Identification of Chalcone Synthase Genes and Their Responses to Salt and Cold Stress in Poncirus trifoliata
Abstract
1. Introduction
2. Results
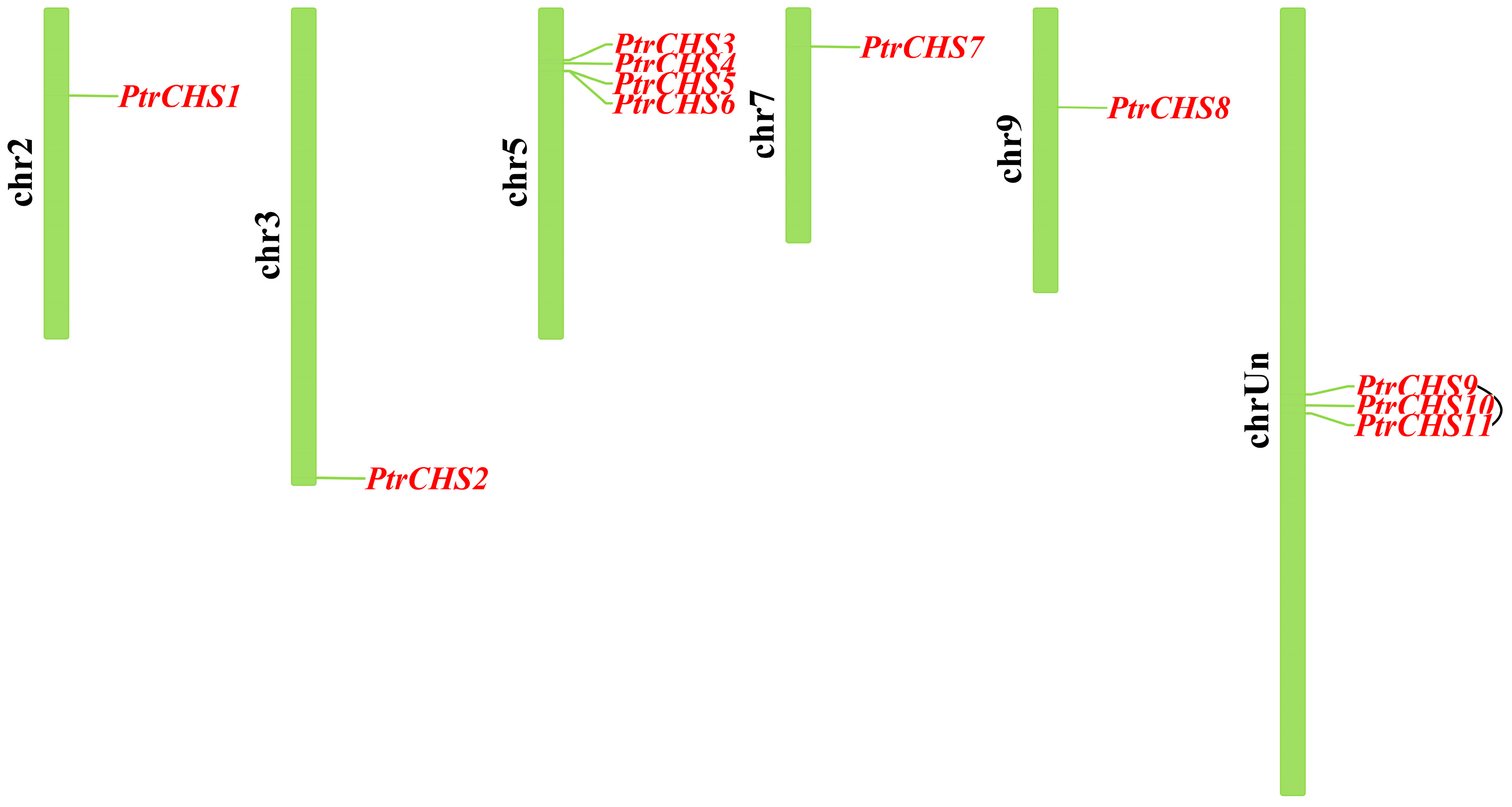
2.1. Identification and Physicochemical Property Prediction of CHS Family Members
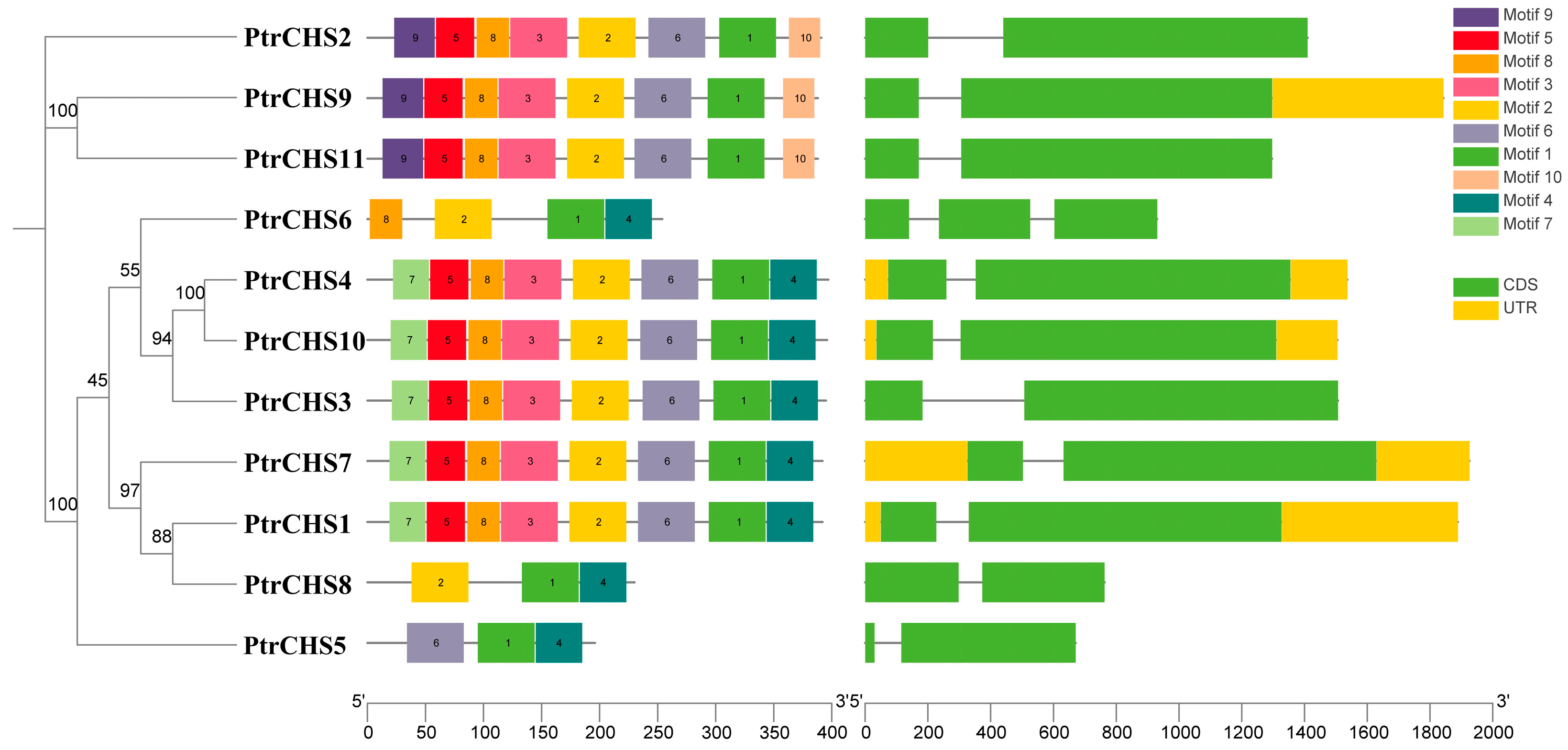
2.2. Gene Structure and Protein Conserved Motif Analysis of the CHS Family in Poncirus trifoliata
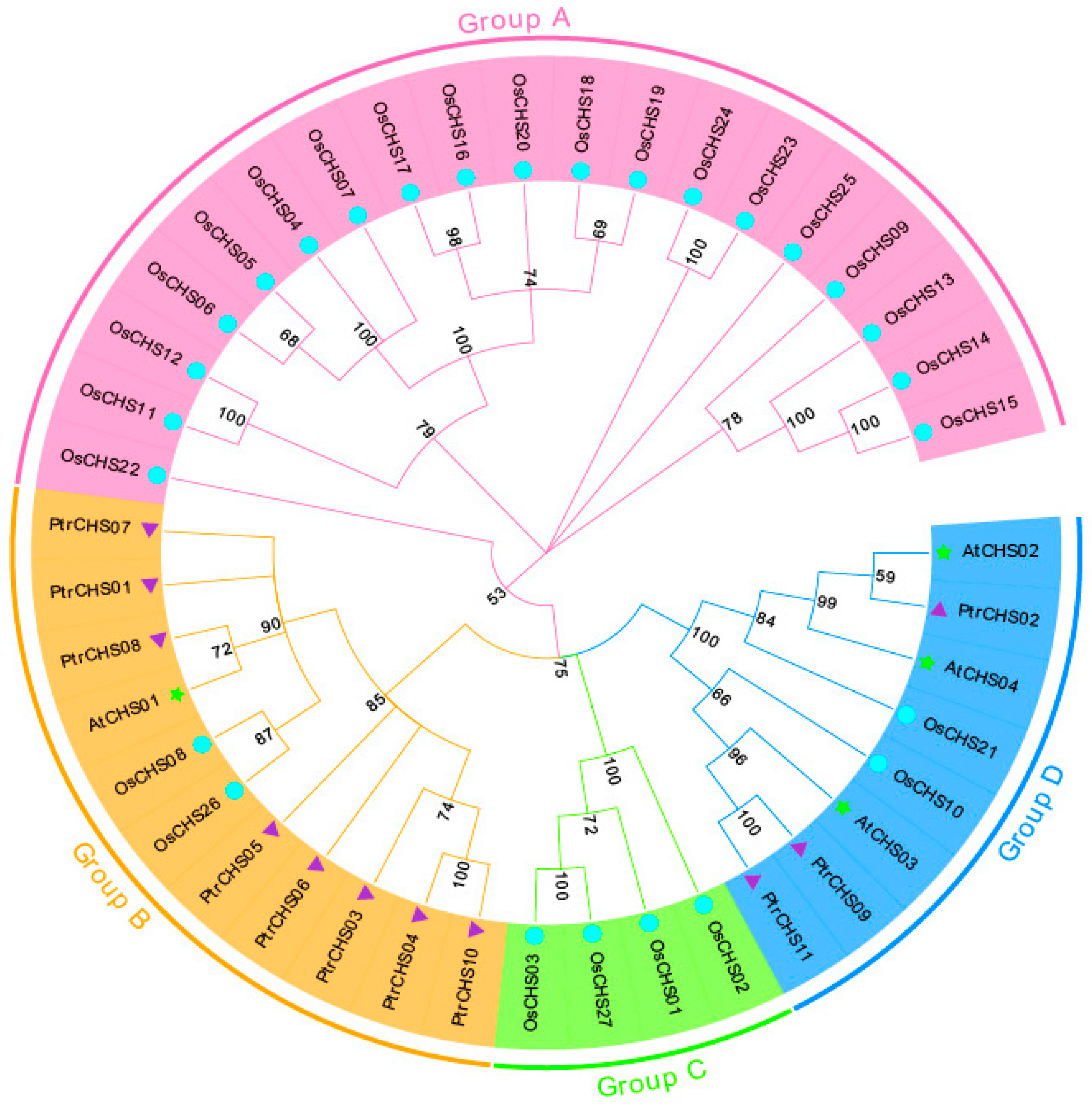
2.3. Phylogenetic Analysis of CHS Family Members in Poncirus trifoliata
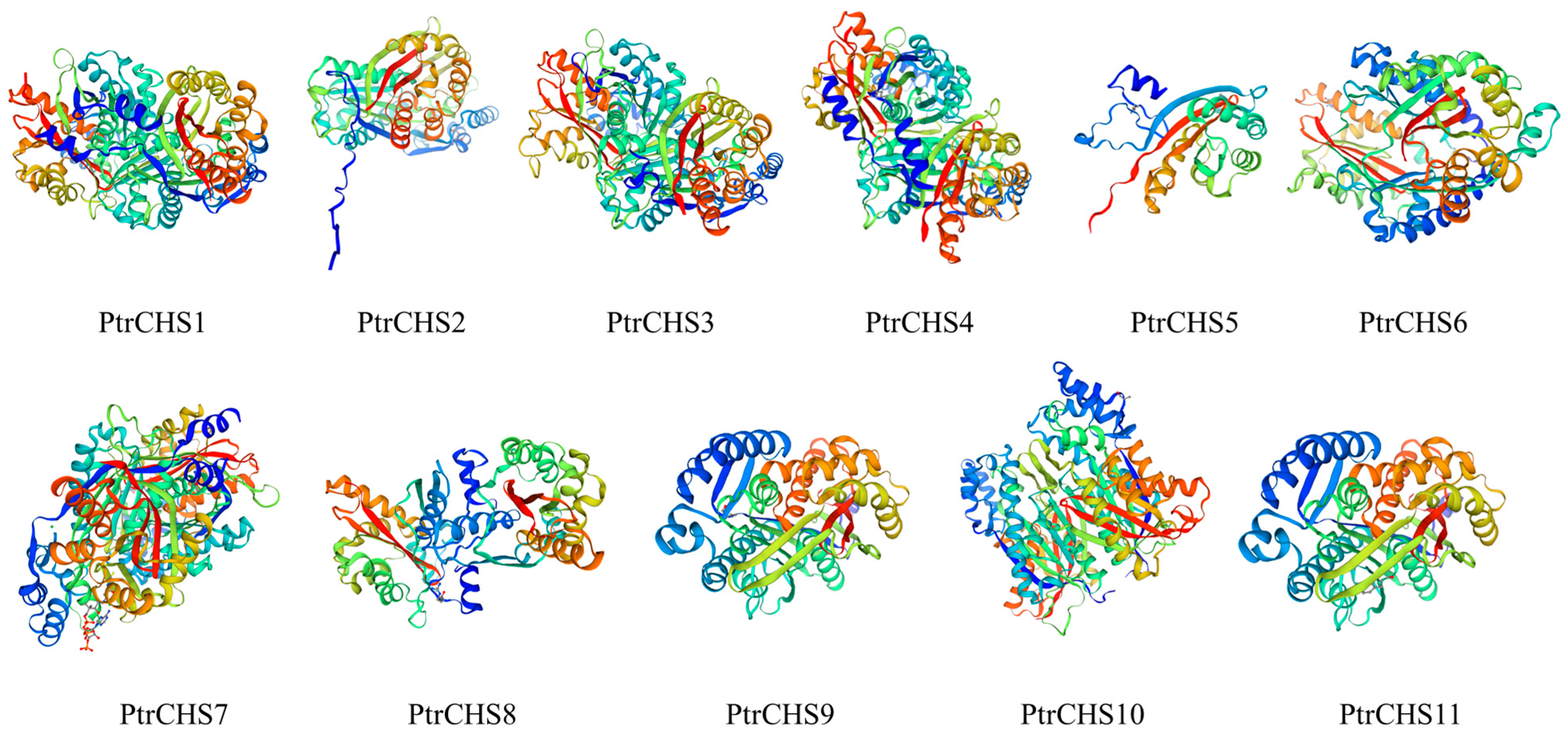
2.4. Secondary and Tertiary Structure Prediction and Phosphorylation Site Analysis of PtrCHS Proteins
2.5. Collinearity Analysis of CHS Gene Family in Poncirus trifoliata
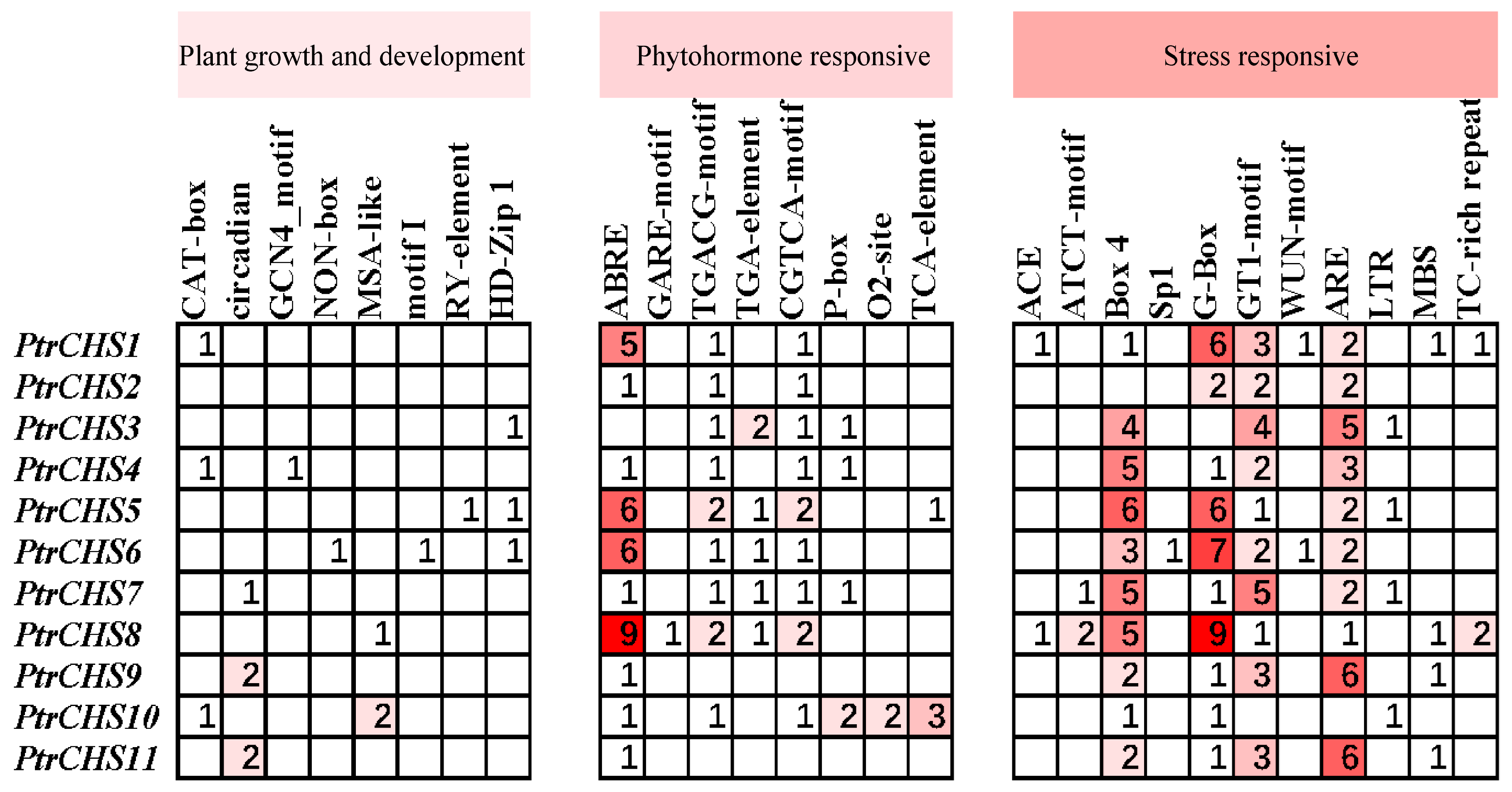
2.6. Cis-Acting Element Analysis of CHS Genes in Poncirus trifoliata
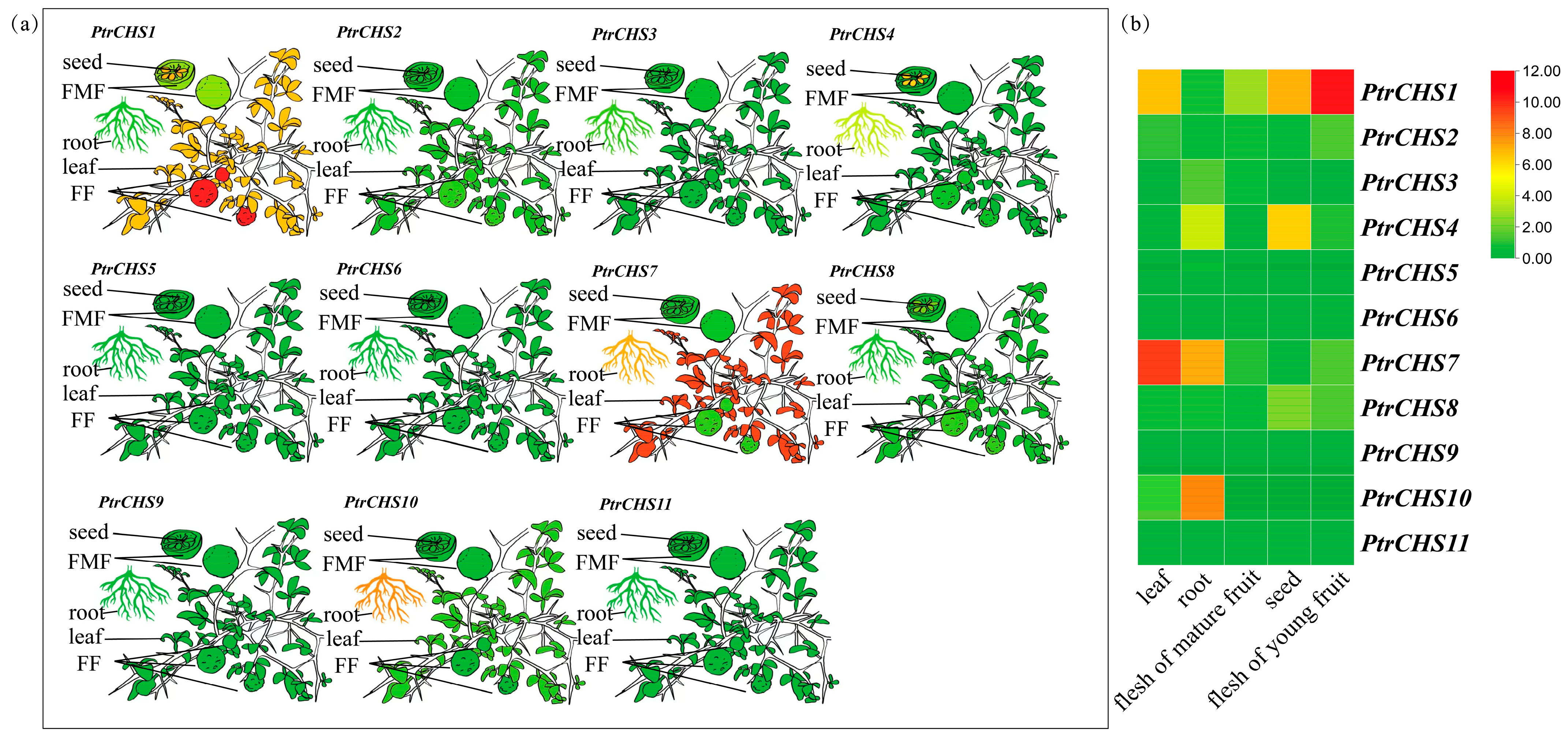
2.7. Tissue Expression Pattern Analysis of CHS Genes in Poncirus trifoliata
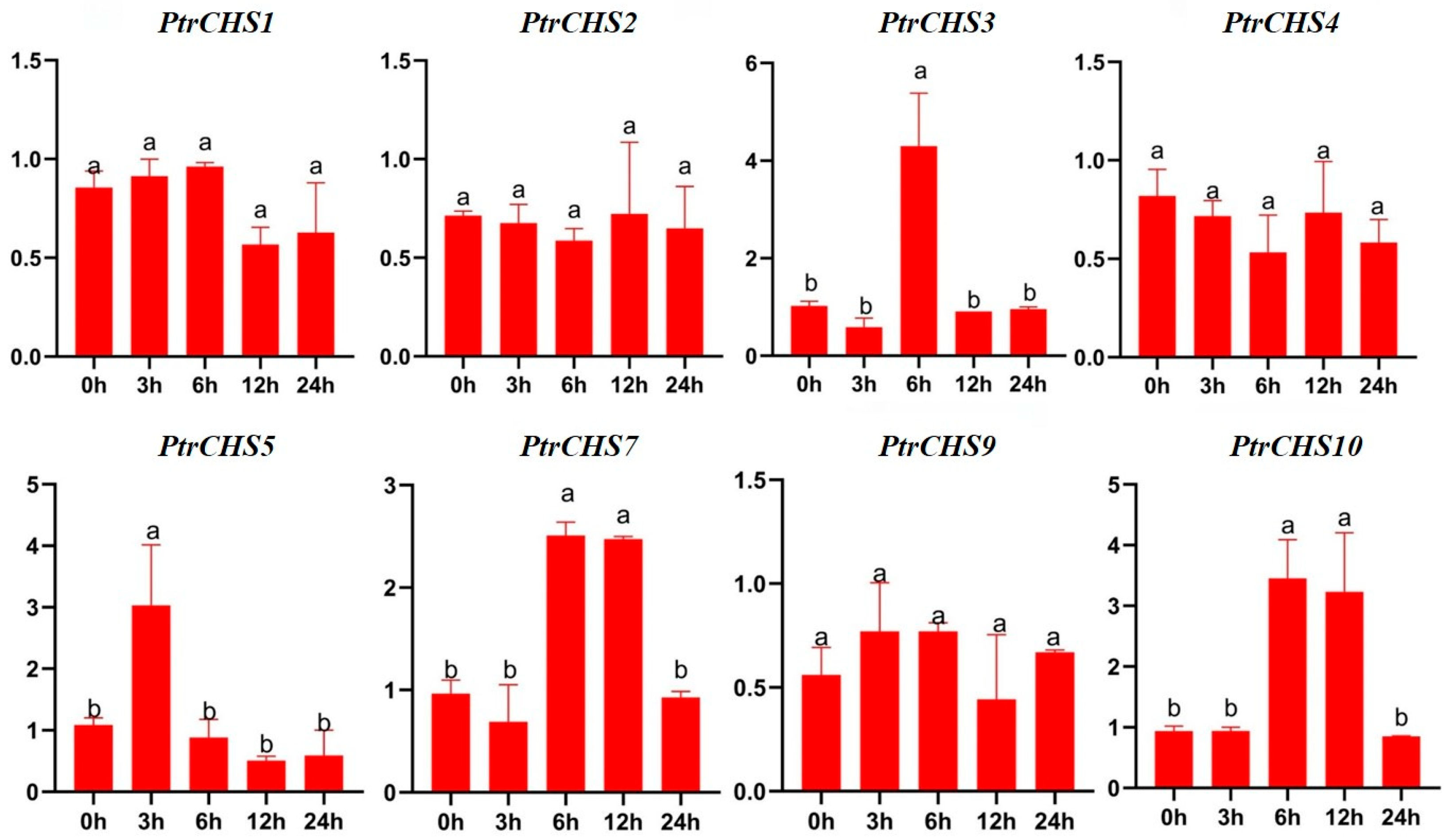
2.8. Expression Pattern Analysis of PtrCHS Under Salt Treatment
2.9. Expression Pattern Analysis of CHS Genes in Poncirus trifoliata Under Cold Treatment
3. Discussion
4. Materials and Methods
4.1. Identification of Chalcone Synthase Gene Family Members in Poncirus trifoliata
4.2. Bioinformatics Analysis of the Chalcone Synthase Gene Family in Poncirus trifoliata
4.3. Expression Analysis of PtrCHSs Under Salt and Cold Stress
4.4. Cloning of PtrCHS7 and PtrCHS10 Genes and Overexpression Vectors Construction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ma, L.; Li, X.; Zhang, J.J.; Yi, D.X.; Li, F.; Wen, H.Y.; Liu, W.H.; Wang, X.M. MsWRKY33 increases alfalfa (Medicago sativa L.) salt stress tolerance through altering the ROS scavenger via activating MsERF5 transcription. Plant Cell Environ. 2023, 46, 3887–3901. [Google Scholar] [CrossRef]
