The Bitter Gourd Transcription Factor McNAC087 Confers Cold Resistance in Transgenic Arabidopsis
Abstract
1. Introduction
2. Results
2.1. Morphological Responses of Momordica charantia L. Under Low-Temperature Stress
2.2. RNA Sequencing
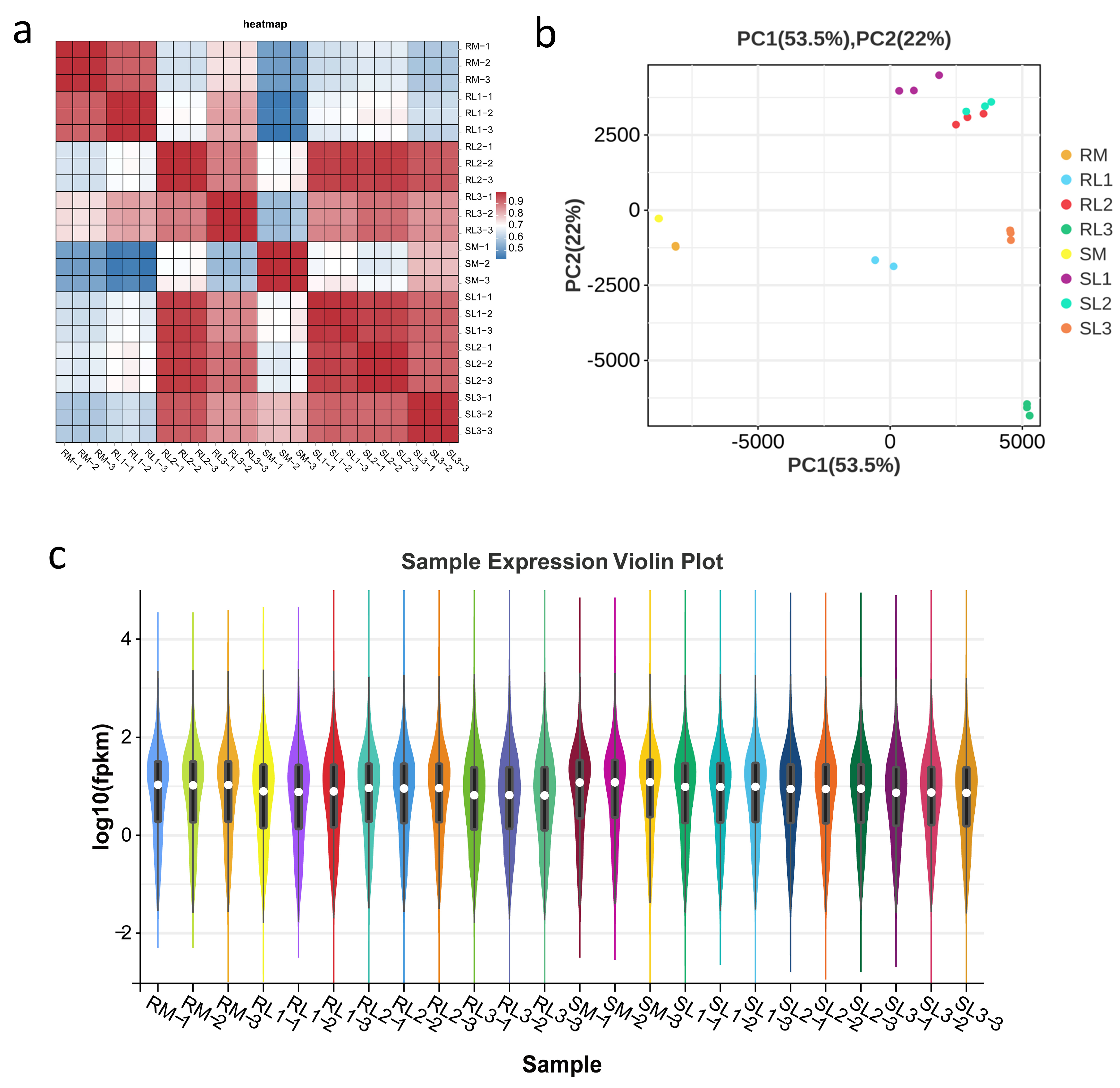
2.3. Sample Analysis
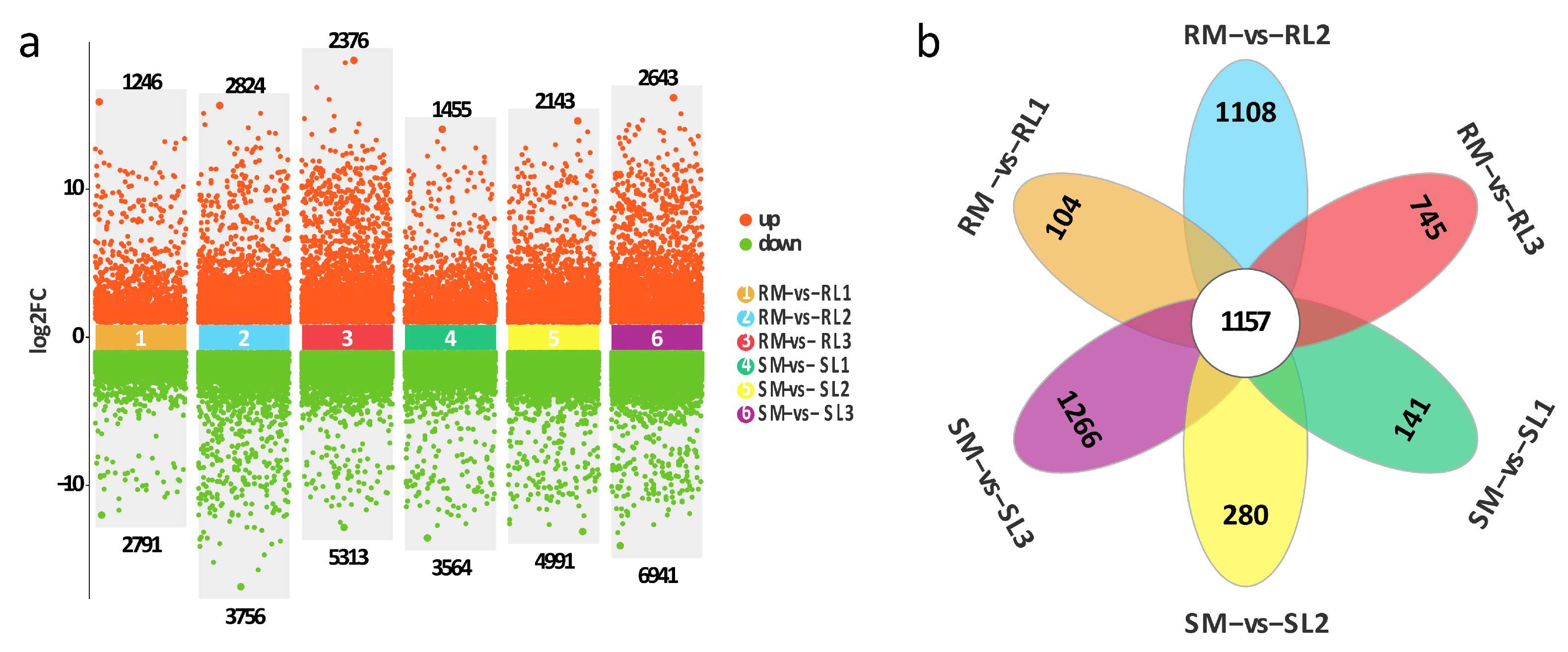
2.4. Statistical Analysis of the Differentially Expressed Gene (DEG)
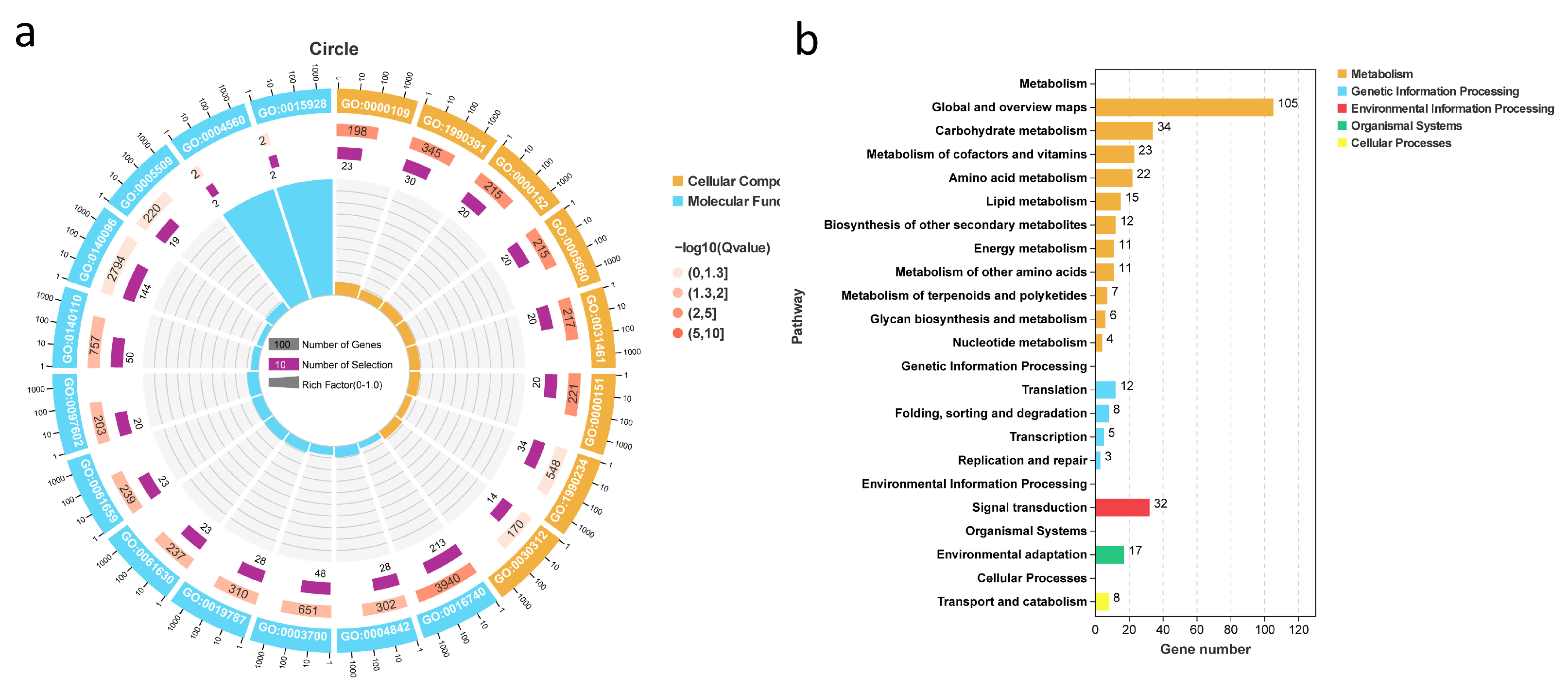
2.5. GO/KEGG Annotation of Cold Stress Genes and Identification of Candidate Gene McNAC087
2.6. Confirmation of DEGs by RT-qPCR
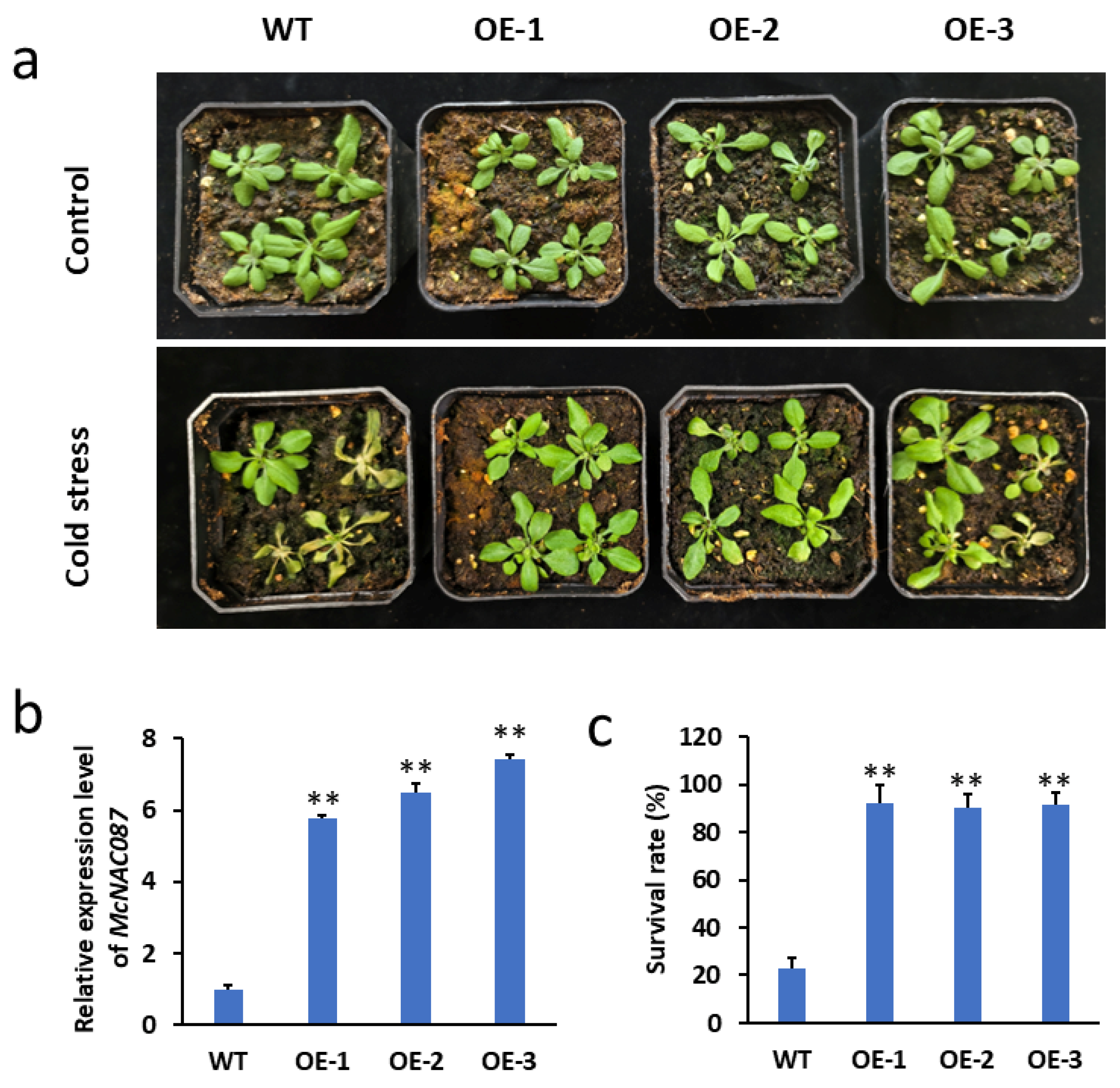
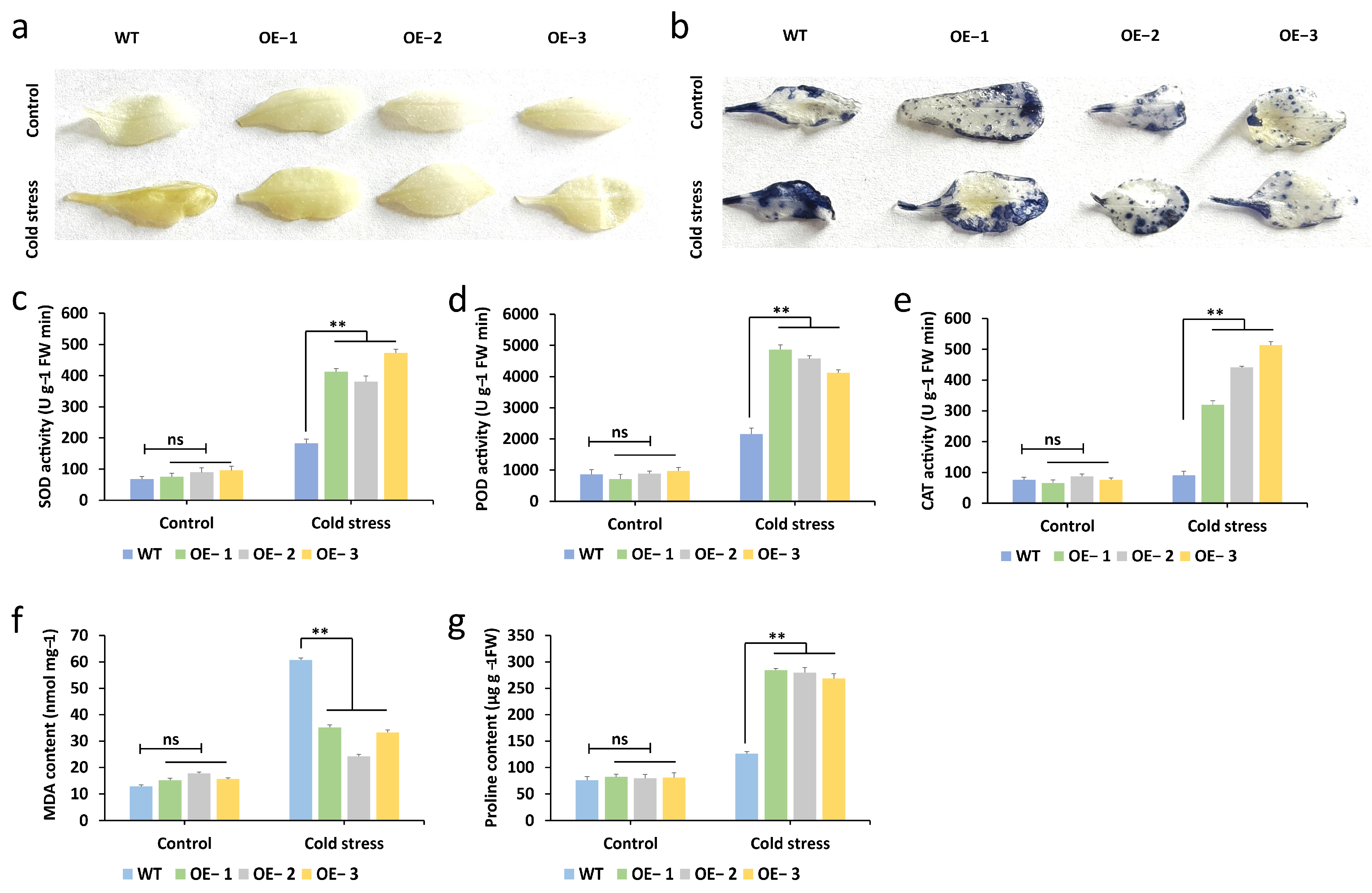
2.7. The Overexpression of McNAC087 Enhances the Cold Resistance of Transgenic Arabidopsis
2.8. Overexpression of the McNAC087 Gene Alters Physiological Traits Under Low-Temperature Stress
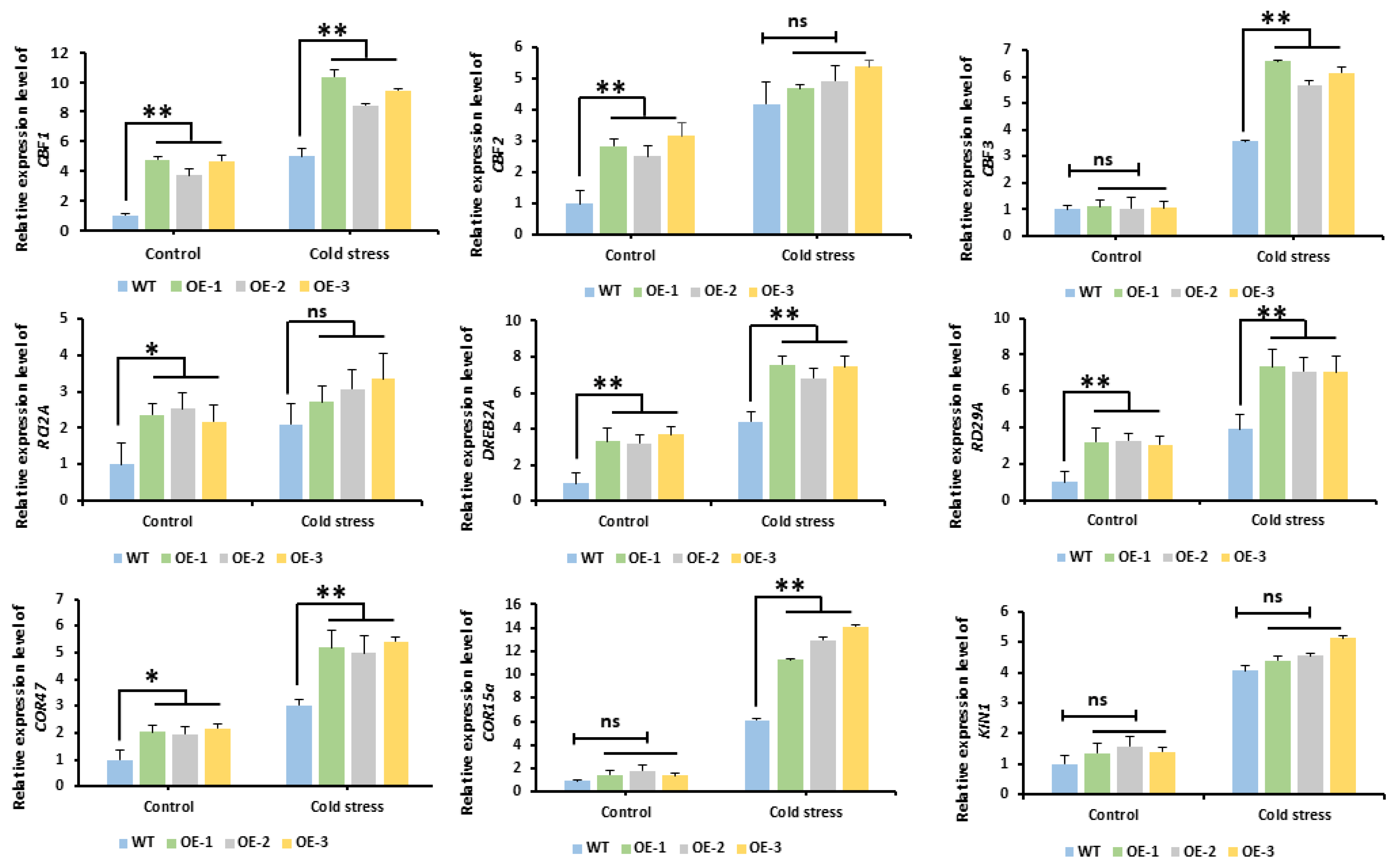
2.9. Analysis of the Expression Patterns of Cold-Responsive Genes in Arabidopsis
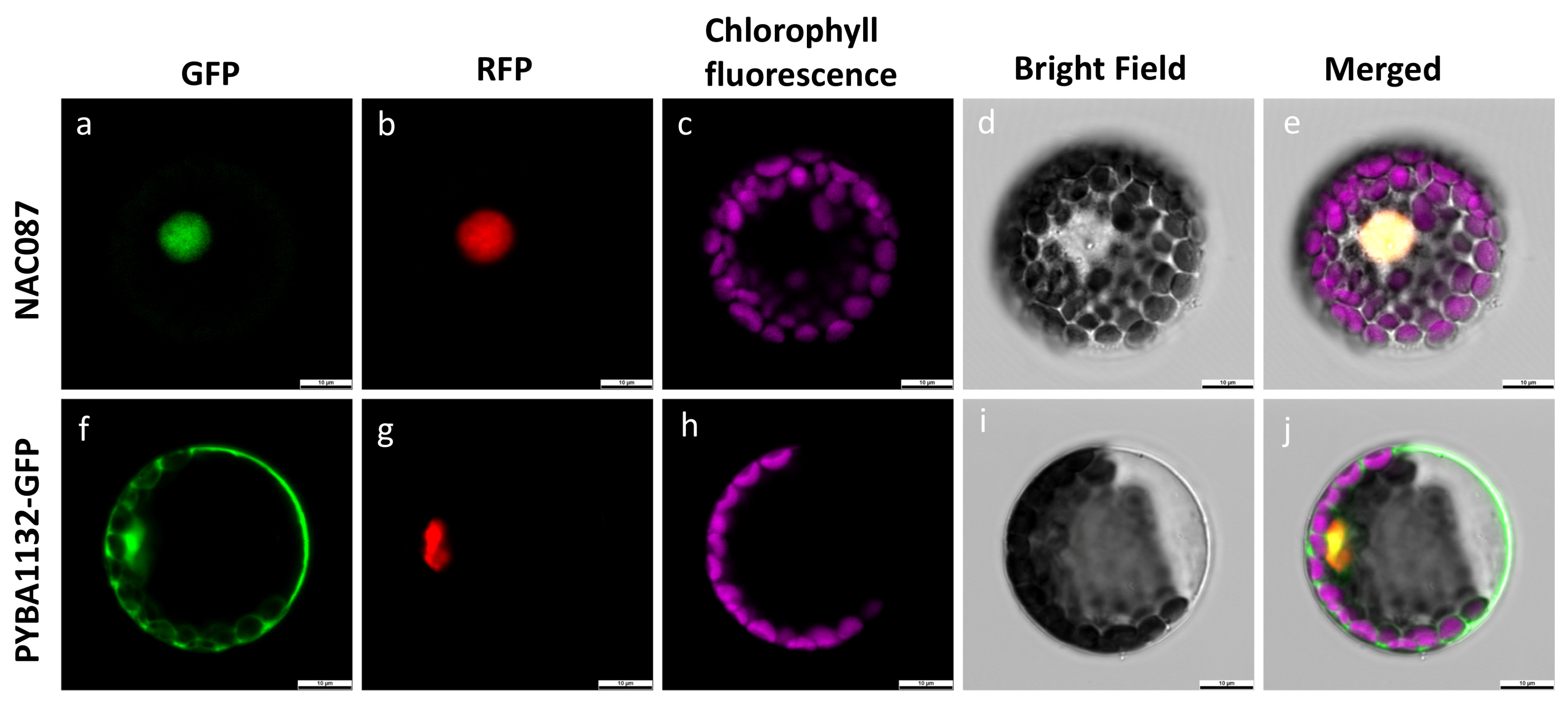
2.10. Subcellular Localization of McNAC087
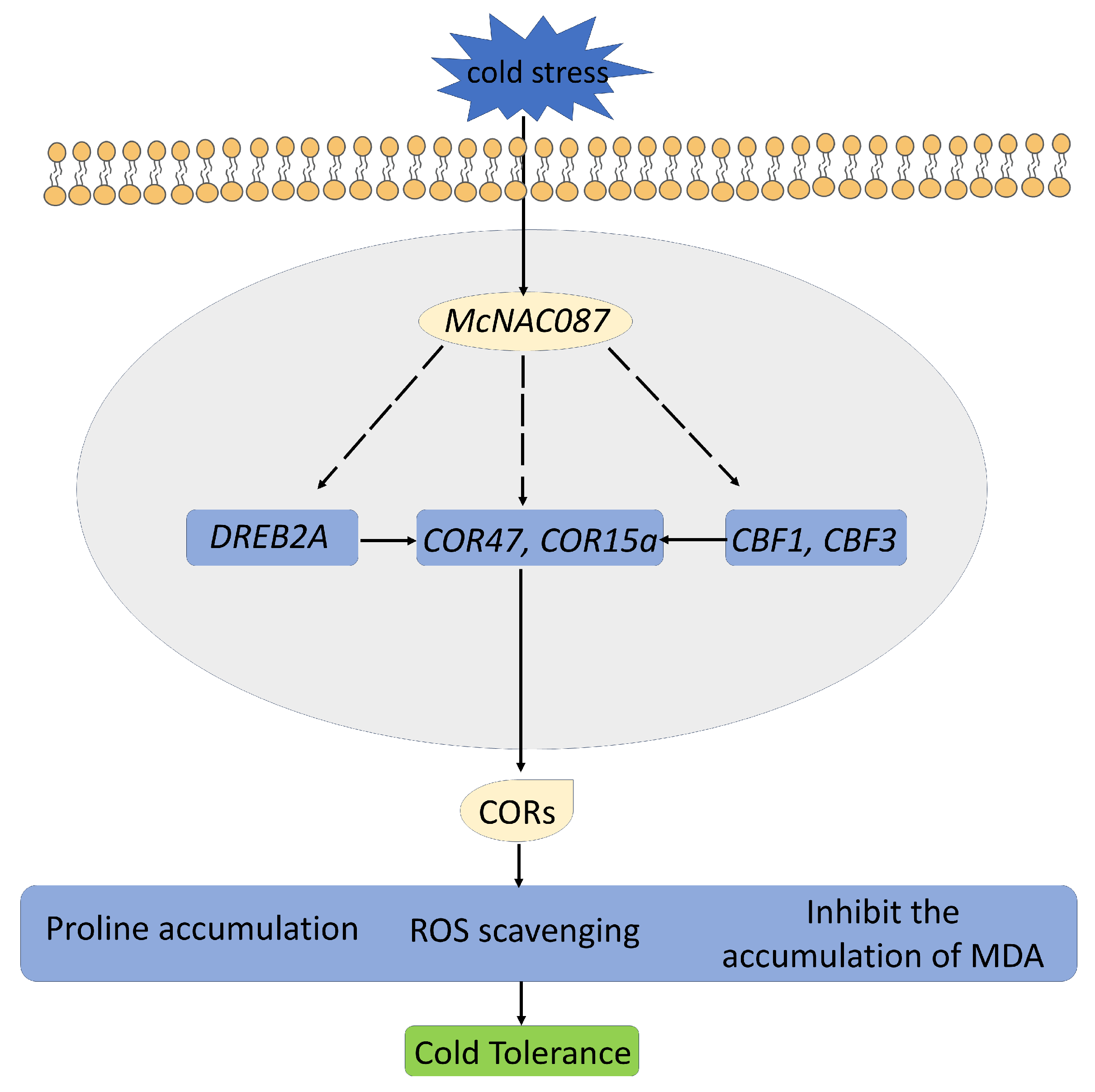
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growing Conditions, and Cold Stress Treatment
4.2. RNA Isolation and Transcriptome Sequencing
4.3. Identification of Differentially Expressed Genes and Bioinformatic Analysis
4.4. Arabidopsis Transformation and Cold Stress Tolerance Evaluation
4.5. RNA Extraction and Quantitative Real-Time PCR Analysis
