Overexpressing BrWRKY22 Delays Flowering and Leaf Senescence via Inhibition of GA Biosynthesis in Brassica rapa
Abstract
1. Introduction
2. Results
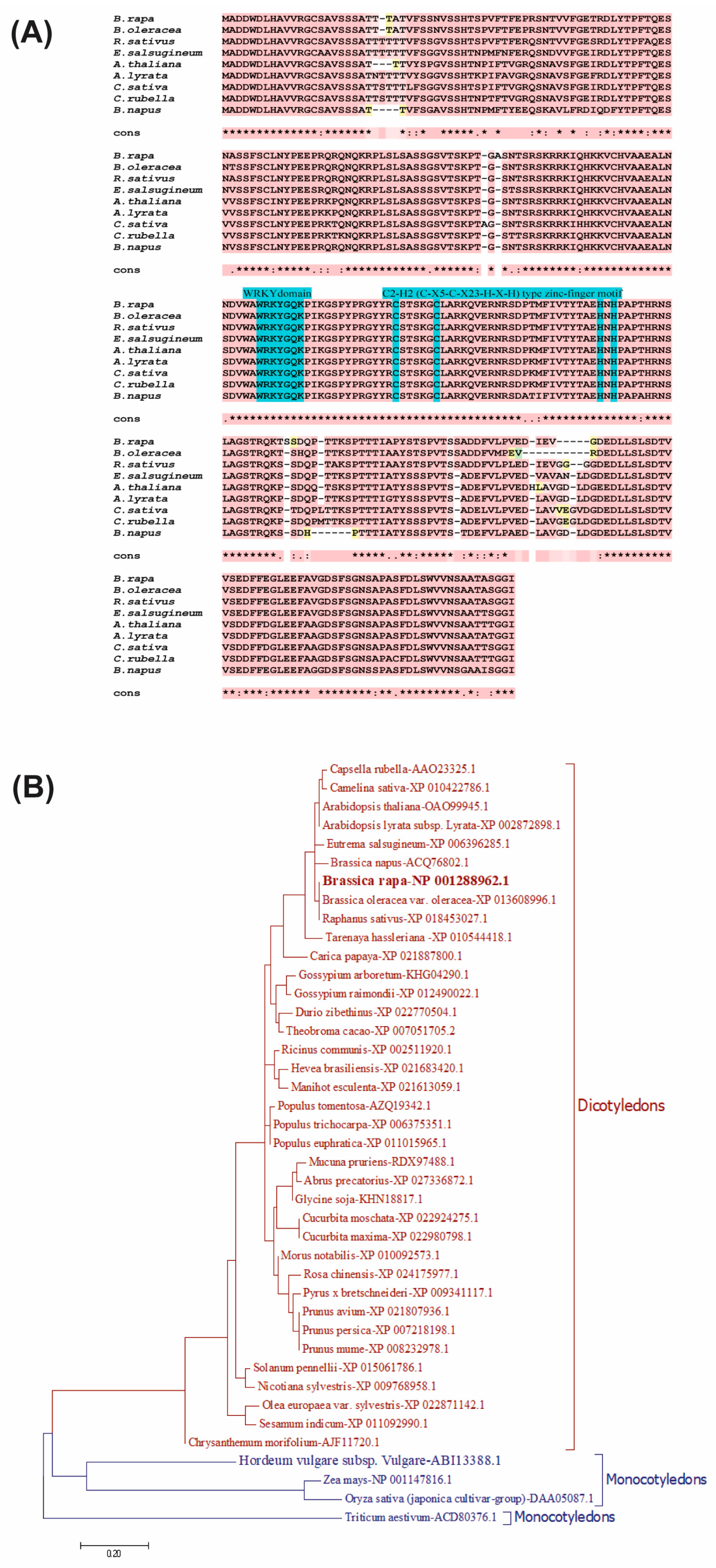
2.1. Sequence Alignment and Phylogenetic Relationship of the BrWRKY22 Protein
