Identification of Stable Meta-QTLs and Candidate Genes Underlying Fiber Quality and Agronomic Traits in Cotton
Abstract
1. Introduction
2. Results
2.1. Identification of QTLs and Their Distribution Across the Cotton Genome
2.2. QTLs Linked to Various Traits and Their Distribution Across the Cotton Genome
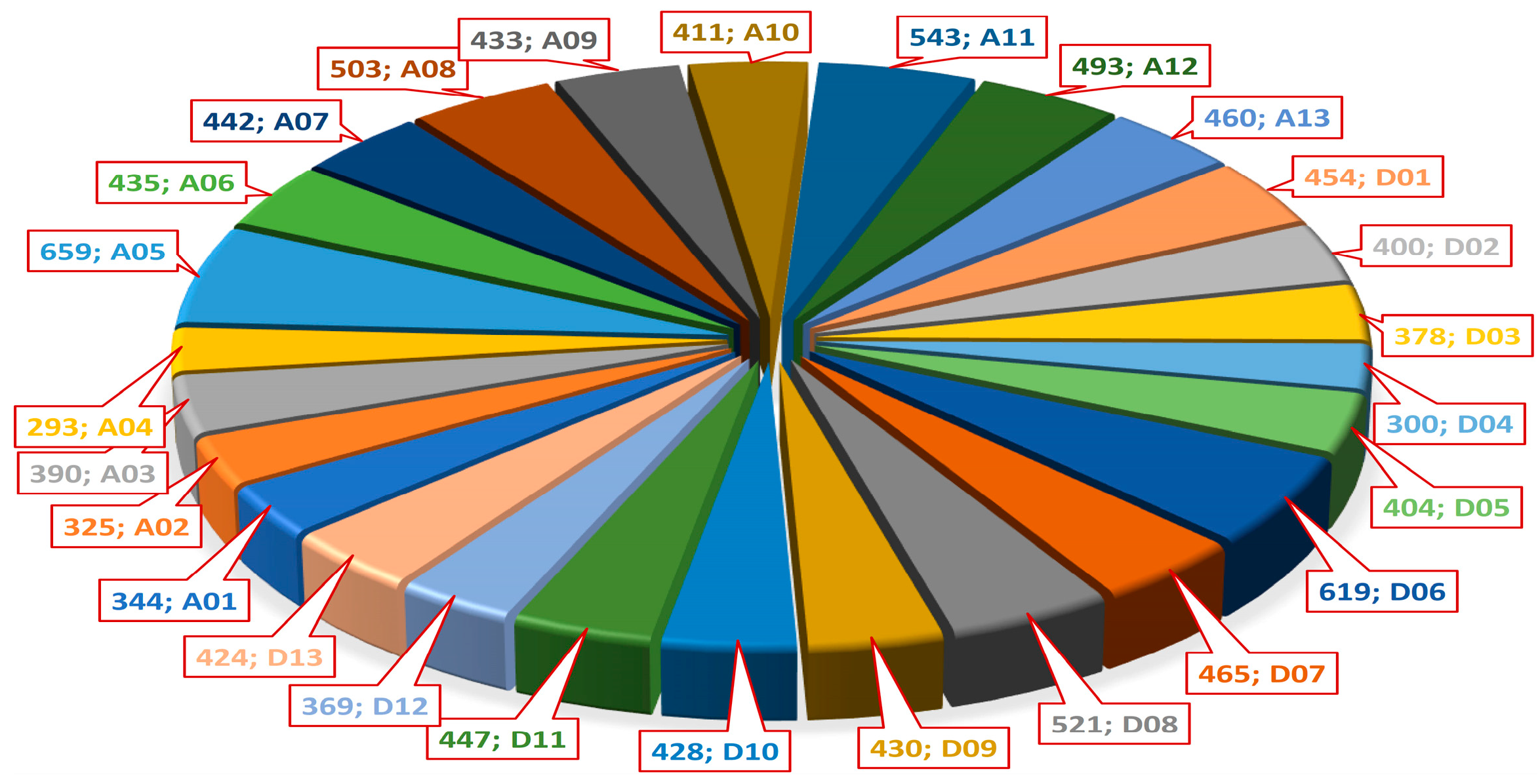
2.3. Meta-QTLs (QTL Clusters) and Their Distribution Across the Genome
2.4. Candidate Gene Identification
2.5. Gene Ontology and KEGG Analysis
3. Discussion
4. Materials and Methods
4.1. Collection of QTL Data
- (a)
- Possession of a well-defined confidence interval (CI) with clear start and stop positions.
- (b)
- An LOD score of ≥2.0.
- (c)
- The value of phenotypic variation (PVE or R2) is indicated.
4.2. Software for MQTL Analysis
4.3. Consensus Map Construction
- (a)
- The maximum interval for each linkage group was set to 1:4.
- (b)
- The accuracy of potential consensus maps was evaluated by comparing their root mean square error (RMSE) values.
- (c)
- The consensus map with the smallest average RMSE value was selected for subsequent analyses.