- Vives-Peris, V.; Pérez-Clemente, R.M.; Gómez-Cadenas, A.; López-Climent, M.F. Involvement of citrus shoots in response and tolerance to abiotic stress. Hortic. Adv. 2024, 2, 3. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Li, J.X.; Yu, Q.H.; Liu, C.; Zhang, N.B.; Xu, W.R. Flavonoids as key players in cold tolerance: Molecular insights and applications in horticultural crops. Hortic. Res. 2025, 12, uhae366. [Google Scholar] [CrossRef]
- Waki, T.; Mameda, R.; Nakano, T.; Yamada, S.; Terashita, M.; Ito, K.; Tenma, N.; Li, Y.B.; Fujino, N.; Uno, K. A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat. Commun. 2020, 11, 870. [Google Scholar] [CrossRef]
- Ahmad, S.; Jeridi, M.; Siddiqui, S.; Shah, A.Z.; Ali, S. Genome-wide identification, characterization, and expression analysis of the Chalcone Synthase gene family in Oryza sativa under Abiotic Stresses. Plant Stress 2023, 9, 100201. [Google Scholar] [CrossRef]
- Han, Y.H.; Ding, T.; Su, B.; Jiang, H.Y. Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. Int. J. Mol. Sci. 2016, 17, 161. [Google Scholar] [CrossRef]
- Vadivel, A.K.A.; Krysiak, K.; Tian, G.; Dhaubhadel, S. Genome-wide identification and localization of chalcone synthase family in soybean (Glycine max [L]Merr). BMC Plant Biol. 2018, 18, 325. [Google Scholar] [CrossRef]
- Wu, X.X.; Zhang, S.M.; Liu, X.H.; Shang, J.; Zhang, A.D.; Zhu, Z.W.; Zha, D.S. Chalcone synthase (CHS) family members analysis from eggplant (Solanum melongena L.) in the flavonoid biosynthetic pathway and expression patterns in response to heat stress. PLoS ONE 2020, 15, e0226537. [Google Scholar] [CrossRef]
- Zhu, L.L.; Ding, Y.Q.; Wang, S.X.; Wang, Z.M.; Dai, L.P. Genome-wide identification, characterization, and expression analysis of CHS gene family members in Chrysanthemum nankingense. Genes 2022, 13, 2145. [Google Scholar] [CrossRef]
- Isiyel, M.; İlhan, E.; Kasapoğlu, A.G.; Muslu, S.; Öner, B.M.; Aygören, A.S.; Yiğider, E.; Aydm, M.; Yıldırım, E. Identification and characterization of Phaseolus vulgaris CHS genes in response to salt and drought stress. Genet. Resour. Crop Evol. 2024, 72, 271–293. [Google Scholar] [CrossRef]