4.6. Determination of Physiological Parameters
4.7. Subcellular Localization Analysis of McNAC087
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| TF | Transcription factor |
| COR | Cold-responsive |
| rRNA | Ribosomal RNA |
| FPKM | Fragment per kilobase of transcript per million mapped reads |
| RSEM | RNA-seq by expectation maximization |
| DEGs | Differentially expressed genes |
| GO | Gene ontology |
| WT | Wild-type |
| DAB | 3,3′-diaminobenzidine |
| NBT | Nitroblue tetrazolium |
| MDA | Malondialdehyde |
| POD | Peroxidase |
| SOD | Superoxide dismutase |
| CAT | Catalase |
| GFP | Green fluorescent protein |
| PEG | Polyethylene glycol |
| PCA | Principal Component Analysis |
| ROS | Reactive oxygen species |
References
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Chen, S. Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef]
- Diao, P.; Chen, C.; Zhang, Y.; Meng, Q.; Lv, W.; Ma, N. The role of NAC transcription factor in plant cold response. Plant Signal. Behav. 2020, 15, 1785668. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.E.; Li, Y.; Labbe, A.; Guevara, D.; Nuin, P.; Whitty, B.; Diaz, C.; Golding, G.B.; Gray, G.R.; Weretilnyk, E.A.; et al. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 2006, 140, 1437–1450. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Z.; Jin, Y.N.; Ding, X.H.; Wang, W.J.; Zhai, S.S.; Bai, L.P.; Guo, Z.F. Gene regulation and signal transduction in the ICE-CBF-COR signaling pathway during cold stress in plants. Biochemistry 2017, 82, 1103–1117. [Google Scholar] [CrossRef]