4.4. QTL Projection and MQTL Identification
4.5. Identification of Candidate Genes
4.6. Gene Ontology Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Safiullina, A.K.; Ernazarova, D.K.; Turaev, O.S.; Rafieva, F.U.; Ernazarova, Z.A.; Arslanova, S.K.; Toshpulatov, A.K.; Oripova, B.B.; Kudratova, M.K.; Khalikov, K.K.; et al. Genetic Diversity and Subspecific Races of Upland Cotton (Gossypium hirsutum L.). Genes 2024, 15, 1533. [Google Scholar] [CrossRef]
- Khidirov, M.T.; Ernazarova, D.K.; Rafieva, F.U.; Ernazarova, Z.A.; Toshpulatov, A.K.; Umarov, R.F.; Kholova, M.D.; Oripova, B.B.; Kudratova, M.K.; Gapparov, B.M.; et al. Genomic and cytogenetic analysis of synthetic polyploids between diploid and tetraploid cotton (Gossypium) species. Plants 2023, 12, 4184. [Google Scholar] [CrossRef]
- Arslanova, S.K.; Ernazarova, Z.A.; Ernazarova, D.K.; Turaev, O.S.; Safiullina, A.K.; Toshpulatov, A.K.; Kholova, M.D.; Azimova, L.A.; Rafiyeva, F.U.; Gapparov, B.M.; et al. Development and Characterization of Synthetic Allotetraploids between Diploid Species Gossypium herbaceum and Gossypium nelsonii for Cotton Genetic Improvement. Plants 2025, 14, 1620. [Google Scholar] [CrossRef]
- Kushanov, F.N.; Komilov, D.J.; Turaev, O.S.; Ernazarova, D.K.; Amanboyeva, R.S.; Gapparov, B.M.; Yu, J.Z. Genetic analysis of mutagenesis that induces the photoperiod insensitivity of wild cotton Gossypium hirsutum subsp. purpurascens. Plants 2022, 11, 3012. [Google Scholar] [CrossRef]
- Umedova, M.E.; Turaev, O.S.; Komilov, D.J.; Amanboyeva, R.S.; Kholova, M.D.; Norov, T.M.; Ernazarova, D.K.; Kushanov, F.N.; Seytnazarova, T.Y.; Rakhmankulov, M.S. Bibliometric analysis of the past research based on mas technology cotton improvement. SABRAO J. Breed. Genet. 2024, 56, 988–1000. [Google Scholar] [CrossRef]
- Rong, J.; Feltus, E.A.; Waghmare, V.N.; Pierce, G.J.; Chee, P.W.; Draye, X.; Saranga, Y.; Wright, R.J.; Wilkins, T.A.; May, O.L.; et al. Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics 2007, 176, 2577–2588. [Google Scholar] [CrossRef] [PubMed]
- Lacape, J.M.; Llewellyn, D.; Jacobs, J.; Arioli, T.; Becker, D.; Calhoun, S.; Al-Ghazi, Y.; Liu ShiMing, L.S.; Palai, O.; Georges, S.; et al. Meta-analysis of cotton fiber quality QTLs across diverse environments in a Gossypium hirsutum × G. barbadense RIL population. BMC Plant Biol. 2010, 10, 132. [Google Scholar] [CrossRef]
- Wang, B.; Guo, W.; Zhu, X.; Wu, Y.; Huang, N.; Zhang, T. QTL mapping of fiber quality in an elite hybrid derived-RIL population of upland cotton. Euphytica 2006, 152, 367–378. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, J.; Chen, X.; Liu, D.J.; Zhang, Z. QTL mapping of fiber quality traits with a composite cross population in upland cotton (Gossypium hirsutum L.). J. Agric. Biotechnol. 2011, 19, 230–235. [Google Scholar]
- Sun, F.D.; Zhang, J.H.; Wang, S.F.; Gong, W.K.; Shi, Y.Z.; Liu, A.Y.; Yuan, Y.L. QTL mapping for fiber quality traits across multiple generations and environments in upland cotton. Mol. Breed. 2012, 30, 569–582. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, J.; Ma, J.; Tang, S.; Liu, D.; Teng, Z.; Zhang, Z. Genetic mapping and quantitative trait locus analysis of fiber quality traits using a three-parent composite population in upland cotton (Gossypium hirsutum L.). Mol. Breed. 2012, 29, 335–348. [Google Scholar] [CrossRef]
- Liang, Q.; Hu, C.; Hua, H.; Li, Z.; Hua, J. Construction of a linkage map and QTL mapping for fiber quality traits in upland cotton (Gossypium hirsutum L.). Chin. Sci. Bull. 2013, 58, 3233–3243. [Google Scholar] [CrossRef]
- Tang, S.; Teng, Z.; Zhai, T.; Fang, X.; Liu, F.; Liu, D.; Zhang, Z. Construction of genetic map and QTL analysis of fiber quality traits for upland cotton (Gossypium hirsutum L.). Euphytica 2015, 201, 195–213. [Google Scholar] [CrossRef]
- Yang, X.; Zhou, X.; Wang, X.; Li, Z.; Zhang, Y.; Liu, H.; Ma, Z. Mapping QTL for cotton fiber quality traits using simple sequence repeat markers, conserved intron-scanning primers, and transcript-derived fragments. Euphytica 2015, 201, 215–230. [Google Scholar] [CrossRef]
- Tan, Z.; Fang, X.; Tang, S.; Zhang, J.; Liu, D.; Teng, Z.; Zhang, Z. Genetic map and QTL controlling fiber quality traits in upland cotton (Gossypium hirsutum L.). Euphytica 2015, 203, 615–628. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Muhammad, J.; Cai, J.; Jia, F.; Shi, Y.; Yuan, Y. High resolution consensus mapping of quantitative trait loci for fiber strength, length and micronaire on chromosome 25 of upland cotton (Gossypium hirsutum L.). PLoS ONE 2015, 10, e0135430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.W.; Zhu, X.F.; Feng, L.C.; Gao, X.; Yang, B.; Zhang, T.Z.; Zhou, B.L. Mapping of fiber quality QTLs reveals useful variation and footprints of cotton domestication using introgression lines. Sci. Rep. 2016, 6, 31954. [Google Scholar] [CrossRef]
- Ma, L.; Zhao, Y.; Wang, Y.; Shang, L.; Hua, J. QTLs analysis and validation for fiber quality traits using maternal backcross population in upland cotton. Front. Plant Sci. 2017, 8, 2168. [Google Scholar] [CrossRef]