- Chen, S.; Wu, F.; Li, Y.; Qian, Y.; Pan, X.; Li, F.; Yang, A. NtMYB4 and NtCHS1 Are Critical Factors in the Regulation of Flavonoid Biosynthesis and Are Involved in Salinity Responsiveness. Front. Plant Sci. 2019, 10, 178. [Google Scholar] [CrossRef]
- Jiang, Y.Q.; Khan, N.M.; Ali, A.; Zhou, G.Z.; Zhou, Y.; Li, P.J.; Wan, Y.L. AcMYB176-Regulated AcCHS5 Enhances Salt Tolerance in Areca catechu by Modulating Flavonoid Biosynthesis and Reactive Oxygen Species Scavenging. Int. J. Mol. Sci. 2025, 26, 3216. [Google Scholar] [CrossRef]
- Wang, X.F.; Wu, J.; Li, H.A.; Zhu, L.; Long, Y. Genome-wide identification, evolution, and expression and metabolic regulation of the maize CHS gene family under abiotic stress. BMC Genom. 2025, 26, 581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Zheng, S.; Liu, Z.J.; Wang, L.G.; Bi, Y.R. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J. Plant Physiol. 2011, 168, 367–374. [Google Scholar] [CrossRef]
- Zhang, Z.R.; Yuan, X.; Liu, J.M.; Guo, C.H.; Kang, C.; Luo, X.; Zhao, H.C.; Quan, S.W.; Niu, J.X. Identification of the walnut CHS and FLS gene families and the role of JrHY5 in flavonoid synthesis. Ind. Crops Prod. 2025, 235, 121698. [Google Scholar] [CrossRef]
- Ubi, B.E.; Honda, C.; Bessho, H.; Kondo, S.; Wada, M.; Kobayashi, S.; Moriguchi, T. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 2006, 170, 571–578. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, Y.C.; Pan, X.H.; Li, Y.T.; Cui, L.J.; Wu, F.Y.; Cao, M.J.; Yang, A.G.; Pan, G.T. Isolation and expression analysis of NtCHS6, a new chalcone synthase gene from Nicotiana tabacum. J. Integr. Agric. 2017, 16, 1443–1450. [Google Scholar] [CrossRef]
- Wang, C.H.; Zhi, S.; Liu, C.Y.; Xu, F.X.; Zhao, A.C.; Wang, X.L.; Tang, X.; Li, Z.A.; Huang, P.; Yu, M.D. Isolation and characterization of a novel chalcone synthase gene family from mulberry. Plant Physiol. Biochem. 2017, 115, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Sugaya, S.; Gemma, H. Decreased anthocyanin biosynthesis in grape berries grown under elevated night temperature condition. Sci. Hortic. 2005, 105, 319–330. [Google Scholar] [CrossRef]