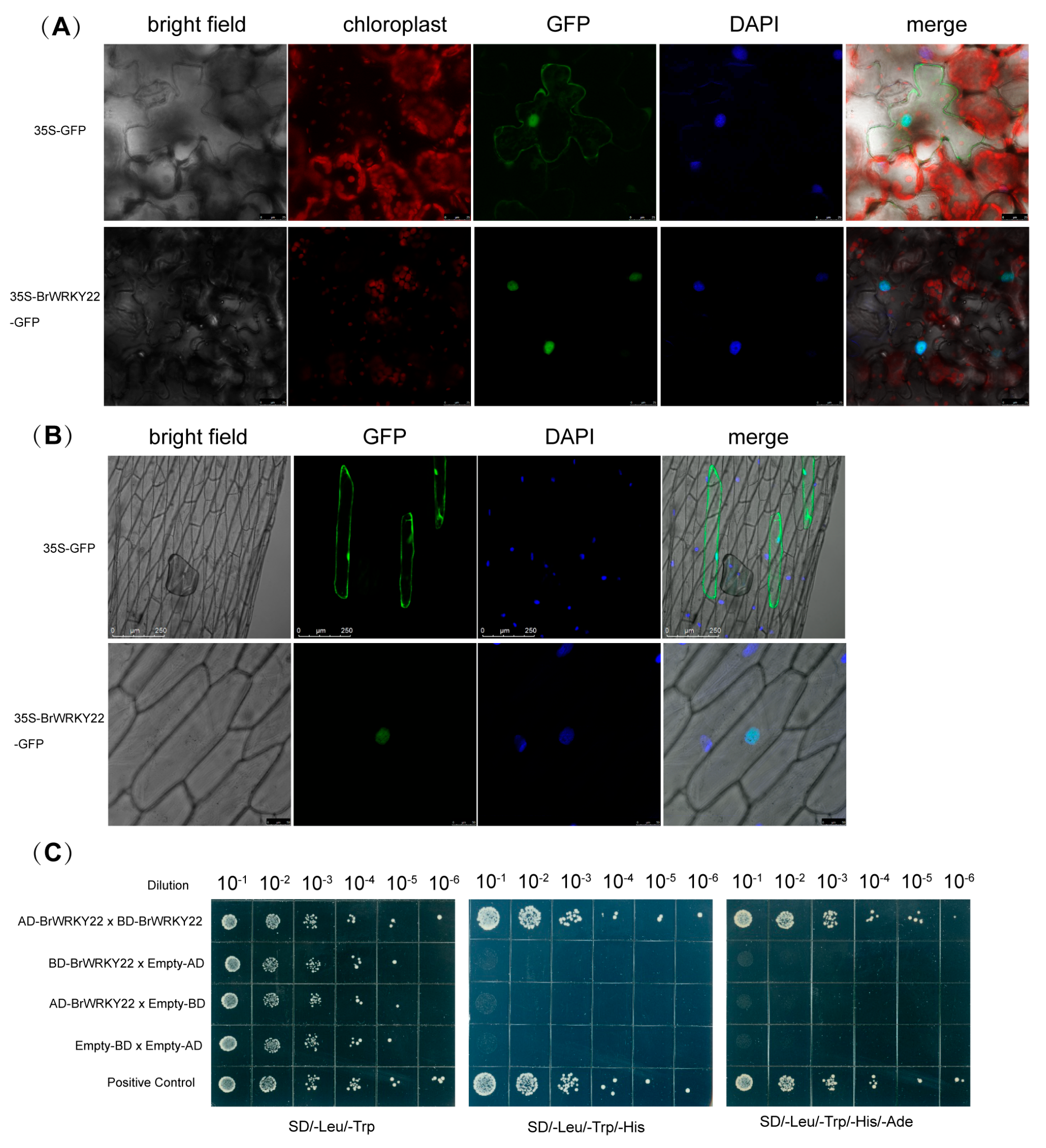
2.2. BrWRKY22 Localizes to the Nucleus and Forms a Homomeric Oligomer
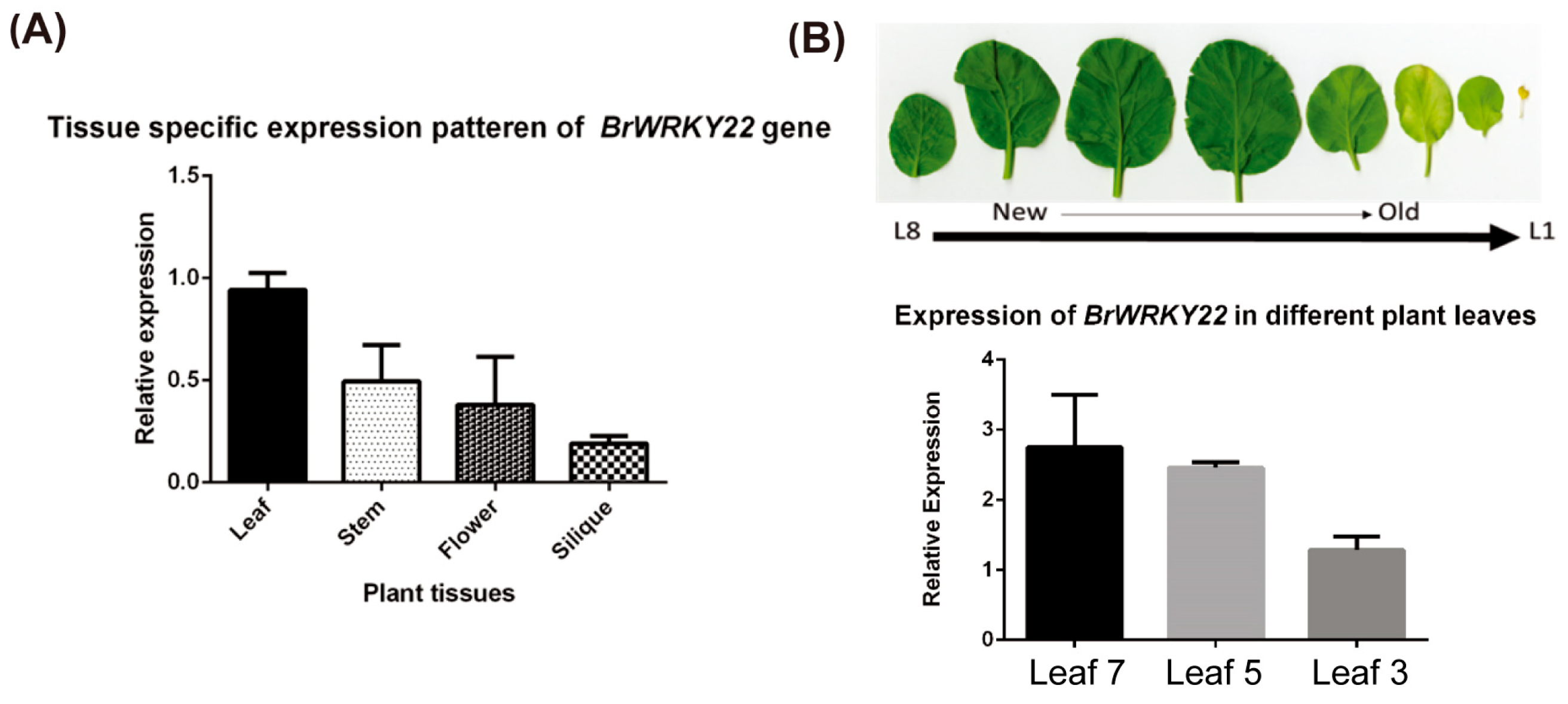
