Endangered with High Dispersal Abilities: Conservation Genetics of Himantoglossum metlesicsianum (Teschner) P. Delforge (Orchidaceae) in the Canary Islands
Abstract
1. Introduction
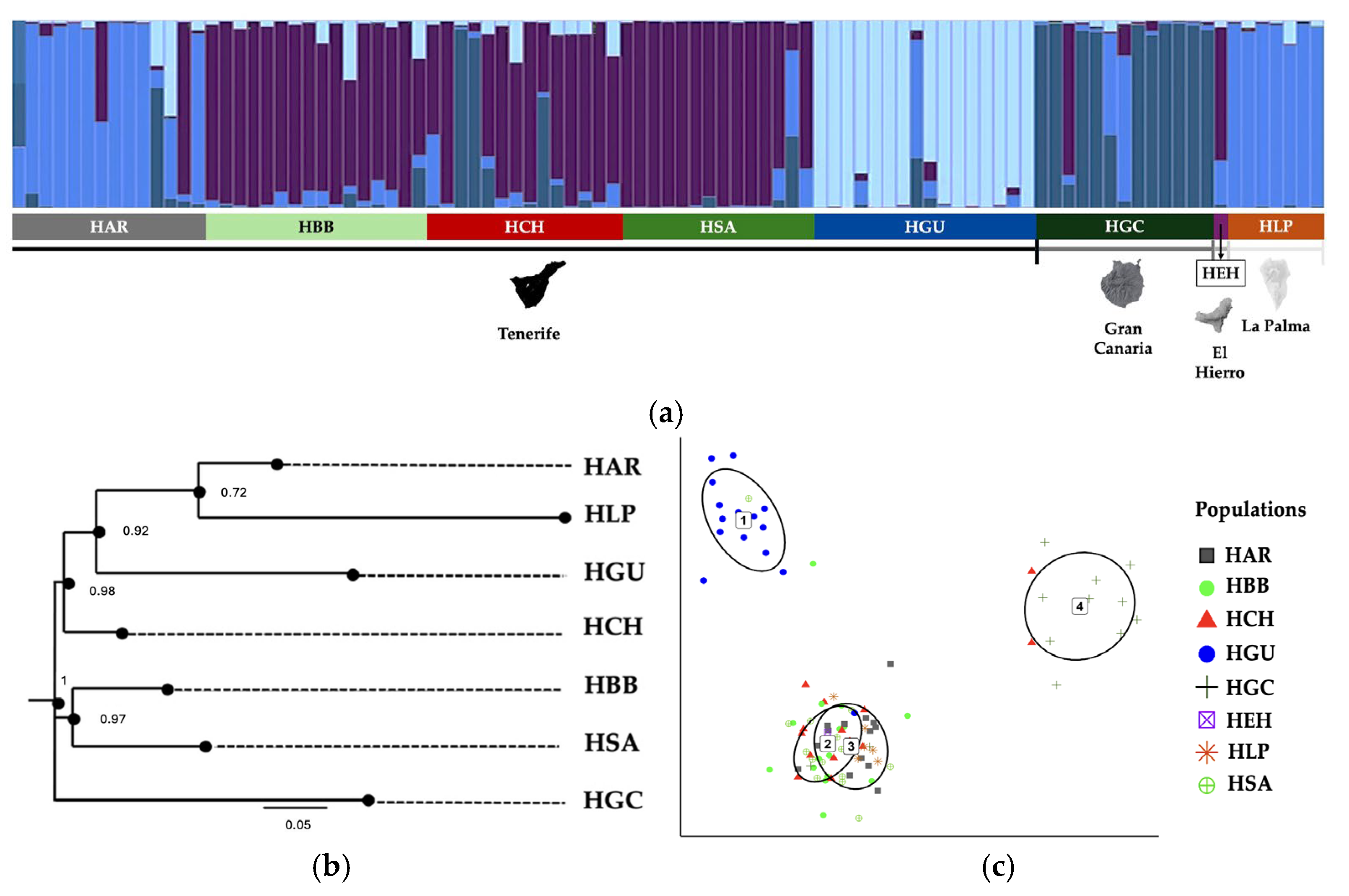
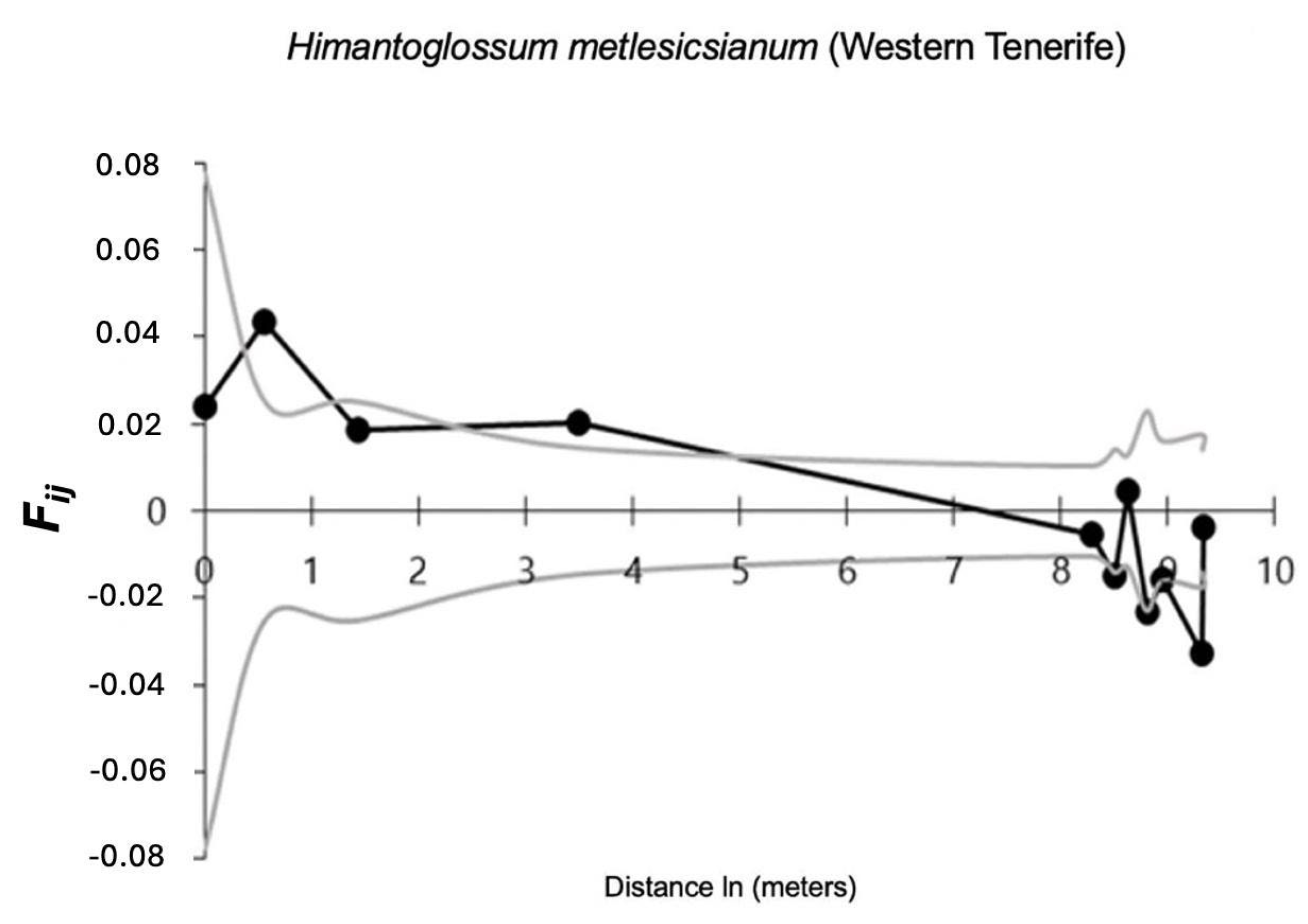
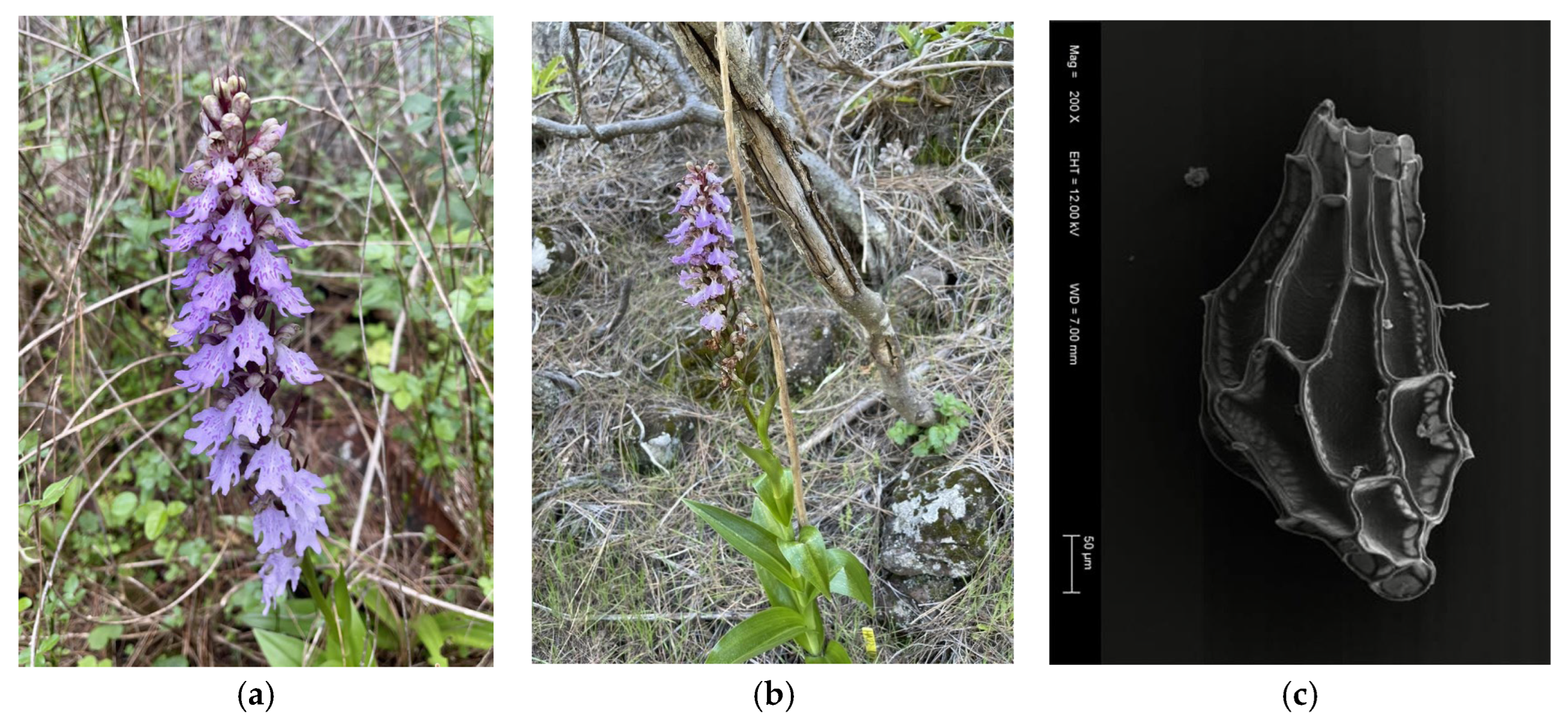
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Genotyping
4.2. Development of Microsatellites
4.3. Genetic Analysis
5. Conclusions
Implications for Conservation
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crawford, D.J.; Whitkus, R.; Stuessy, T.F. Plant evolution and speciation on oceanic islands. In Patterns of Differentiation in Higher Plants; Urbanska, E.K., Ed.; Academic Press: New York, NY, USA, 1987; pp. 183–199. [Google Scholar]
- Adsersen, H. Research on islands: Classic, recent and prospective approaches. In Islands: Biological Diversity and Ecosystem Function; En, P.M., Vitousek, L., Loope, L., Adsersen, H., Eds.; Springer: Berlin/Heidelberg, Germany, 1995; pp. 7–21. [Google Scholar] [CrossRef]
- Crawford, D.J.; Stuessy, T.F. Plant speciation on oceanic islands. In Evolution and Diversification of Land Plants; Iwatsuki, E.K., Raven, P.H., Eds.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 249–267. [Google Scholar]
- Juan, C.; Emerson, B.C.; Oromí, P.; Hewitt, G.M. Colonization and diversification: Towards a phylogeographic synthesis for the Canary Islands. Trends Ecol. Evol. 2000, 15, 104–109. [Google Scholar] [CrossRef] [PubMed]
- García-Verdugo, C.; Mairal, M.; Monroy, P.; Sajeva, M.; Caujapé-Castells, J. Do island plant populations really have lower genetic variation than mainland populations? Effects of selection and distribution range on genetic diversity estimates. Mol. Ecol. 2015, 24, 726–741. [Google Scholar] [CrossRef]
- Fernández-Mazuecos, M.; Vargas, P. Genetically depauperate in the continent but rich in oceanic islands: Cistus monspeliensis (Cistaceae) in the Canary Islands. PLoS ONE 2011, 6, e17172. [Google Scholar] [CrossRef]
- Warren, B.; Simberloff, D.; Ricklefs, R.; Aguilée, R.; Condamine, F.; Gravel, D.; Morlon, H.; Mouquet, N.; Rosindell, J.; Casquet, J.; et al. Islands as model systems in ecology and evolution: Prospects fifty years after MacArthur-Wilson. Ecol. Lett. 2015, 18, 200–217. [Google Scholar] [CrossRef]
- Medail, F.; Quezel, P. Hot-Spots Analysis for Conservation of Plant Biodiversity in the Mediterranean Basin. Ann. Mo. Bot. Gard. 1997, 84, 112–127. [Google Scholar] [CrossRef]