- Ma, Q.; Tan, L.Y.; Zhou, D.H.; Wang, X.N.; Sun, H.L.; Wang, Y.H.; Tian, L.; Shi, C.; Wei, A.Z.; Fei, X.T. Exogenous methyl jasmonate induces CHS and promotes flavonoid accumulation in Zanthoxylum bungeanum. Ind. Crops Prod. 2024, 222, 119682. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.J.; Wang, Y.X.; Geng, Z.Q.; Liu, S.H.; Chen, C.W.; Chen, S.M.; Jiang, J.F.; Chen, F.D. CmMYB9a activates floral coloration by positively regulating anthocyanin biosynthesis in chrysanthemum. Plant Mol. Biol. 2022, 108, 51–63. [Google Scholar] [CrossRef]
- Wu, G.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2024, 32, 656–662. [Google Scholar] [CrossRef]
- Sahin-Çevik, M. Identification and expression analysis of early cold-induced genes from cold-hardy citrus relative Poncirus trifoliata (L.) Raf. Gene 2013, 512, 536–545. [Google Scholar] [CrossRef]
- Ding, Y.L.; Shi, Y.T.; Yang, S.H. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Dahro, B.; Li, C.L.; Liu, J.H. Overlapping responses to multiple abiotic stresses in citrus: From mechanism understanding to genetic improvement. Hortic. Adv. 2023, 1, 4. [Google Scholar] [CrossRef]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Zhu, X.; Duan, N.; Liu, J.H. PtrBAM1, a β-amylase-coding gene of Poncirus trifoliata, is a CBF regulon member with function in cold tolerance by modulating soluble sugar levels. Plant Cell Environ. 2014, 37, 2754–2767. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, W.; Wang, M.; Khan, M.; Liu, J.H. A C2H2-type zinc finger protein ZAT12 of Poncirus trifoliata acts downstream of CBF1 to regulate cold tolerance. Plant J. 2024, 117, 1317–1329. [Google Scholar] [CrossRef]
- Xu, Y.Y.; Hui, Q.L.; Li, M.; Peng, H.X.; He, Y.Z.; Chun, C.P.; Peng, L.Z.; Fu, X.Z. Global analysis of basic leucine zipper transcription factors in trifoliate orange and the function identification of PtbZIP49 in salt tolerance. Hortic. Plant J. 2024, 10, 115–130. [Google Scholar] [CrossRef]
