmDof63 in Response to Phytophthora sojae Infection. Plants, 14(23), 3621. https://doi.org/10.3390/plants14233621