2.3. Expression Analysis of BrWRKY22 in Different Tissues
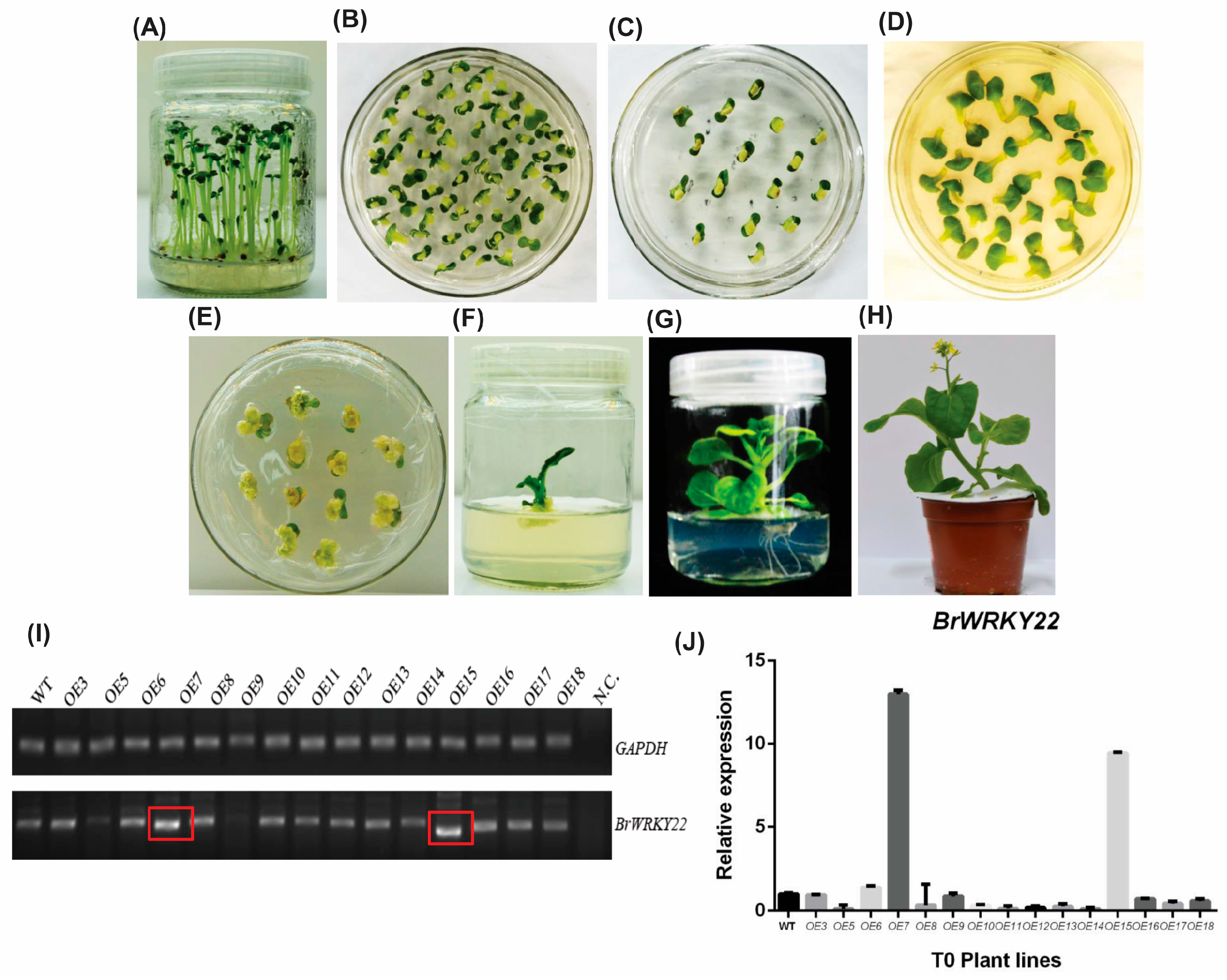
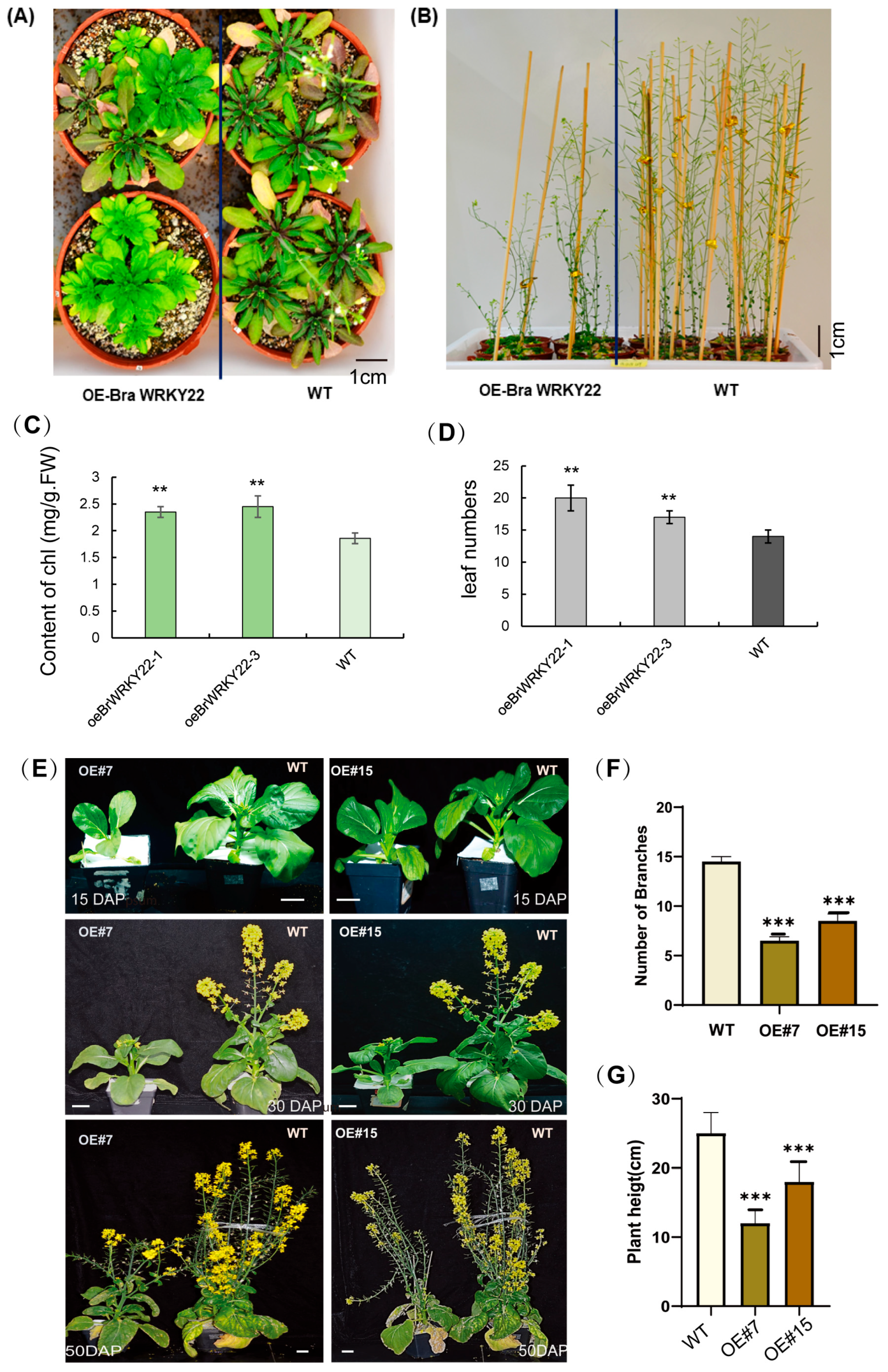