- Yu, H.; Kong, X.; Huang, H.; Wu, W.; Park, J.; Yun, D.J.; Lee, B.H.; Shi, H.; Zhu, J.K. STCH4/REIL2 confers cold stress tolerance in Arabidopsis by promoting rRNA processing and CBF protein translation. Cell Rep. 2020, 30, 229–242.e225. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Tang, J.; Wang, S.; Cheng, X.; Wang, H.; Ou, S.; Gao, S.; Li, B.; Qian, Y.; Gao, C.; et al. Antagonistic control of seed dormancy in rice by two bHLH transcription factors. Nat. Genet. 2022, 54, 1972–1982. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Han, K.; Zhao, Y.; Sun, Y.; Li, Y. NACs, generalist in plant life. Plant Biotechnol. J. 2023, 21, 2433–2457. [Google Scholar] [CrossRef]
- Xiong, H.; He, H.; Chang, Y.; Miao, B.; Liu, Z.; Wang, Q.; Dong, F.; Xiong, L. Multiple roles of NAC transcription factors in plant development and stress responses. J. Integr. Plant Biol. 2025, 67, 510–538. [Google Scholar] [CrossRef]
- Wang, T.; Ma, X.; Chen, Y.; Wang, C.; Xia, Z.; Liu, Z.; Gao, L.; Zhang, W. SlNAC3 suppresses cold tolerance in tomatoes by enhancing ethylene biosynthesis. Plant Cell Environ. 2024, 47, 3132–3146. [Google Scholar] [CrossRef]
- Li, R.; Song, Y.; Wang, X.; Zheng, C.; Liu, B.; Zhang, H.; Ke, J.; Wu, X.; Wu, L.; Yang, R.; et al. OsNAC5 orchestrates OsABI5 to fine-tune cold tolerance in rice. J. Integr. Plant Biol. 2024, 66, 660–682. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K.; et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ. 2014, 37, 2116–2127. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Liu, Z.; He, H.; Han, X.; Qi, Z.; Yang, Y. Gene expression and metabolic changes of Momordica charantia L. seedlings in response to low temperature stress. PLoS ONE 2020, 15, e0233130. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef]
- Hou, X.M.; Zhang, H.F.; Liu, S.Y.; Wang, X.K.; Zhang, Y.M.; Meng, Y.C.; Luo, D.; Chen, R.G. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef] [PubMed]
- Dorjee, T.; Cui, Y.; Zhang, Y.; Liu, Q.; Li, X.; Sumbur, B.; Yan, H.; Bing, J.; Geng, Y.; Zhou, Y.; et al. Characterization of NAC gene family in Ammopiptanthus mongolicus and functional analysis of AmNAC24, an osmotic and cold-stress-induced NAC gene. Biomolecules 2024, 14, 182. [Google Scholar] [CrossRef]
- Huang, L.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Ma, F.; Huang, D.; Xing, W.; Wu, B.; Sun, P.; Xu, B.; Song, S. Characterization of the NAC transcription factor in Passion Fruit (Passiflora edulis) and functional identification of PeNAC-19 in cold stress. Plants 2023, 12, 1393. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Chang, Y.; Chu, G.; Wang, M. Low-temperature tolerance and transcriptome analyses during seed germination of Anabasis aphylla. J. Plant Interact. 2019, 14, 254–264. [Google Scholar] [CrossRef]