- Ma, L.; Su, Y.; Nie, H.; Cui, Y.; Cheng, C.; Ijaz, B.; Hua, J. QTL and genetic analysis controlling fiber quality traits using paternal backcross population in upland cotton. J. Cotton Res. 2020, 3, 22. [Google Scholar] [CrossRef]
- Wang, X.Q.; Yu, Y.Y.; Sang, J.; Wu, Q.Z.; Zhang, X.L.; Lin, Z.X. Intraspecific linkage map construction and QTL mapping of yield and fiber quality of Gossypium barbadense. Aust. J. Crop Sci. 2013, 7, 1252–1261. [Google Scholar] [CrossRef]
- Zhang, T.; Qian, N.; Zhu, X.; Chen, H.; Wang, S.; Mei, H.; Zhang, Y. Variations and transmission of QTL alleles for yield and fiber qualities in upland cotton cultivars developed in China. PLoS ONE 2013, 8, e57220. [Google Scholar] [CrossRef]
- Yu, J.Z.; Ulloa, M.; Hoffman, S.M.; Kohel, R.J.; Pepper, A.E.; Fang, D.D.; Burke, J.J. Mapping genomic loci for cotton plant architecture, yield components, and fiber properties in an interspecific (Gossypium hirsutum L. × G. barbadense L.) RIL population. Mol. Genet. Genom. 2014, 289, 1347–1367. [Google Scholar] [CrossRef]
- Liu, D.; Liu, F.; Shan, X.; Zhang, J.; Tang, S.; Fang, X.; Liu, D. Construction of a high-density genetic map and lint percentage and cottonseed nutrient trait QTL identification in upland cotton (Gossypium hirsutum L.). Mol. Genet. Genom. 2015, 290, 1683–1700. [Google Scholar] [CrossRef]
- Wang, H.; Huang, C.; Guo, H.; Li, X.; Zhao, W.; Dai, B.; Lin, Z. QTL mapping for fiber and yield traits in upland cotton under multiple environments. PLoS ONE 2015, 10, e0130742. [Google Scholar] [CrossRef] [PubMed]
- Kushanov, F.N.; Buriev, Z.T.; Shermatov, S.E.; Turaev, O.S.; Norov, T.M.; Pepper, A.E.; Abdurakhmonov, I.Y. QTL mapping for flowering-time and photoperiod insensitivity of cotton Gossypium darwinii Watt. PLoS ONE 2017, 12, e0186240. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Teng, Z.; Wang, J.; Wu, T.; Zhang, Z.; Deng, X.; Zhang, Z. Enriching an intraspecific genetic map and identifying QTL for fiber quality and yield component traits across multiple environments in upland cotton (Gossypium hirsutum L.). Mol. Genet. Genom. 2017, 292, 1281–1306. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, A.; Li, J.; Zhang, J.; Zhang, B.; Ge, Q.; Yuan, Y. Dissecting the genetic basis of fiber quality and yield traits in inter-specific backcross populations of Gossypium hirsutum × Gossypium barbadense. Mol. Genet. Genom. 2019, 294, 1385–1402. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pan, Z.; He, S.; Jia, Y.; Geng, X.; Chen, B.; Du, X. QTL mapping of agronomic and economic traits for four F2 populations of upland cotton. J. Cotton Res. 2021, 4, 3. [Google Scholar] [CrossRef]
- Oluoch, G.; Zheng, J.; Wang, X.; Khan, M.K.R.; Zhou, Z.; Cai, X.; Wang, K. QTL mapping for salt tolerance at seedling stage in the interspecific cross of Gossypium tomentosum with Gossypium hirsutum. Euphytica 2016, 209, 223–235. [Google Scholar] [CrossRef]
- Diouf, L.; Pan, Z.; He, S.P.; Gong, W.F.; Jia, Y.H.; Magwanga, R.O.; Du, X. High-density linkage map construction and mapping of salt-tolerant QTLs at seedling stage in upland cotton using genotyping by sequencing (GBS). Int. J. Mol. Sci. 2017, 18, 2622. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Thyssen, G.N.; Fang, D.D.; Jenkins, J.N.; McCarty, J.C.; Wedegaertner, T.; Zhang, J. GWAS reveals consistent QTL for drought and salt tolerance in a MAGIC population of 550 lines derived from intermating of 11 upland cotton (Gossypium hirsutum) parents. Mol. Genet. Genom. 2021, 296, 119–129. [Google Scholar] [CrossRef]
- Guo, A.H.; Su, Y.; Huang, Y.; Wang, Y.M.; Nie, H.S.; Zhao, N.; Hua, J.P. QTL controlling fiber quality traits under salt stress in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 2021, 134, 661–685. [Google Scholar] [CrossRef]
- Guo, A.; Su, Y.; Nie, H.; Li, B.; Ma, X.; Hua, J. Identification of candidate genes involved in salt stress response at germination and seedling stages by QTL mapping in upland cotton. G3 Genes Genomes Genet. 2022, 12, jkac099. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhao, J.; Zhou, L.; Guo, W.; Zhang, T. Molecular mapping of Verticillium wilt resistance QTL clustered on chromosomes D7 and D9 in upland cotton. Sci. China Life Sci. 2009, 52, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Sattar, M.N.; Javed, M.; Hussain, S.B.; Babar, M.; Chee, P.W.; Iqbal, Z.; Al-Hashedi, S.A. Mapping of quantitative trait loci (QTLs) controlling cotton leaf curl disease (CLCuD) resistance in Upland cotton. Plant Breed. 2023, 142, 247–257. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Zhu, Y.; Zhang, J. Quantitative trait locus mapping for Fusarium wilt race 4 resistance in a recombinant inbred line population of Pima cotton (Gossypium barbadense). Pathogens 2022, 11, 1143. [Google Scholar] [CrossRef] [PubMed]
- Goffinet, B.; Gerber, S. Quantitative trait loci: A meta-analysis. Genetics 2000, 155, 463–473. [Google Scholar] [CrossRef]