2.4. BrWRKY22 Overexpressed Transgenic Lines in Arabidopsis and in Brassica rapa Exhibit a Delay in Flowering Time and Leaf Senescence
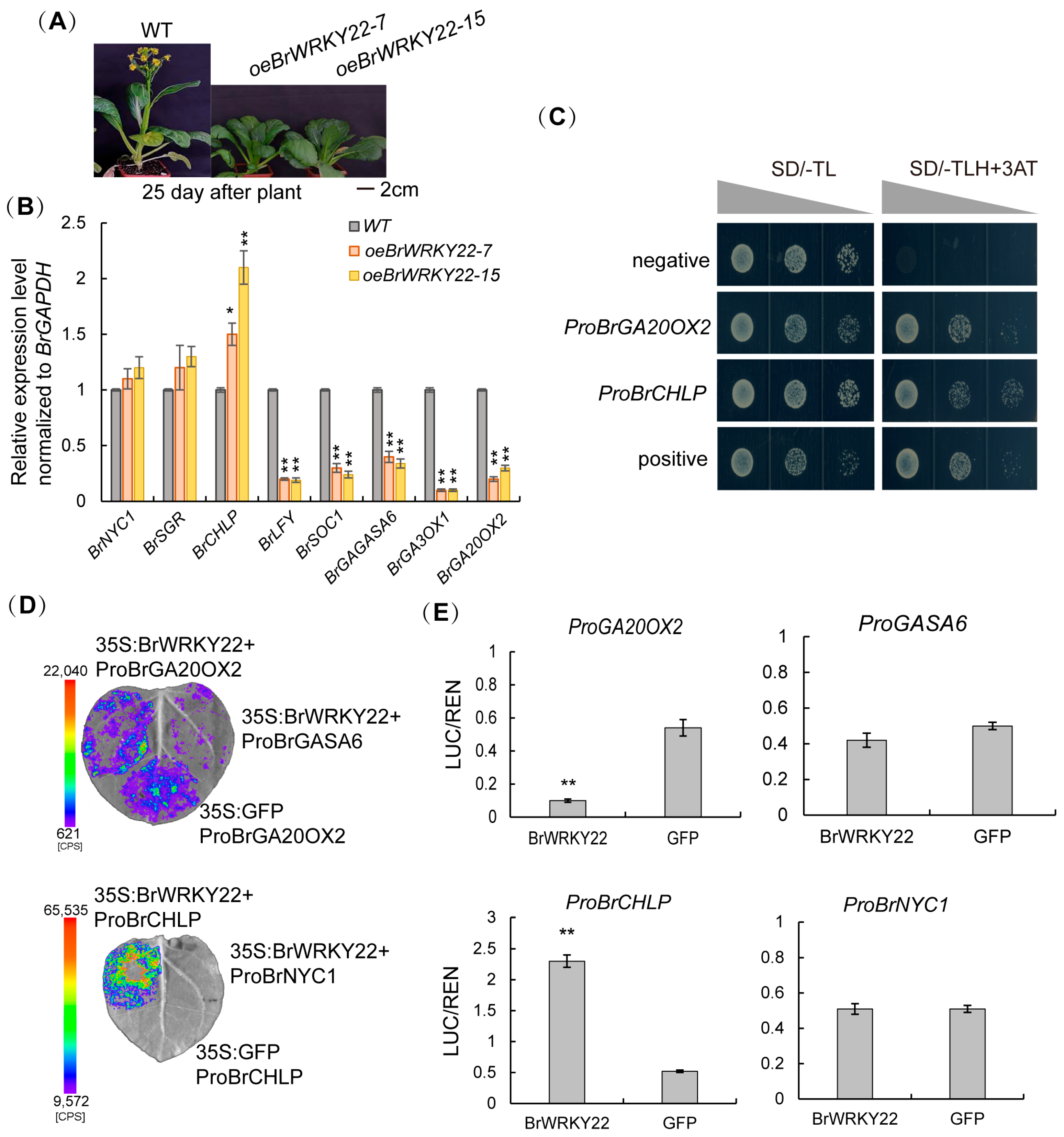
2.5. BrWRKY22 Directly Represses BrGA20OX2 Transcription and Activates BrCHLP Transcription in the