- Caujapé-Castells, J. General GTP conclusions: Endemism, speciation and conservation. In The Biology of Island Floras; Caujapé-Castells, J., Ed.; Cambridge University Press: Cambridge, UK, 2011; pp. 313–340. [Google Scholar]
- Caujapé-Castells, J.; García-Verdugo, C.; Marrero-Rodríguez, Á.; Fernández-Palacios, J.M.; Crawford, D.J.; Mort, M.E. Island ontogenies, syngameons, and the origins and evolution of genetic diversity in the Canarian endemic flora. Perspect. Plant Ecol. Evol. Syst. 2017, 27, 9–22. [Google Scholar] [CrossRef]
- Bateman, R.M. Circumscribing genera in the European orchid flora: A subjective critique of recent contributions. Ber. Arbeitskrs. Heim. Orch. Beiheft 2012, 8, 94–126. [Google Scholar]
- Frankham, R. Inbreeding and extinction: Island populations. Conserv. Biol. 1998, 12, 665–675. [Google Scholar] [CrossRef]
- Bouzat, J.L. Conservation genetics of population bottlenecks: The role of chance, selection, and history. Conserv. Genet. 2010, 11, 463–478. [Google Scholar] [CrossRef]
- Silva, J.M.; González-Pérez, M.A.; Nogales, M.; Rumeu, B. Reduced reproductive fitness of an endemic insular juniper population: Implications for conservation. Biol. Conserv. 2006, 129, 469–479. [Google Scholar]
- Hedrick, P. Genetics of Populations, 4th ed.; Jones and Bartlett: Boston, MA, USA, 2010. [Google Scholar]
- Carroll, S.P.; Fox, C.W. (Eds.) Conservation Biology: Evolution in Action; Oxford University Press: Oxford, UK, 2008. [Google Scholar] [CrossRef]
- Fay, M.F. Orchid conservation: A global perspective. Orchid Rev. 2020, 128, 3–10. [Google Scholar]
- IUCN. The IUCN Red List of Threatened Species; Versión 2020; IUCN: Gland, Switzerland, 2020. [Google Scholar]
- IUCN. The IUCN Red List of Threatened Species; Versión 2023.1; IUCN: Gland, Switzerland, 2023. [Google Scholar]
- Seaton, P.; Hu, H.; Perner, H.; Pritchard, H. Ex Situ Conservation of Orchids in a Warming World. Bot. Rev. 2010, 76, 193–203. [Google Scholar] [CrossRef]
- Godo, T.; Komori, M.; Nakaoki, E.; Yukawa, T.; Miyoshi, K. Germination of mature seeds of Calanthe tricarinata Lindl., an endangered terrestrial orchid, by asymbiotic culture in vitro. Vitr. Cell. Dev. Biol. Anim. 2010, 46, 323–328. [Google Scholar] [CrossRef]
- Wotavová, K.; Balounová, Z.; Kindlmann, P. Factors affecting persistence of terrestrial orchids in wet meadows and implications for their conservation in a changing agricultural landscape. Biol. Conserv. 2004, 118, 271–279. [Google Scholar] [CrossRef]