- Veyrieras, J.B.; Goffinet, B.; Charcosset, A. MetaQTL: A package of new computational methods for the meta-analysis of QTL mapping experiments. BMC Bioinform. 2007, 8, 49. [Google Scholar] [CrossRef]
- Said, J.I.; Lin, Z.; Zhang, X.; Song, M.; Zhang, J. A comprehensive meta-QTL analysis for fiber quality, yield, yield-related and morphological traits, drought tolerance, and disease resistance in tetraploid cotton. BMC Genom. 2013, 14, 766. [Google Scholar] [CrossRef]
- Gupta, M.; Choudhary, M.; Singh, A.; Sheoran, S.; Singla, D.; Rakshit, S. Meta-QTL analysis for mining of candidate genes and constitutive gene network development for fungal disease resistance in maize (Zea mays L.). Crop J. 2023, 11, 511–522. [Google Scholar] [CrossRef]
- Wang, W.; Ren, Z.; Li, L.; Du, Y.; Zhou, Y.; Zhang, M.; Li, Z.; Yi, F.; Duan, L. Meta-QTL analysis explores the key genes, especially hormone-related genes, involved in the regulation of grain water content and grain dehydration rate in maize. BMC Plant Biol. 2022, 22, 346. [Google Scholar] [CrossRef]
- Saini, D.K.; Srivastava, P.; Pal, N.; Araus, J.L.; Parida, S.K.; Gupta, P.K. Meta-QTLs, ortho-meta-QTLs and candidate genes for grain yield and associated traits in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2022, 135, 1049–1081. [Google Scholar] [CrossRef] [PubMed]
- Khahani, B.; Tavakol, E.; Shariati, V.; Rossini, L. Meta-QTL and ortho-MQTL analyses identified genomic regions controlling rice yield, yield-related traits, and root architecture under water deficit conditions. Sci. Rep. 2021, 11, 6942. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.; Soe, Y.P.; Palanog, A.; Hore, T.K.; Nha, C.T.; Calayugan, M.I.; Inabangan-Asilo, M.A.; Amparado, A.; Pandey, I.D.; Cruz, P.C.S.; et al. Meta-QTLs and haplotypes for efficient zinc biofortification of rice. Plant Genome 2023, 16, e20315. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Wu, J.; Islam, M.S.; Sun, C.; Lu, B.; Wei, P.; Liu, D.; Chen, C. Genome-wide meta-analysis of QTL for morphological related traits of flag leaf in bread wheat. PLoS ONE 2022, 17, e0276602. [Google Scholar] [CrossRef]
- Vasconcellos, R.C.; Oraguzie, O.B.; Soler, A.; Arkwazee, H.; Myers, J.R.; Ferreira, J.J.; Song, Q. Meta-QTL for resistance to white mold in common bean. PLoS ONE 2017, 2, e0171685. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Liu, F.; Song, M.; Zhang, J.F. A meta-analysis of quantitative trait loci for abiotic and biotic stress resistance in tetraploid cotton. Mol. Genet. Genom. 2017, 292, 1221–1235. [Google Scholar] [CrossRef]
- Guo, Y.; McCarty, J.C.; Jenkins, J.N.; Saha, S. QTLs for node of first fruiting branch in a cross of an upland cotton, Gossypium hirsutum L., cultivar with primitive accession Texas 701. Euphytica 2008, 163, 113–122. [Google Scholar] [CrossRef]
- Zhiyuan, N.; Chen, H.; Mei, H.; Zhang, T. Molecular tagging of QTLs for fiber quality and yield in the upland cotton cultivar Acala-Prema. Euphytica 2014, 195, 143–156. [Google Scholar] [CrossRef]
- Wang, M.; Li, C.; Wang, Q. Quantitative trait loci mapping and genetic dissection for lint percentage in upland cotton (Gossypium hirsutum). J. Genet. 2014, 93, 371–378. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, C.; Liu, G.; Chen, Y.; Zhang, J.; Qiao, Q.; Zhang, J. Phenotypic variation analysis and QTL mapping for cotton (Gossypium hirsutum L.) fiber quality grown in different cotton-producing regions. Euphytica 2016, 211, 169–183. [Google Scholar] [CrossRef]
- Shang, L.; Liu, F.; Wang, Y.; Abduweli, A.; Cai, S.; Wang, K.; Hua, J. Dynamic QTL mapping for plant height in Upland cotton (Gossypium hirsutum). Plant Breed. 2015, 134, 703–712. [Google Scholar] [CrossRef]
- Sankeshwar, M.; Jadhav, M.P.; Adiger, S.; Patil, R.S.; Katageri, I.S. Mapping of QTLs for traits related to leaf pubescence, jassid resistance and yield in cotton (Gossypium spp.). Indian J. Genet. Plant Breed. 2018, 78, 252–260. [Google Scholar] [CrossRef]
- Liu, X.F.; Li, J.W.; Yu, X.K.; Shi, Y.Z.; Jia, F.; Sun, D.F.; Yuan, Y.L. Identification of QTL for cottonseed oil and protein content in Upland cotton (Gossypium hirsutum L.) based on a RIL population. Mol. Plant Breed. 2013, 11, 520–528. [Google Scholar]
- Yu, J.; Zhang, K.; Li, S.; Yu, S.; Zhai, H.; Wu, M.; Zhang, J. Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Theor. Appl. Genet. 2013, 126, 275–287. [Google Scholar] [CrossRef]
- Zhe, X.I.A.; Zhang, X.; Liu, Y.Y.; Jia, Z.F.; Zhao, H.H.; Li, C.Q.; Wang, Q.L. Major gene identification and quantitative trait locus mapping for yield-related traits in upland cotton (Gossypium hirsutum L.). J. Integr. Agric. 2014, 13, 299–309. [Google Scholar] [CrossRef]
- Shang, L.; Cai, S.; Ma, L.; Wang, Y.; Abduweli, A.; Wang, M.; Hua, J. Seedling root QTLs analysis on dynamic development and upon nitrogen deficiency stress in Upland cotton. Euphytica 2016, 207, 645–663. [Google Scholar] [CrossRef]
- Oin, Y.; Liu, R.; Mei, H.; Zhang, T.; Guo, W. QTL mapping for yield traits in Upland cotton (Gossypium hirsutum L.). Acta Agron. Sin. 2009, 35, 1812–1821. [Google Scholar] [CrossRef]