- Geng, J.J.; Liu, J.H. The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene. J. Exp. Bot. 2018, 69, 2677–2692. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.J.; Wei, T.L.; Wang, Y.; Huang, X.S.; Liu, J.H. Overexpression of PtrbHLH, a basic helixloop-helix transcription factor from Poncirus trifoliata, confers enhanced cold tolerance in pummelo (Citrus grandis) by modulation of H2O2 level via regulating a CAT gene. Tree Physiol. 2019, 39, 2045–2054. [Google Scholar] [PubMed]
- Huang, X.S.; Wang, W.; Zhang, Q.; Liu, J.H. A Basic Helix-Loop-Helix Transcription Factor, PtrbHLH, of Poncirus trifoliata Confers Cold Tolerance and Modulates Peroxidase-Mediated Scavenging of Hydrogen Peroxide. Plant Physiol. 2013, 162, 1178–1194. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.S.; Zhang, Q.H.; Zhu, D.X.; Fu, X.Z.; Wang, M.; Zhang, Q.; Moriguchi, T.; Liu, J.H. ICE1 of Poncirus trifoliata functions in cold tolerance by modulating polyamine levels through interacting with arginine decarboxylase. J. Exp. Bot. 2015, 66, 3259–3274. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Dai, W.S.; Du, J.; Ming, R.H.; Dahro, B.; Liu, J.H. ERF109 of trifoliate orange (Poncirus trifoliata (L.) Raf.) contributes to cold tolerance by directly regulating expression of Prx1 involved in antioxidative process. Plant Biotechnol. J. 2019, 17, 1316–1332. [Google Scholar] [CrossRef]
- Singh, S.; Chaudhary, C.; Bharsakale, R.D.; Gazal, S.; Verma, L.; Tarannum, Z.; Behere, G.T.; Giri, J.; Germain, H.; Ghosh, D.K.; et al. PRpnp, a novel dual activity PNP family protein improves plant vigour and confers multiple stress tolerance in Citrus aurantifolia. Plant Biotechnol. J. 2023, 21, 726–741. [Google Scholar] [CrossRef]
- Zhang, Y.; Ming, R.; Khan, M.; Wang, Y.; Dahro, B.; Xiao, W.; Li, C.L.; Liu, J.H. ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione S-transferase U17 gene. Plant Biotechnol. J. 2022, 20, 183–200. [Google Scholar] [CrossRef]