- Pfeifer, M.; Jetschke, G. Influence of Geographical Isolation on Genetic Diversity of Himantoglossum hircinum (Orchidaceae). Folia Geobot. 2006, 41, 3–20. [Google Scholar] [CrossRef]
- Fay, M.F.; Bone, R.; Cook, P.; Kahandawala, I.; Greensmith, J.; Harris, S.; Pedersen, H.A.; Ingrouille, M.J.; Lexer, C. Genetic Diversity in Cypripedium Calceolus (Orchidaceae) with a Focus on North-western Europe, as Revealed by Plastid DNA Length Polymorphisms. Ann. Bot. 2009, 104, 517–525. [Google Scholar] [CrossRef]
- Malanson, G.; Armstrong, M. Dispersal probability and forest diversity in a fragmented landscape. Ecol. Model. 1996, 87, 91–102. [Google Scholar] [CrossRef]
- Cain, M.; Milligan, B.; Strand, A. Long-distance seed dispersal in plant populations. Am. J. Bot. 2000, 87, 1217–1227. [Google Scholar] [CrossRef]
- Baldauf, S.; Engqvist, L.; Weissing, F. Diversifying evolution of competitiveness. Nat. Commun. 2014, 5, 5233. [Google Scholar] [CrossRef]
- Helsen, K.; Jacquemyn, H.; Honnay, O. Hidden founder effects: Small-scale spatial genetic structure in recently established populations of the grassland specialist plant Anthyllis vulneraria. Mol. Ecol. 2015, 24, 2715–2728. [Google Scholar] [CrossRef]
- Arditti, J.; Ghani, A.K.A. Numerical and physical properties of orchid seeds and their biological implications. New Phytol. 2000, 145, 367–421. [Google Scholar] [CrossRef] [PubMed]
- Jacquemyn, H.; Vandepitte, K.; Brys, R.; Honnay, O.; Roldán-Ruiz, I. Fitness variation and genetic diversity in small remnant populations of the food deceptive orchid Orchis purpurea. Biol. Conserv. 2007, 139, 203–210. [Google Scholar] [CrossRef]
- Chung, M.Y. Low levels of genetic variation within populations of the four rare orchids Gymnadenia cucullata, Gymnadenia camtschatica, Amitostigma gracile, and Pogonia minor in South Korea: Indication of genetic drift and implications for conservation. Plant Syst. Evol. 2009, 281, 65–76. [Google Scholar] [CrossRef]
- Brzosko, E.; Ostrowiecka, B.; Kotowicz, J.; Bolesta, M.; Gromotowicz, A.; Gromotowicz, M.; Orzechowska, A.; Orzołek, J.; Wojdalska, M. Seed dispersal in six species of terrestrial orchids in Biebrza National Park (NE Poland). Acta Soc. Bot. Pol. 2017, 86, E1. [Google Scholar] [CrossRef]