- Zheng, J.Y.; Oluoch, G.; Khan, M.K.R.; Wang, X.X.; Cai, X.Y.; Zhou, Z.L.; Wang, K.B. Mapping QTLs for drought tolerance in an F2:3 population from an inter-specific cross between Gossypium tomentosum and Gossypium hirsutum. Genet. Mol. Res. 2016, 15, 1–14. [Google Scholar] [CrossRef]
- Song, W.; Wang, M.; Su, W.; Lu, Q.; Xiao, X.; Cai, J.; Yuan, Y. Genetic and phenotypic effects of chromosome segments introgressed from Gossypium barbadense into Gossypium hirsutum. PLoS ONE 2017, 12, e0184882. [Google Scholar] [CrossRef]
- Rani, S.; Baber, M.; Naqqash, T.; Malik, S.A. Identification and genetic mapping of potential QTLs conferring heat tolerance in cotton (Gossypium hirsutum L.) by using microsatellite markers approach. Agronomy 2022, 12, 1381. [Google Scholar] [CrossRef]
- Lu, Q.; Li, P.; Yang, R.; Xiao, X.; Li, Z.; Wu, Q.; Yuan, Y. QTL mapping and candidate gene prediction for fiber yield and quality traits in a high-generation cotton chromosome substitution line with Gossypium barbadense segments. Mol. Genet. Genom. 2022, 297, 287–301. [Google Scholar] [CrossRef]
- Shukla, R.P.; Tiwari, G.J.; Joshi, B.; Song-Beng, K.; Tamta, S.; Boopathi, N.M.; Jena, S.N. GBS-SNP and SSR based genetic mapping and QTL analysis for drought tolerance in upland cotton. Physiol. Mol. Biol. Plants 2021, 27, 1731–1745. [Google Scholar] [CrossRef]
- Si, Z.; Jin, S.; Chen, J.; Wang, S.; Fang, L.; Zhu, X.; Hu, Y. Construction of a high-density genetic map and identification of QTLs related to agronomic and physiological traits in an interspecific (Gossypium hirsutum × Gossypium barbadense) F2 population. BMC Genom. 2022, 23, 307. [Google Scholar] [CrossRef]
- Diouf, L.; Magwanga, R.O.; Gong, W.; He, S.; Pan, Z.; Jia, Y.H.; Kirungu, J.N.; Du, X. QTL Mapping of Fiber quality and Yield-Related Traits in an Intra-Specific Upland Cotton Using Genotype by Sequencing (GBS). Int. J. Mol. Sci. 2018, 19, 441. [Google Scholar] [CrossRef] [PubMed]
- Gowda, S.A.; Katageri, I.S.; Patil, R.S.; Kumar, P.S.; Tiwari, G.J.; Jena, S.N.; Sawant, S.V. 63 K and 50 K SNP array based high-density genetic mapping and QTL analysis for productivity and fiber quality traits in cotton. Euphytica 2022, 218, 93. [Google Scholar] [CrossRef]
- Jamshed, M.; Jia, F.; Gong, J.; Palanga, K.K.; Shi, Y.; Li, J.; Shang, H.; Liu, A.; Chen, T.; Zhang, Z.; et al. Identification of stable quantitative trait loci (QTLs) for fiber quality traits across multiple environments in Gossypium hirsutum recombinant inbred line population. BMC Genom. 2016, 17, 197. [Google Scholar] [CrossRef]
- Jia, X.; Pang, C.; Wei, H.; Wang, H.; Ma, Q.; Yang, J.; Cheng, S.; Su, J.; Fan, S.; Song, M.; et al. High-density linkage map construction and qTL analysis for earliness-related traits in Gossypium hirsutum L. BMC Genom. 2016, 17, 909. [Google Scholar] [CrossRef]
- Shen, X.; Guo, W.; Lu, Q.; Zhu, X.; Yuan, Y.; Zhang, T. Genetic mapping of quantitative trait loci for fiber quality and yield trait by RIL approach in Upland cotton. Euphytica 2007, 155, 371–380. [Google Scholar] [CrossRef]
- Wang, Y.; Ning, Z.; Hu, Y.; Chen, J.; Zhao, R.; Chen, H.; Ai, N.; Guo, W.; Zhang, T. Molecular Mapping of Restriction-Site Associated DNA Markers in Allotetraploid Upland Cotton. PLoS ONE 2015, 10, e0124781. [Google Scholar] [CrossRef]
- Liu, R.; Xiao, X.; Gong, J.; Li, J.; Zhang, Z.; Liu, A.; Lu, Q.; Shang, H.; Shi, Y.; Ge, Q.; et al. QTL mapping for plant height and fruit branch number based on RIL population of upland cotton. J. Cotton Res. 2020, 3, 5. [Google Scholar] [CrossRef]
- Li, C.; Dong, Y.; Zhao, T.; Li, L.; Li, C.; Yu, E.; Mei, L.; Daud, M.K.; He, Q.; Chen, J.; et al. Genome-Wide SNP Linkage Mapping and QTL Analysis for Fiber Quality and Yield Traits in the Upland Cotton Recombinant Inbred Lines Population. Front. Plant Sci. 2016, 7, 1356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ge, Q.; Liu, A.; Li, J.; Gong, J.; Shang, H.; Shi, Y.; Chen, T.; Wang, Y.; Palanga, K.K.; et al. Construction of a High-Density Genetic Map and its Application to QTL Identification for Fiber Strength in Upland Cotton. Crop Sci. 2017, 57, 774. [Google Scholar] [CrossRef]
- Said, J.I.; Song, M.; Wang, H.; Lin, Z.; Zhang, X.; Fang, D.; Zhang, J. A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol Genet Genom. 2015, 290, 1003–1025. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Li, Y.; Zhang, W.; Ju, J.; Guo, X.; Yang, J.; Su, J. Pinpointing MQTLs and candidate genes related to early maturity in upland cotton through the integration of meta analysis, RNA-seq, and VIGS approaches. Ind. Crops Prod. 2025, 223, 120195. [Google Scholar] [CrossRef]
- Xu, S.; Pan, Z.; Yin, F.; Lin, Z.; Wen, T.; Zhu, L.; Zhang, D.; Nie, X. Identification of candidate genes controlling fiber quality traits in upland cotton through integration of meta_QTL significant SNP and transcriptomic data. J. Cotton Res. 2020, 3, 34. [Google Scholar] [CrossRef]