Chlorophyll and GA Biosynthesis Pathway
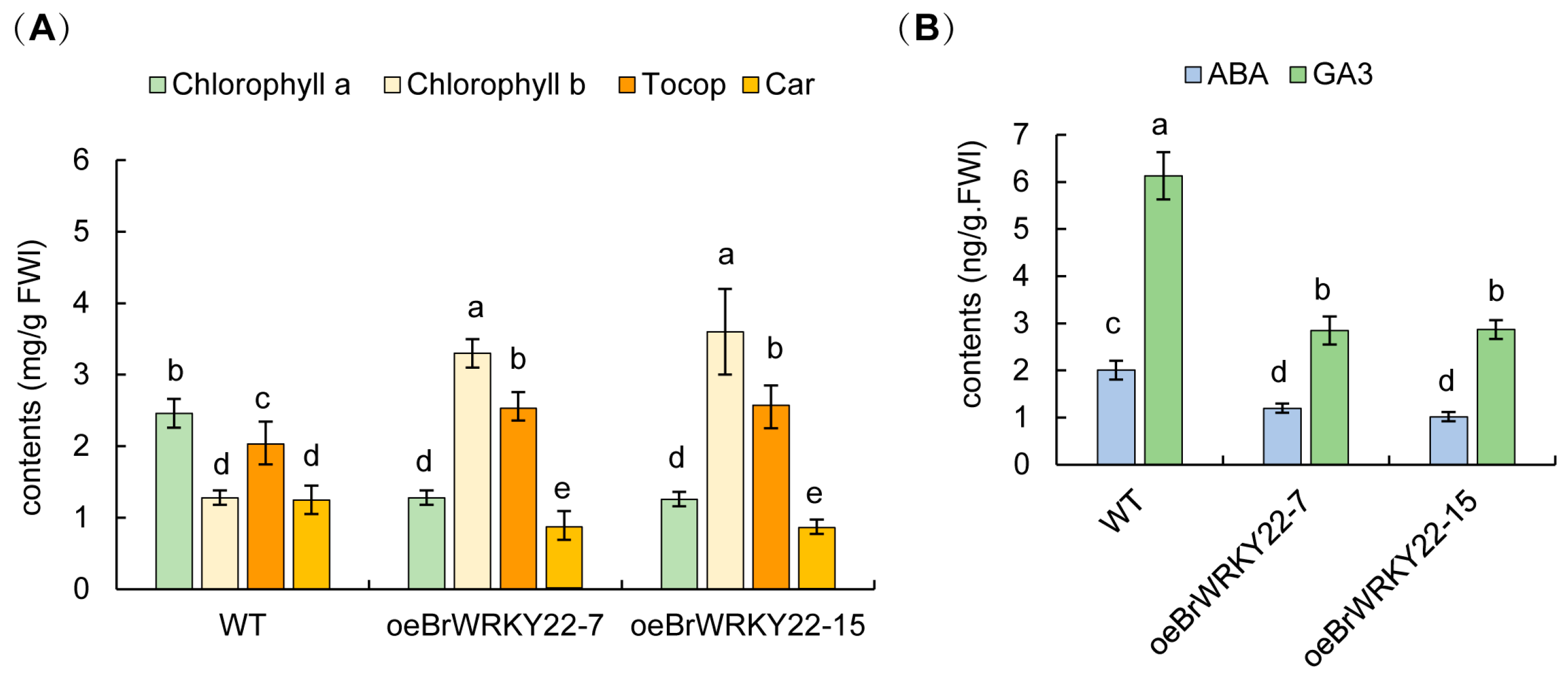
2.6. Overexpressing BrWRKY22 Plants Decreased the GA Content