- Phillips, R.D.; Dixon, K.W.; Peakall, R. Low population genetic differentiation in the Orchidaceae: Implications for the diversification of the family. Mol. Ecol. 2012, 21, 5208–5220. [Google Scholar] [CrossRef]
- Johnson, S.; Edwards, T. The structure and function of orchid pollinaria. Plant Syst. Evol. 2000, 222, 243–269. [Google Scholar] [CrossRef]
- Nilsson, L.A. Mimesis of bellflower (Campanula) by the red helleborine orchid Cephalanthera rubra. Nature 1983, 305, 799–800. [Google Scholar] [CrossRef]
- Peter, C.I.; Johnson, S.D. Mimics and magnets: The importance of color and ecological facilitation in floral deception. Ecology 2008, 89, 1583–1595. [Google Scholar] [CrossRef]
- Jersáková, J.; Jürgens, A.; Šmilauer, P.; Johnson, S.D. The evolution of floral mimicry: Identifying traits that visually attract pollinators. Funct. Ecol. 2012, 26, 1381–1389. [Google Scholar] [CrossRef]
- Cozzolino, S.; Widmer, A. Orchid Diversity: An Evolutionary Consequence of Deception? Trends Ecol. Evol. 2005, 20, 487–494. [Google Scholar] [CrossRef]
- Claessens, J.; Kleynen, J. The Flower of the European Orchid: Form and Function; Claessens & Kleynen: Geulle, The Netherlands, 2011; ISBN 978-90-9025556-9. [Google Scholar]
- Bateman, R.M.; Rudall, P.J.; Hawkins, J.A.; Sramkó, G. Morphometric, molecular, ontogenetic and demographic observations on selected populations of the lizard orchid. Himantoglossum hircinum. New J. Bot. 2013, 3, 122–140. [Google Scholar] [CrossRef]
- Bateman, R.; Hollingsworth, P.; Preston, J.; Yi-Bo, L.; Pridgeon, A.; Chase, M. Molecular phylogenetics and evolution of Orchidinae and selected Habenariinae (Orchidaceae). Bot. J. Linn. Soc. 2003, 142, 1–40. [Google Scholar] [CrossRef]
- Sramkó, G.; Attila, M.; Hawkins, J.; Bateman, R. Molecular phylogeny and evolutionary history of the Eurasiatic orchid genus Himantoglossum s. l. (Orchidaceae). Ann. Bot. 2014, 114, 1609–1626. [Google Scholar] [CrossRef]
- Martín Osorio, V.E.; Wildpret Martín, W.; González Negrín, R.; De la Torre, W. Study of the current vegetation of the historical lava flows of the Arafo Volcano, Tenerife, Canary Islands, Spain. Mediterr. Bot. 2020, 41, 193–212. [Google Scholar] [CrossRef]
- Claessens, J. Himantoglossum metlesicsianum in Northern Tenerife: An endangered orchid. J. Hardy Orchid. Soc. 2015, 12, 75. [Google Scholar]
- Acevedo Rodríguez, A.; Mesa Coello, R. Chorological additions of Himantoglossum metlesicsianum (W.P. Teschner) P. Delforge (Orchidaceae): First record for the island of La Palma (Canary Islands). Botánica Macaronésica 2013, 28, 123–128. [Google Scholar]