- Huo, W.Q.; Zhang, Z.Q.; Ren, Z.Y.; Zhao, J.J.; Song, C.X.; Wang, X.X.; Pei, X.Y.; Liu, Y.G.; He, K.L.; Zhang, F.; et al. Unraveling genomic regions and candidate genes for multiple disease resistance in upland cotton using meta-QTL analysis. Heliyon 2023, 9, e18731. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ahlawat, Y.K.; Biswal, A.K.; Harun, S.; Harman-Ware, A.E.; Doeppke, C.; Sharma, N.; Hankoua, B.B. Heterologous expression of Arabidopsis laccase2, laccase4 and peroxidase52 driven under developing xylem specific promoter DX15 improves saccharification in populus. Biotechnol. Biofuels Bioprod. 2024, 17, 5. [Google Scholar] [CrossRef]
- Balasubramanian, V.K.; Rai, K.M.; Thu, S.W.; Hii, M.M.; Mendu, V. Genome-wide identification of multifunctional laccase gene family in cotton (Gossypium spp.); expression and biochemical analysis during fiber development. Sci. Rep. 2016, 6, 34309. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Li, M.; Wu, L.; Rao, W.; Peng, X.; Jiang, H. The ZmHSF08-ZmUGT92A1 module regulates heat tolerance by altering reactive oxygen species levels in maize. Crop J. 2024, 12, 1437–1446. [Google Scholar] [CrossRef]
- Chen, Y.; Cao, Y.; Duan, Y.; Deng, D.; Gao, Q.; Shen, Q.; Fang, W.; Zhu, X. The effects of overexpressing UDP-Glycosyltransferases genes on the plant response to abiotic stress: A meta-analysis. Beverage Plant Res. 2023, 3, 28. [Google Scholar] [CrossRef]
- Yu, J.; Hu, F.; Dossa, K.; Wang, Z.; Ke, T. Genome-wide analysis of UDP-glycosyltransferase superfamily in Brassica rapa and Brassica oleracea reveals its evolutionary history and functional characterization. BMC Genom. 2017, 18, 474. [Google Scholar] [CrossRef]
- Zhang, D.; Hrmova, M.; Wan, C.H.; Wu, C.; Balzen, J.; Cai, W.; Wang, J.; Densmore, L.; Fincher, G.B.; Zhang, H.; et al. Members of a new group of chitinase-like genes are expressed preferentially in cotton cells with secondary walls. Plant Mol. Biol. 2004, 54, 353–372. [Google Scholar] [CrossRef] [PubMed]
- Mokshina, N.; Gorshkova, T.; Deyholos, M.K. Chitinase-Like (CTL) and Cellulose Synthase (CESA) Gene Expression in Gelatinous-Type Cellulosic Walls of Flax (Linum usitatissimum L.) Bast Fibers. PLoS ONE 2014, 9, e97949. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Yao, H.; Wang, J.; Wang, J.; Xue, H.; Zuo, K. GhLTPG1, a cotton GPI-anchored lipid transfer protein, regulates the transport of phosphatidylinositol monophosphates and cotton fiber elongation. Sci Rep. 2016, 6, 26829. [Google Scholar] [CrossRef]
- Hou, K.; Wu, W.; Gan, S.S. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef]
- Czarnocka, W.; Fichman, Y.; Bernacki, M.; Różańska, E.; Sańko-Sawczenko, I.; Mittler, R.; Karpiński, S. FMO1 Is Involved in Excess Light Stress-Induced Signal Transduction and Cell Death Signaling. Cells 2020, 9, 2163. [Google Scholar] [CrossRef]
- Zhao, H.; Li, D.; Liu, Y.; Zhang, T.; Zhao, X.; Su, H.; Li, J. Flavin-containing monooxygenases FMOGS-OXs integrate flowering transition and salt tolerance in Arabidopsis thalian. Physiol. Plant. 2024, 176, e14287. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cao, X.; Wang, H.; Liu, Y.; Liu, W.; Zhang, H.; Kuang, M.; Peng, J. Comprehensive identification of polygalacturonases in cotton: Genomic analysis, potential regulatory mechanisms and expression patterns in anthers. Ind. Crops Prod. 2023, 200, 116874. [Google Scholar] [CrossRef]
- Arcade, A.; Labourdette, A.; Falque, M.; Mangin, B.; Chardon, F.; Charcosset, A.; Joets, J. BioMercator: Integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 2004, 20, 2324–2326. [Google Scholar] [CrossRef]
- Guo, W.; Cai, C.; Wang, C.; Zhao, L.; Wang, L.; Zhang, T. A preliminary analysis of genom structure and composition in Gossypium hirstum. BMC Genom. 2008, 9, 314. [Google Scholar] [CrossRef]
- Yu, Y.; Yuan, D.; Liang, S.; Li, X.; Lin, Z.; Zhang, X. Genome structure of cotton revealed by genome-wide SSR genetic map constructed from a BC1 population between G. hirsutum and G. barbadense. BMC Genom. 2011, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Khan, M.; Zhou, Z.; Wang, X.; Cai, X.; Ilyas, M.; Wang, C.; Wang, Y.; Li, Y.; Liu, F.; et al. A high-density SSR genetic map constructed from a F2 population of Gossypium hirsutum and Gossypium darwinii. Gene 2015, 574, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Endrizzi, J.E. In Cotton, Fang, D.D., Percy, R.G., Eds.; Cytology and Cytogenetics. Agronomy Monograph 57, 2nd ed.; ASA, CSSA, and SSSA: Madison, WI, USA, 2015; pp. 129–154. [Google Scholar] [CrossRef]
- Venske, E.; Dos Santos, R.S.; Farias, D.D.R.; Rother, V.; da Maia, L.C.; Pegoraro, C.; Costa de Oliveira, A. Meta-analysis of the QTLome of fusarium head blight resistance in bread wheat: Refining the current puzzle. Front. Plant Sci. 2019, 10, 727. [Google Scholar] [CrossRef]


| No. | Population Type and Size | No. of QTLs | Traits | Ref. |
|---|---|---|---|---|
| 1. | F2 (4Su-271; 4I-248; SgJ-276; Sg4-304) | 50 | PH, BW, LP, FL, FS, FU, FE, FM, | [28] |
| 2. | F2, 251 | 5 | NFB | [48] |
| 3. | RIL, 180 | 113 | BW, LP, SI, FL, FU, FM, FE, FS | [26] |
| 4. | F2, 270 | 79 | LP, CP, CO, LA, OA, PA | [23] |