- Khan, M.; Hu, J.B.; Dahro, B.; Ming, R.H.; Zhang, Y.; Wang, Y.; Alhag, A.; Li, C.L.; Liu, J.H. ERF108 from Poncirus trifoliata (L.) Raf. functions in cold tolerance by modulating raffinose synthesis through transcriptional regulation of PtrRafS. Plant J. 2021, 108, 705–724. [Google Scholar] [CrossRef]
- Peng, Z.; Bredeson, J.V.; Wu, G.A.; Shu, S.Q.; Rawat, N.; Du, D.L.; Parajuli, S.; Yu, Q.B.; You, Q.; Rokhsar, D.S.; et al. A chromosome-scale reference genome of trifoliate orange (Poncirus trifoliata) provides insights into disease resistance, cold tolerance and genome evolution in Citrus. Plant J. 2020, 104, 1215–1232. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, Y.T.; Jiang, X.L.; Yu, H.W.; Jia, H.H.; Tan, C.M.; Hu, G.; Hu, Y.B.; Rao, M.J.; Deng, X.X.; et al. Genome of a citrus rootstock and global DNA demethylation caused by heterografting. Hortic. Res. 2021, 8, 69. [Google Scholar] [CrossRef]
- Ji, C.Y.; Song, F.; He, C.; An, J.Y.; Huang, S.Y.; Yu, H.M.; Lu, H.; Xiao, S.Y.; Bucher, M.; Pan, Z.Y. Integrated miRNA–mRNA analysis reveals candidate miRNA family regulating arbuscular mycorrhizal symbiosis of Poncirus trifoliata. Plant Cell Environ. 2023, 46, 1805–1821. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.E.; Spelt, C.E.; Mol, J.N.M. The chalcone synthase multigene family of Petunia hybrida (V30): Differential, light-regulated expression during flower development and UV light induction. Plant Mol. Biol. 1989, 12, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Leyva, A.; Jarillo, J.A.; Salinas, J.; Martinez-Zapater, J.M. Low Temperature Induces the Accumulation of Phenylalanine Ammonia-Lyase and Chalcone Synthase mRNAs of Arabidopsis thaliana in a Light-Dependent Manner. Plant Physiol. 1995, 108, 39–46. [Google Scholar] [CrossRef]
- Singh, N.; Kumaria, S. Molecular cloning and characterization of chalcone synthase gene from Coelogyne ovalis Lindl. and its stress-dependent expression. Gene 2020, 762, 145104. [Google Scholar] [CrossRef]