- Marrero, Á.; Claessens, M.; González, D.; Santiago, C.; Claessens, J. Chorological additions and distribution of the native orchids of Gran Canaria. Botánica Macaronésica 2019, 30, 65–88. [Google Scholar]
- González Negrín, R. Study of the populations of Himantoglossum metlesicsianum (Teschner) P. Delforge (Orchidaceae) in the Canary Islands. Doctoral Dissertation, University of La Laguna, San Cristóbal de La Laguna, Spain, 2025. [Google Scholar]
- Martín Osorio, V.E.; González Negrín, R.; Wildpret Martín, W.; Wildpret de La Torre, W. The potencial vegetation of Himantoglossum metlesicsianum (W.P. Teschner) P. Delforge, (Orchidaceae) Critically Endangered species in the Canary Islands. Int. J. Geobot. Res. 2022, 11, 89–104. [Google Scholar]
- Mesa Coello, R. Seguimiento de Poblaciones de Especies Amenazadas (2006). Himantoglossum metlesicsianum (W. P. Techner) P. Delforge. Banco de datos de Biodiversidad de Canarias.Viceconsejería de Medio Ambiente. Gobierno de Canarias. 2006; 19p, Unpublished document. [Google Scholar]
- Pfeifer, M.; Schatz, B.; Xavier Picó, F.; Passalacqua, N.; Fay, M.; Carey, P.; Jeltsch, F. Phylogeography and genetic structure of the orchid Himantoglossum hircinum (L.) Spreng. across its European central-marginal gradient. J. Biogeogr. 2009, 36, 2353–2365. [Google Scholar] [CrossRef]
- Chung, M.Y.; Nason, J.D.; Chung, M.G. Spatial genetic structure in populations of the terrestrial orchid Cephalanthera longibracteata (Orchidaceae). Am. J. Bot. 2004, 91, 52–57. [Google Scholar] [CrossRef]
- Loiselle, B.A.; Sork, V.L.; Nason, J.; Graham, C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 1995, 82, 1420–1425. [Google Scholar] [CrossRef]
- De Paz, J.; Caujapé-Castells, J. Review of the Allozyme Data Set for the Canarian Endemic Flora: Causes of the High Genetic Diversity Levels and Implications for Conservation. Ann. Bot. 2013, 111, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Hamrick, J.L. Isozymes and the analysis of genetic structure in plant populations. In Isozymes in Plant Biology; Soltis, E.D.E., Soltis, P.S., Eds.; Dioscorides Press: Portland, OR, USA, 1989; pp. 87–105. [Google Scholar]