| 5. | RIL, 180 | 86 | LP, SY, SI, BW, FE, FL, FS, FSCI, FBN | [49] |
| 6. | F2:3 | 5 | LP | [50] |
| 7. | Ils, 115 | 60 | FL, FS, MIC, FU, FE | [17] |
| 8. | RIL | 27 | FL, FS, FM | [51] |
| 9. | RIL, 180 | 33 | FL, FS, FM, FMAT, FR, FB, FE, FSFI | [8] |
| 10. | RIL, 180 | 62 | FL, FU, FM, FE, FS | [13] |
| 11. | BC1F2, 115 | 44 | FL, FU, FM, FE, FS | [14] |
| 12. | Composite cross-population, 172 | 63 | FE, FL, FM, FU, FS | [11] |
| 13. | BC2F1, 133 | 153 | FL, FS, FM, BW, SI, LI | [27] |
| 14. | RIL, 177 | 41 | PH | [52] |
| 15. | RIL, 180 | 59 | FL, FU, FS, FE, FM | [15] |
| 16. | F2:3, 188 | 11 | RL, SFW, RFW, SDW, RDW, CHL, SH | [29] |
| 17. | F2:3, 155; RIL, 190 | 50 | FS, FL, FM, FU, FE | [10] |
| 18. | F2, 150 | 15 | CL | [35] |
| 19. | RIL, 200 | 11 | LFMP, JI, SCY | [53] |
| 20. | F2:3, 229 | 41 | VR | [34] |
| 21. | RIL, 178 | 134 | FL, FU, MIC, FE, FS, SCW, LW, LP, SI, BN | [24] |
| 22. | composite cross-population | 11 | FL, FS, FU | [9] |
| 23. | F2, 124 | 33 | SW, LW, LP, LI, SI, MV, FE, FS, FUHML, HSW | [20] |
| 24. | a four-way cross-mapping population (4WC), 239 | 74 | PB, NB, BW, LP, LI, SI, FL, FS, FM, FU, FE | [21] |
| 25. | RIL, 196 | 8 | Oil, Pro | [54] |
| 26. | RIL, 177 | 55 | FL, FU, FS, FE, FM | [18] |
| 27. | BIL, 146 | 67 | FL, FS, FM, FE, FU, BW, LP, LY, SCY | [55] |
| 28. | F2:3 | 50 | SY, LY, LP, BN, BS, LI, SI | [56] |
| 29. | RIL, 177 | 34 | MRL, PH, RL, RSA, NRT, NRF, SW, RW, RV | [57] |
| 30. | F2 | 43 | PH, FN, BS, SY, SI, LY | [58] |
| 31. | RIL, 163 | 9 | FOV | [36] |
| 32. | F2:3, 173 | 39 | FL, FU, FM, FS, FE | [12] |
| 33. | F2:3, 188 | 8 | Bla, Fcc, Bcc, Fbbw | [59] |
| 34. | F2, 347 | 18 | LP, BW, FL, FM, FS | [60] |
| 35. | F2, 96 | 17 | FSH, SNH, PBS, TNSa, TNN, NOB, TNB, LOB, LOS, LOp | [61] |
| 36. | F2, 123 | 17 | BW, LP, FL, FU, FM, FE | [62] |
| 37. | RIL, 177 | 24 | FL, FS, FU, FE, FM | [19] |
| 38. | F5, 122 | 19 | PH, RWC, CSI, PC, TCC, NRA | [63] |
| 39. | RIL, 196 | 37 | FL, FM, FS | [16] |
| 40. | RIL, 186 | 16 | PA, YC, FP | [22] |
| 41. | F2, 249 | 112 | PH, CNH, FTLH, STLH, SLA, SPn, TPn, Sci, Tci, Scond, Tcond, STr, TTr, Chla, FBN, SCY, LY, BW, SI, LP, LI, FL, FS, MC | [64] |
| 42 | 277 F2:3, | 88 | BW, FE, FL, FM, FR, FS, FU, FY, LP, SCI | [65] |
| 43 | RIL, 178 | 170 | BN, FE, FM, FMAT, FU, FUHML, LI, PH, SCW, SCY, SI, TNMB, TNSB | [66] |
| 44 | RIL, 196 | 104 | FE, FL, FM, FU, FS | [67] |
| 45 | RIL, 137 | 280 | FS, FT, FBP, PH, NFFB, VR | [68] |
| 46 | RIL, - | 49 | FE, FL, FM, FS, LP, SCW, SCY, SI | [69] |
| 47 | RIL, 161 | 20 | VR, FS | [70] |
| 48 | RIL, 231 | 45 | FBN, PH | [71] |
| 49 | RIL, 188 | 171 | FU, FL, FS, FM, LP, SCW | [72] |
| 50 | RIL, 196 | 104 | FS | [73] |
| Chr. | MQTL Name | No. of QTLs | Position | CI | Trait | Reference |
|---|---|---|---|---|---|---|
| A01 | MQTLchr1-1 | 10 | 14.28 | 0.8 | FM, FL, PH, LI, FU, FBN, FE | [39,74] |
| MQTLchr1-2 | 9 | 27.69 | 2.24 | FM, FU, FL, PH, VR, CP, | - | |
| MQTLchr1-3 | 6 | 44.02 | 2.05 | FM, FBN, PH, TPn, Sci, | [39] | |
| MQTLchr1-4 | 5 | 52.21 | 0.2 | FM, FL, PH | - | |
| A02 | MQTLchr2-1 | 12 | 3.05 | 0.49 | FS, Fl, FE, PH, SCI, FU, Oil | [39,74] |
| A03 | MQTLchr3-1 | 17 | 11.55 | 1.21 | FS, FM, FU, FL, LP, SY, PH, TNS, | [39,47,74] |
| MQTLchr3-2 | 9 | 44.29 | 1.53 | FL, FM, LP, BN | [39] | |
| MQTLchr3-3 | 4 | 100.46 | 1.16 | LP, FL, NFFB, BN | [75] | |
| MQTLchr3-4 | 4 | 128.9 | 0.09 | FL, FT, NFFB | - | |
| A04 | MQTLchr4-1 | 12 | 10.33 | 1.04 | [47] | FE, LI, FM, FU, Oil, FUHML, NB, LP |
| MQTLchr4-2 | 7 | 85.1 | 1.9 | FS, BW | - | |
| A05 | MQTLchr5-1 | 12 | 2.72 | 2.6 | PH, BW, NFFB, RV, LP, SI, FE, Oil, FBP | [39,47,74] |
| MQTLchr5-2 | 13 | 12.62 | 2.6 | FBP, FM, FS, LP, FBN, RV, FT, BW, FB | [39,47,74,75,76] | |
| MQTLchr5-3 | 11 | 37.93 | 2.56 | FU, LP, FM, FU, FS, BW, FB | [39,47,74] | |
| MQTLchr5-4 | 5 | 82.83 | 0.56 | RL, PH, FS, RSA, BW | [76] | |
| A06 | MQTLchr6-1 | 9 | 13.73 | 1.6 | FS, OA, FU, NB, BW | [39,47,74,76] |
| MQTLchr6-2 | 5 | 47.51 | 0.79 | LI, FMAT, FU, BW, FM | [77] | |
| A07 | MQTLchr7-1 | 15 | 6.12 | 0.53 | FE, PH, LY, FS, SCY | [39,47,74] |
| MQTLchr7-2 | 7 | 28.99 | 1.69 | SCY, FBN, FM, FE, LP, FU | [39,74,75] | |
| A08 | MQTLchr8-1 | 22 | 32.34 | 1.52 | FM, FS, FL, FU | [39,74] |
| MQTLchr8-2 | 8 | 48.66 | 1.51 | LY, FS, SI, FM | [74,75,77] | |
| MQTLchr8-3 | 10 | 52.88 | 1.48 | VR, FS, SI, LY | [74] | |
| MQTLchr8-4 | 12 | 84.01 | 1.52 | FM, LP, FB, FM, SI | - | |
| A09 | MQTLchr9-1 | 18 | 2.05 | 0.42 | FS, PH, FL, FU, FBP, VR | [39,74,75] |
| MQTLchr9-2 | 14 | 18.2 | 0.4 | FU, PH, FS, SI, FSCI, FU, FM, SCY | [39,74] | |
| MQTLchr9-3 | 7 | 32.08 | 1.33 | SI, SCW, Bcc, FT, FS, LP | [74] | |
| MQTLchr9-4 | 8 | 46.44 | 0.73 | FU, Bcc, SCW, FM | - | |
| A10 | MQTLchr10-1 | 10 | 16.98 | 3.2 | SCW, FMAT, LP, FMIC, FE, FR, PH, NB | [39,74] |
| MQTLchr10-2 | 5 | 36.16 | 0.36 | NB, SCY, LP, FU, FS | [74] | |
| MQTLchr10-3 | 5 | 58.17 | 0.76 | FM, PA, FT | [77] | |
| A11 | MQTLchr11-1 | 14 | 0.01 | 0.06 | CL, PB, PH, FE, LP, FL, SI, FB, FM | [39,47,74,77] |
| MQTLchr11-2 | 10 | 5.35 | 0.7 | FU, FE, Cl, FB, FL, PH, PB | [47,74,77] | |