- Barrero-Gil, J.; Huertas, R.; Rambla, J.L.; Granell, A.; Salinas, J. Tomato plants increase their tolerance to low temperature in a chilling acclimation process entailing comprehensive transcriptional and metabolic adjustments. Plant Cell Environ. 2016, 39, 2303–2318. [Google Scholar] [CrossRef]
- Li, S.; Yang, Y.; Zhang, Q.; Liu, N.; Xu, Q.; Hu, L. Differential physiological and metabolic response to low temperature in two zoysiagrass genotypes native to high and low latitude. PLoS ONE 2018, 13, e0198885. [Google Scholar] [CrossRef]
- Liu, A.; Hu, Z.; Bi, A.; Fan, J.; Gitau, M.M.; Amombo, E.; Chen, L.; Fu, J. Photosynthesis, antioxidant system and gene expression of bermudagrass in response to low temperature and salt stress. Ecotoxicology 2016, 25, 1445–1457. [Google Scholar] [CrossRef]
- Khedr, A.H.; Abbas, M.A.; Wahid, A.A.; Quick, W.P.; Abogadallah, G.M. Proline induces the expression of salt-stress-responsive proteins and may improve the adaptation of Pancratium maritimum L. to salt-stress. J. Exp. Bot. 2003, 54, 2553–2562. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z.; et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signaling in rice. Plant Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Ma, K.; Zhao, Y.; Mei, C.; Mamat, A.; Wang, J.; Qin, L.; He, T. The cold-stress responsive gene DREB1A involved in low-temperature tolerance in Xinjiang wild walnut. PeerJ 2022, 10, e14021. [Google Scholar] [CrossRef]