- Francisco-Ortega, J.; Santos-Guerra, A.; Kim, S.-C.; Crawford, D.J. Plant Genetic Diversity in the Canary Islands: A Conservation Perspective. Am. J. Bot. 2000, 87, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Sosa, P.A.; González-Pérez, M.A.; González-González, E.A.; Rivero, E. Genetic diversity of Canarian endemisms revealed by microsatellites: Knowledge after one decade of analysis. In Proceedings of the Amurga international Conferences on Island Biodiversity, Maspalomas, Gran Canaria, March 2013; pp. 94–100. [Google Scholar]
- Gholami, S.; Vafaee, Y.; Nazari, F.; Ghorbani, A. Molecular characterization of endangered Iranian terrestrial orchids using ISSR markers and association with floral and tuber-related phenotypic traits. Physiol. Mol. Biol. Plants 2021, 27, 53–68. [Google Scholar] [CrossRef]
- Hartl, D.L.; Clark, A.G. Principles of Population Genetics, 3rd ed.; Sinauer Associates: Sunderland, MA, USA, 1997. [Google Scholar]
- Carey, P.D. Cambios en la distribución y abundancia de Himantoglossum hircinum (L.) Sprengel (Orchidaceae) durante los últimos 100 años. Watsonia 1999, 22, 353–364. [Google Scholar]
- García-Verdugo, C.; Fay, M.F. Ecology and evolution on oceanic islands: Broadening the botanical perspective. Bot. J. Linn. Soc. 2014, 174, 271–275. [Google Scholar] [CrossRef][Green Version]
- Hamrick, J.L.; Godt, M.J.W. Allozyme Diversity in Plant Species. In Plant Population Genetics, Breeding, and Genetic Resources; Brown, A.H.D., Clegg, M.T., Kahler, A.L., Weir, B.S., Eds.; Sinauer Associates: Sunderland, UK, 1990; pp. 43–63. [Google Scholar]
- Nybom, H.; Bartish, I.V. Effects of Life History Traits and Sampling Strategies on Genetic Diversity Estimates Obtained with RAPD Markers in Plants. Perspectives in Plant Ecology. Evol. Syst. 2000, 3, 93–164. [Google Scholar]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Scopece, G.; Cozzolino, S.; Johnson, S.D.; Schiestl, F.P. Pollination efficiency and the evolution of specialized deceptive pollination systems. Am. Nat. 2009, 175, 98–105. [Google Scholar] [CrossRef]
- Kraus, F.B.; Wolf, S.; Moritz, R.F.A. Male flight distance and population substructure in the bumblebee Bombus terrestris. J. Anim. Ecol. 2009, 78, 247–252. [Google Scholar] [CrossRef]
- Yang, Q.; Fu, Y.; Wang, Y.; Wang, Y.; Zhang, W.; Li, X.; Zhang, J. Genetic diversity and differentiation in the critically endangered orchid (Amitostigma hemipilioides): Implications for conservation. Plant Syst. Evol. 2014, 300, 871–879. [Google Scholar] [CrossRef]
- Yun, S.; Son, H.; Im, H.; Kim, S. Genetic diversity and population structure of the endangered orchid Pelatantheria scolopendrifolia (Orchidaceae) in Korea. PLoS ONE 2020, 15, E0237546. [Google Scholar] [CrossRef]