| MQTLchr11-3 | 8 | 13.21 | 1.71 | FU, SI, FB, FL, FE, PH, PB | [47,74,76,77] | |
| MQTLchr11-4 | 5 | 47.97 | 1.77 | NFFB, FU, FL, TCi, PH | [75,77] | |
| A12 | MQTLchr12-1 | 11 | 18.94 | 2.37 | STr, SA, FU, TTr, LP, FS, BW, FE, RWC | [74,77] |
| MQTLchr12-2 | 5 | 51.58 | 0.75 | FU, FBN, LP, RWC | [74,77] | |
| MQTLchr12-3 | 7 | 71.65 | 0.85 | FBP, TNMB, CO, OA, CP | [75] | |
| A13 | MQTLchr13-1 | 14 | 10.34 | 1.26 | FL, NRT, RL, NFFB, PH, RSA, BW, LP, FE, NRF, PB, FU | [39,74,76] |
| MQTLchr13-2 | 9 | 38.38 | 1.51 | FE, RSA, NFBB, RL, PH, TNSB | [74] | |
| D02 | MQTLchr14-1 | 23 | 1.02 | 0.38 | BW, LP, FMIC, FMAT, FU, PH, LY, SCY, PH, SI, FE, FR, FS, LI, RL, FB | [39,47,74,76,77] |
| MQTLchr14-2 | 8 | 16.99 | 0.67 | FBP, NRF, FU, PH, FM | [39,47,74,76] | |
| MQTLchr14-3 | 10 | 39.74 | 0.43 | FU, LA, FT, CP, FBP, BW, NFFB, FS, FOV | - | |
| MQTLchr14-4 | 7 | 55.04 | 0.72 | FBN, STr, VR, FS, PH, FM | [39] | |
| D01 | MQTLchr15-2 | 19 | 3.19 | 0.67 | MV, NFFB, FE, SI, SH, FL, SDW, RFW, PH, FUHML, NFFB, LI, SFW | [39,47,74] |
| MQTLchr15-1 | 9 | 51.62 | 1.03 | FT, NFB, STLH, MIC, PH, Oil | [47,74,77] | |
| D03 | MQTLchr17-1 | 15 | 86.9 | 0.21 | FT, NFFB, SI, FM, PH | [77] |
| MQTLchr17-2 | 16 | 95.88 | 0.86 | FT, NFFB, PH, FBP, SI | - | |
| MQTLchr17-3 | 7 | 67.61 | 0.49 | FS, NFFB | [39] | |
| D13 | MQTLchr18-1 | 18 | 13.03 | 1.47 | BW, FE, FL, SI, LOS, OA, BS, NB, LA | [39,74] |
| D05 | MQTLchr19-1 | 9 | 2.06 | 0.28 | FL, FS, FU, BW, BS, SI, PH, CL, SW | [39,47,74,76] |
| MQTLchr19-2 | 14 | 19.75 | 0.41 | FL, FS, PH, FU, BW, PH, LP, BS | [39,47,74] | |
| MQTLchr19-3 | 8 | 31.53 | 0.54 | BS, FM, TNSB, FS, LP, FOV, SI, LI | - | |
| D10 | MQTLchr20-1 | 10 | 2.01 | 0.42 | FL, FS, BS, FE, FBN, LY, NFFB | [39,47,74,76] |
| MQTLchr20-2 | 10 | 28.59 | 0.56 | FBN, SCY, FS, BN, FS, PH | [39] | |
| MQTLchr20-3 | 10 | 47.65 | 0.49 | PH, FM, FS, FU, PH, LI, FE | - | |
| MQTLchr20-4 | 6 | 67.46 | 0.35 | FE, FS, SI, PH, BN | [75] | |
| D11 | MQTL21-1 | 11 | 0.47 | 0.23 | CL, FM, FL, FMAT, LP | [74,77] |
| MQTLchr21-2 | 6 | 39.76 | 0.24 | FL, FOV, BW, FMAT, FBN | - | |
| MQTL21-3 | 5 | 74.38 | 0.08 | FM, FBN, FL | [39,77] | |
| MQTLchr21-4 | 7 | 83.99 | 0.25 | FM, PH, FBN, SA, FL, NFFB | - | |
| D04 | MQTLchr22-1 | 7 | 11.88 | 1.72 | NFFB, FU, FS, FB, FT, FM | [39,74,77] |
| MQTLchr22-2 | 6 | 53.94 | 1.27 | LP, FM, FS, HSW, SI, FS | [74] | |
| MQTLchr22-3 | 6 | 87.17 | 0.06 | FS, Sci, CP, Tci, Tcond, FE | - | |
| D09 | MQTLchr23-1 | 11 | 41.85 | 0.37 | VR, FR, FS, NFFB, LI, FU, LP | [39,47,74,76,77] |
| MQTLchr23-2 | 8 | 56.03 | 1.56 | CO, LI, FL, NFFB, FL, FS, PH, CP | [39,74,77] | |
| D08 | MQTLchr24-1 | 15 | 47.24 | 0.13 | FBP, LY, SI, NFFB, LP, SCY, FE, SCW, FN, FU | [74,77] |
| MQTLchr24-2 | 10 | 64.77 | 0.31 | BW, SI, RV, FS, LP, RL, LA, NFFB | [39,77] | |
| MQTLchr24-3 | 9 | 32.36 | 0.48 | LP, FN, FM, LY, FM, FE, SCW | [74] | |
| D06 | MQTLchr25-1 | 10 | 75.32 | 0.42 | FS, SCW, FL, FE | [74,75] |
| MQTLchr25-2 | 6 | 31.88 | 0.75 | FU, FS, SCY, BW, FBP | [74] | |
| MQTLchr25-3 | 4 | 86.68 | 1.03 | FS | ||
| MQTLchr25-4 | 14 | 8.33 | 2.05 | FM, FMIC, FE, FL, Oil, FMAT, PH, FL, FSFI | [39,74,76] | |
| D12 | MQTLchr26-1 | 8 | 24.97 | 0.48 | FL, SDW, FM, FU, MV, LY, SCI | [47] |
| MQTLchr26-2 | 8 | 35.1 | 0.3 | FM, SI, FT, PH, FE, FBN, FL | [39,74] | |
| MQTLchr26-3 | 6 | 51.43 | 1.64 | FL, BS, BW, SCY, CO, FU | [76] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toshpulatov, A.K.; Turaev, O.S.; Iskandarov, A.A.; Khalikov, K.K.; Arslanova, S.K.; Safiullina, A.K.; Kudratova, M.K.; Oripova, B.B.; Rafieva, F.U.; Kholova, M.D.; et al. Identification of Stable Meta-QTLs and Candidate Genes Underlying Fiber Quality and Agronomic Traits in Cotton. Plants 2025, 14, 3252. https://doi.org/10.3390/plants14213252
Toshpulatov AK, Turaev OS, Iskandarov AA, Khalikov KK, Arslanova SK, Safiullina AK, Kudratova MK, Oripova BB, Rafieva FU, Kholova MD, et al. Identification of Stable Meta-QTLs and Candidate Genes Underlying Fiber Quality and Agronomic Traits in Cotton. Plants. 2025; 14(21):3252. https://doi.org/10.3390/plants14213252
Chicago/Turabian StyleToshpulatov, Abdulqahhor Kh., Ozod S. Turaev, Abdulloh A. Iskandarov, Kuvandik K. Khalikov, Sevara K. Arslanova, Asiya K. Safiullina, Mukhlisa K. Kudratova, Barno B. Oripova, Feruza U. Rafieva, Madina D. Kholova, and et al. 2025. "Identification of Stable Meta-QTLs and Candidate Genes Underlying Fiber Quality and Agronomic Traits in Cotton" Plants 14, no. 21: 3252. https://doi.org/10.3390/plants14213252
APA StyleToshpulatov, A. K., Turaev, O. S., Iskandarov, A. A., Khalikov, K. K., Arslanova, S. K., Safiullina, A. K., Kudratova, M. K., Oripova, B. B., Rafieva, F. U., Kholova, M. D., Ernazarova, D. K., Kodirov, D. M., Gapparov, B. M., Komilov, D. J., Togaeva, M. A., Kurbanov, A. K., Erjigitov, D. S., Khidirov, M. T., Yu, J. Z., & Kushanov, F. N. (2025). Identification of Stable Meta-QTLs and Candidate Genes Underlying Fiber Quality and Agronomic Traits in Cotton. Plants, 14(21), 3252. https://doi.org/10.3390/plants14213252