- Liu, W.; Wang, T.; Wang, Y.; Liang, X.; Han, J.; Han, D. MbMYBC1, a M. baccata MYB transcription factor, contribute to cold and drought stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 2023, 14, 1141446. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liang, X.; Cai, W.; Wang, H.; Liu, X.; Cheng, L.; Song, P.; Luo, G.; Han, D. Isolation and functional analysis of VvWRKY28, a Vitis vinifera WRKY transcription factor gene, with functions in tolerance to cold and salt stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 3418. [Google Scholar] [CrossRef]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Sample | Total Raw Reads | Total Clean Reads (%) | Total Clean Bases | Clean Reads Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|
| RM-1 | 40,018,692 | 39,893,848 (99.69%) | 5,892,853,624 | 5,735,816,993 (97.34%) | 2,721,951,784 (46.19%) |
| RM-2 | 42,835,104 | 42,663,970 (99.60%) | 6,294,026,844 | 6,122,549,196 (97.28%) | 2,904,925,895 (46.15%) |
| RM-3 | 39,476,622 | 39,366,156 (99.72%) | 5,810,570,919 | 5,671,334,032 (97.60%) | 2,686,534,839 (46.24%) |
| RL1-1 | 40,893,740 | 40,743,182 (99.63%) | 6,022,858,080 | 5,824,480,038 (96.71%) | 2,745,037,914 (45.58%) |
| RL1-2 | 48,058,064 | 47,896,750 (99.66%) | 7,073,938,218 | 6,849,945,058 (96.83%) | 3,226,081,644 (45.61%) |
| RL1-3 | 39,433,994 | 39,265,546 (99.57%) | 5,830,950,164 | 5,654,256,539 (96.97%) | 2,681,993,642 (46.00%) |
| RL2-1 | 37,975,582 | 37,825,924 (99.61%) | 5,597,834,381 | 5,444,217,924 (97.26%) | 2,546,110,109 (45.48%) |