- Vekemans, X.; Hardy, O. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol. 2004, 13, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, S.; Cafasso, D.; Pellegrino, G.; Musacchio, A.; Widmer, A. Molecular Evolution of a Plastid Tandem Repeat Locus in an Orchid Lineage. J. Mol. Evol. 2003, 57, S41–S49. [Google Scholar] [CrossRef]
- Brzosko, E.; Wróblewska, A. Genetic diversity of nectar-rewarding Platanthera chlorantha and nectarless Cephalanthera rubra. Bot. J. Linn. Soc. 2013, 171, 751–763. [Google Scholar] [CrossRef]
- Forrest, A.; Hollingsworth, M.; Hollingsworth, P.; Sydes, C.; Bateman, R. Population genetic structure in European populations of Spiranthes romanzoffiana set in the context of other genetic studies on orchids. Heredity 2004, 92, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Jacquemyn, H.; Brys, R.; Honnay, O.; Hutchings, M.J. Biological flora of the British Isles: Orchis mascula (L.) L. Funct. Ecol. 2009, 23, 1–9. [Google Scholar] [CrossRef]
- Dellaporta, S.; Wood, J.; Hicks, J.B. A Plant DNA Minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar]
- Blacket, M.; Robin, C.; Good, R.; Lee, S.F.; Miller, A.D. Universal primers for fluorescent labelling of PCR fragments: An efficient and cost-effective approach to genotyping by fluorescence. Mol. Ecol. Resour. 2012, 12, 456–463. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete re-implementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Guo, S.W.; Thompson, E.A. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 1992, 48, 361–372. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P. GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Kalinowski, S.T. HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2004, 4, 6–7. [Google Scholar] [CrossRef]
- David, P.; Pujol, B.; Viard, F.; Castella, V.; Goudet, J. Reliable selfing rate estimates from imperfect population genetic data. Mol. Ecol. 2007, 16, 1181–1194. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.; VonHoldt, B. Structure harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Langella, O. Populations 1.2 Software. 2002. Available online: https://bioinformatics.org/populations/ (accessed on 25 February 2024).
- Rambaut, A. FigTree Software; v1.4.4; The University of Edinburgh: Edinburgh, UK, 2007; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 6 March 2024).