- Deng, Y.X.; Li, C.L.; Li, H.Q.; Lu, S.F. Identification and Characterization of Flavonoid Biosynthetic Enzyme Genes in Salvia miltiorrhiza (Lamiaceae). Molecules 2018, 23, 1467. [Google Scholar] [CrossRef]
- Harker, C.L.; Ellis, T.H.N.; Coen, E.S. Identification and Genetic Regulation of the Chalcone Synthase Multigene Family in Pea. Plant Cell 1990, 2, 185–194. [Google Scholar]
- Jiang, M.Y.; Zhang, J.H. Effect of Abscisic Acid on Active Oxygen Species, Antioxidative Defence System and Oxidative Damage in Leaves of Maize Seedlings. Plant Cell Physiol. 2001, 42, 1265–1273. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, T.T.; Li, Y.X.; Kang, Y.Q.; Wang, P.; Liu, W.; Wang, Y.J.; Tian, L.B.; Dai, J.; Zhou, Y. Genome-Wide Identification and Expression Analysis of the Chalcone Synthase (CHS) Gene Family in Dendrobium catenatum. Agronomy 2023, 13, 1488. [Google Scholar] [CrossRef]
- Sadowska-Bartosz, I.; Bartosz, G. Antioxidant Activity of Anthocyanins and Anthocyanidins: A Critical Review. Int. J. Mol. Sci. 2024, 25, 12001. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | CPBD Number | Chromosome | Number of Amino Acids (aa) | Molecular Weight/kDa | Isoelectric Point | The Instability Index | In Silico Subcellular Localization Predicted |
|---|---|---|---|---|---|---|---|
| PtrCHS1 | Pt2g009170.1 | Chr2 | 391 | 42.62 | 6.29 | 35.00 | Cytosol |
| PtrCHS2 | Pt3g040440.1 | Chr3 | 390 | 42.89 | 5.81 | 48.96 | Cytosol |
| PtrCHS3 | Pt5g007170.1 | Chr5 | 394 | 43.42 | 6.80 | 42.09 | Cytosol |
| PtrCHS4 | Pt5g007600.1 | Chr5 | 396 | 43.35 | 6.33 | 37.25 | Cytosol |
| PtrCHS5 | Pt5g008530.1 | Chr5 | 195 | 20.88 | 4.62 | 29.90 | Cytosol |
| PtrCHS6 | Pt5g008550.1 | Chr5 | 253 | 27.62 | 5.90 | 36.37 | Cytosol |
| PtrCHS7 | Pt7g002450.1 | Chr7 | 391 | 42.76 | 5.61 | 38.31 | Cytosol |
| PtrCHS8 | Pt9g010550.1 | Chr9 | 229 | 24.69 | 5.57 | 38.80 | Cytosol |
| PtrCHS9 | PtUn017340.1 | ChrUn | 387 | 42.44 | 5.87 | 43.29 | Cytosol |