| RL2-2 | 43,237,410 | 43,027,214 (99.51%) | 6,367,927,150 | 6,162,603,972 (96.78%) | 2,882,536,185 (45.27%) |
| RL2-3 | 42,298,414 | 42,158,916 (99.67%) | 6,270,987,686 | 6,109,302,688 (97.42%) | 2,862,340,959 (45.64%) |
| RL3-1 | 41,215,956 | 41,046,658 (99.59%) | 6,039,075,711 | 5,864,527,892 (97.11%) | 2,803,041,196 (46.42%) |
| RL3-2 | 36,423,022 | 36,315,076 (99.70%) | 5,346,884,016 | 5,211,124,486 (97.46%) | 2,483,918,080 (46.46%) |
| RL3-3 | 45,698,842 | 45,556,060 (99.69%) | 6,708,210,242 | 6,539,270,593 (97.48%) | 3,116,849,459 (46.46%) |
| SM-1 | 41,163,672 | 41,035,338 (99.69%) | 6,066,036,632 | 5,911,629,664 (97.45%) | 2,810,644,738 (46.33%) |
| SM-2 | 40,644,408 | 40,503,714 (99.65%) | 5,989,237,980 | 5,836,799,898 (97.45%) | 2,777,363,713 (46.37%) |
| SM-3 | 38,901,736 | 38,791,550 (99.72%) | 5,733,378,673 | 5,595,459,985 (97.59%) | 2,656,090,816 (46.33%) |
| SL1-1 | 44,059,370 | 43,907,724 (99.66%) | 6,508,407,863 | 6,302,246,629 (96.83%) | 2,931,273,138 (45.04%) |
| SL1-2 | 43,099,768 | 42,949,204 (99.65%) | 6,353,584,714 | 6,184,131,820 (97.33%) | 2,907,200,197 (45.76%) |
| SL1-3 | 39,855,366 | 39,681,250 (99.56%) | 5,881,590,044 | 5,693,476,583 (96.80%) | 2,666,575,152 (45.34%) |
| SL2-1 | 42,366,614 | 42,212,362 (99.64%) | 6,236,239,419 | 6,076,107,478 (97.43%) | 2,846,297,902 (45.64%) |
| SL2-2 | 44,085,818 | 43,874,656 (99.52%) | 6,486,341,791 | 6,278,963,701 (96.80%) | 2,938,327,609 (45.30%) |
| SL2-3 | 43,235,248 | 43,092,714 (99.67%) | 6,380,155,079 | 6,217,591,803 (97.45%) | 2,915,883,323 (45.70%) |