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; McInnes, K.H. Introduction to Conservation Genetics; Illustrated by Karina H. McInnes; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar] [CrossRef]

| Population | N | Na | Ar | Rar | PA | NTA | Ho | He | FIS | Sr | P (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HAR | 14 | 4.929 | 3.52 | 0.19 | 2 | 69 | 0.509 | 0.612 | 0.000 * | 0.188 | 100% |
| HBB | 15 | 5.643 | 4.13 | 0.41 | 4 | 79 | 0.686 | 0.695 | 0.012 ns | - | 93% |
| HCH | 15 | 5.572 | 3.99 | 0.26 | 4 | 78 | 0.644 | 0.677 | 0.0069 ns | 0.040 | 100% |
| HGU | 15 | 4.286 | 3.4 | 0.11 | 1 | 60 | 0.657 | 0.617 | 0.0096 ns | 0.136 | 100% |
| HSA | 15 | 5.571 | 4.03 | 0.46 | 6 | 78 | 0.667 | 0.694 | 0.0103 ns | 0.200 | 100% |
| HGC | 13 | 5.50 | 4.01 | 0.43 | 4 | 77 | 0.533 | 0.619 | 0.000 ** | 0.087 | 100% |
| HLP | 7 | 3.714 | 3.37 | 0.14 | 1 | 52 | 0.503 | 0.568 | 0.0004 ** | - | 93% |
| Total | 94 | - | - | 39 | 128 |
| Source of Variation | Degrees of Freedom | Sum of Squares | Components of Variance | Percentage of Variation | F-Statistics |
|---|---|---|---|---|---|
| TENERIFE, LA PALMA AND GRAN CANARIA | |||||
| Between islands | 2 | 45.619 | 0.341 | 7.81 | FCT = 0.067 *** |
| Between populations within islands | 4 | 58.491 | 0.370 | 8.46 | FSC = 0.116 *** |
| Within populations | 181 | 663.251 | 3.664 | 83.73 | FST = 0.175 *** |
| Total | 187 | 767.362 | 4.376 | ||
| TENERIFE [HAR, HBB, HCH, HGU, HSA] | |||||
| Between groups (West vs. East Tenerife) | 1 | 22.487 | 0.127 | 2.76 | FCT = 0.097 *** |
| Between populations within groups | 3 | 40.858 | 0.319 | 6.94 | FSC = 0.071 *** |
| Within populations | 143 | 593.743 | 4.152 | 90.30 | FST = 0.02 *** |
| Total | 147 | 657.088 | 4.598 | ||
| Statistical significance: *** indicates p < 0.001 | |||||
| Population | Population Code | UTM Coordinates | Total Number Individuals | Number of Individuals Analysed |
|---|---|---|---|---|
| Arafo | HAR | 357000 3134500 | 21 | 14 |
| Barranco Bermejo | HBB | 329500 3123000 | 20 | 15 |
| Chío | HCH | 325500 3126500 | 41 | 15 |
| Santiago del Teide | HSA | 323000 3132000 | 84 | 15 |
| Güímar | HGU | 357000 3134000 | 116 | 15 |
| La Palma | HLP | 213000 3181500 | 7 | 7 |
| Gran Canaria | HGC | 429500 3104000 | 14 | 13 |
| El Hierro | HEH | 199500 3069500 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González Negrín, R.; Martín Osorio, V.E.; Sosa, P.A.; Rodríguez-Rodríguez, P. Endangered with High Dispersal Abilities: Conservation Genetics of Himantoglossum metlesicsianum (Teschner) P. Delforge (Orchidaceae) in the Canary Islands. Plants 2025, 14, 1862. https://doi.org/10.3390/plants14121862
González Negrín R, Martín Osorio VE, Sosa PA, Rodríguez-Rodríguez P. Endangered with High Dispersal Abilities: Conservation Genetics of Himantoglossum metlesicsianum (Teschner) P. Delforge (Orchidaceae) in the Canary Islands. Plants. 2025; 14(12):1862. https://doi.org/10.3390/plants14121862
Chicago/Turabian StyleGonzález Negrín, Rocío, Victoria Eugenia Martín Osorio, Pedro A. Sosa, and Priscila Rodríguez-Rodríguez. 2025. "Endangered with High Dispersal Abilities: Conservation Genetics of Himantoglossum metlesicsianum (Teschner) P. Delforge (Orchidaceae) in the Canary Islands" Plants 14, no. 12: 1862. https://doi.org/10.3390/plants14121862
APA StyleGonzález Negrín, R., Martín Osorio, V. E., Sosa, P. A., & Rodríguez-Rodríguez, P. (2025). Endangered with High Dispersal Abilities: Conservation Genetics of Himantoglossum metlesicsianum (Teschner) P. Delforge (Orchidaceae) in the Canary Islands. Plants, 14(12), 1862. https://doi.org/10.3390/plants14121862