| PtrCHS10 | PtUn017780.1 | ChrUn | 395 | 43.50 | 6.17 | 32.10 | Cytosol |
| PtrCHS11 | PtUn018300.1 | ChrUn | 387 | 42.42 | 5.87 | 43.29 | Cytosol |
| Protein Name | Secondary Structure | Phosphorylation Amino Acid Number | |||||
|---|---|---|---|---|---|---|---|
| Alpha Helix (%) | Extended Strand (%) | Beta Turn (%) | Random Coil (%) | Serine | Threonine | Tyrosine | |
| PtrCHS1 | 38.87 | 14.83 | 0.00 | 46.29 | 16 | 13 | 5 |
| PtrCHS2 | 38.21 | 13.59 | 0.00 | 48.21 | 17 | 14 | 3 |
| PtrCHS3 | 40.61 | 14.47 | 0.00 | 44.92 | 17 | 13 | 6 |
| PtrCHS4 | 38.89 | 14.65 | 0.00 | 46.46 | 19 | 9 | 5 |
| PtrCHS5 | 35.38 | 16.92 | 0.00 | 47.69 | 9 | 6 | 1 |
| PtrCHS6 | 36.36 | 17.39 | 0.00 | 46.25 | 11 | 6 | 1 |
| PtrCHS7 | 40.41 | 15.09 | 0.00 | 44.50 | 14 | 12 | 3 |
| PtrCHS8 | 42.36 | 15.28 | 0.00 | 42.36 | 8 | 4 | 0 |
| PtrCHS9 | 39.79 | 14.21 | 0.00 | 45.99 | 16 | 12 | 5 |
| PtrCHS10 | 39.49 | 14.68 | 0.00 | 45.82 | 14 | 7 | 5 |
| PtrCHS11 | 40.83 | 15.5 | 0.00 | 43.67 | 15 | 12 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Sheng, Y.; Song, C.; Liu, T.; Sheng, S.; Xu, X. Identification of Chalcone Synthase Genes and Their Responses to Salt and Cold Stress in Poncirus trifoliata. Plants 2025, 14, 3003. https://doi.org/10.3390/plants14193003
Jiang L, Sheng Y, Song C, Liu T, Sheng S, Xu X. Identification of Chalcone Synthase Genes and Their Responses to Salt and Cold Stress in Poncirus trifoliata. Plants. 2025; 14(19):3003. https://doi.org/10.3390/plants14193003
Chicago/Turabian StyleJiang, Lijuan, Yu Sheng, Chengyang Song, Teng Liu, Shuangyu Sheng, and Xiaoyong Xu. 2025. "Identification of Chalcone Synthase Genes and Their Responses to Salt and Cold Stress in Poncirus trifoliata" Plants 14, no. 19: 3003. https://doi.org/10.3390/plants14193003
APA StyleJiang, L., Sheng, Y., Song, C., Liu, T., Sheng, S., & Xu, X. (2025). Identification of Chalcone Synthase Genes and Their Responses to Salt and Cold Stress in Poncirus trifoliata. Plants, 14(19), 3003. https://doi.org/10.3390/plants14193003