| SL3-1 | 45,315,638 | 45,165,830 (99.67%) | 6,686,262,990 | 6,520,401,992 (97.52%) | 3,074,245,023 (45.98%) |
| SL3-2 | 38,973,144 | 38,818,152 (99.60%) | 5,753,672,630 | 5,581,677,280 (97.01%) | 2,647,618,786 (46.02%) |
| SL3-3 | 40,610,978 | 40,486,400 (99.69%) | 5,970,974,264 | 5,823,732,363 (97.53%) | 2,746,837,661 (46.00%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Wang, K.; Guan, F.; Shi, B.; Xie, Y.; Du, C.; Tang, T.; Yang, Z.; Ma, S.; Wan, X. The Bitter Gourd Transcription Factor McNAC087 Confers Cold Resistance in Transgenic Arabidopsis. Plants 2025, 14, 3440. https://doi.org/10.3390/plants14223440
Yang X, Wang K, Guan F, Shi B, Xie Y, Du C, Tang T, Yang Z, Ma S, Wan X. The Bitter Gourd Transcription Factor McNAC087 Confers Cold Resistance in Transgenic Arabidopsis. Plants. 2025; 14(22):3440. https://doi.org/10.3390/plants14223440
Chicago/Turabian StyleYang, Xuetong, Kai Wang, Feng Guan, Bo Shi, Yuanyuan Xie, Chang Du, Tong Tang, Zheng Yang, Shijie Ma, and Xinjian Wan. 2025. "The Bitter Gourd Transcription Factor McNAC087 Confers Cold Resistance in Transgenic Arabidopsis" Plants 14, no. 22: 3440. https://doi.org/10.3390/plants14223440
APA StyleYang, X., Wang, K., Guan, F., Shi, B., Xie, Y., Du, C., Tang, T., Yang, Z., Ma, S., & Wan, X. (2025). The Bitter Gourd Transcription Factor McNAC087 Confers Cold Resistance in Transgenic Arabidopsis. Plants, 14(22), 3440. https://doi.org/10.3390/plants14223440