but Increased the Chlorophyll b and Tocopherol Contents
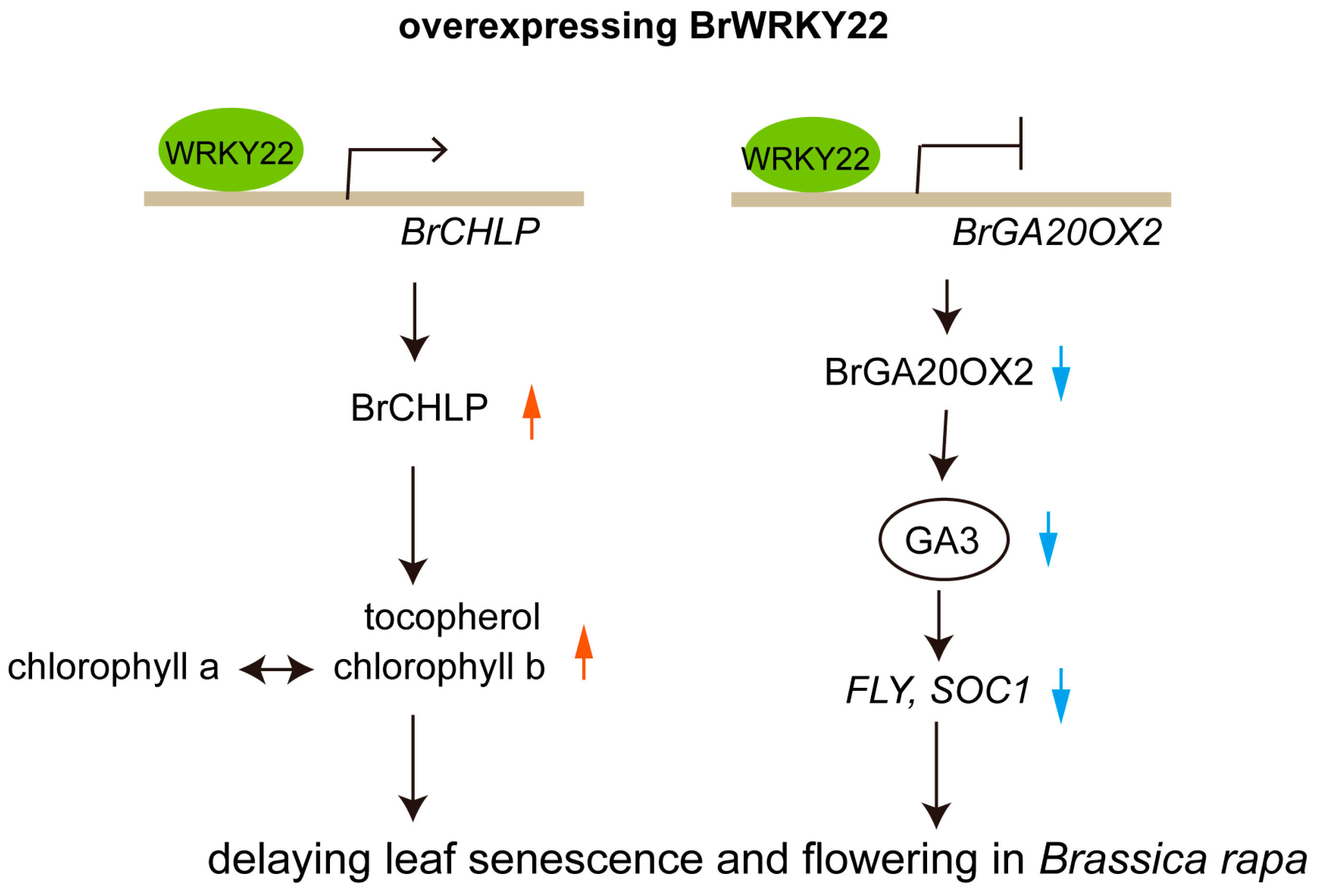
3. Discussion
4. Materials and Methods
4.1. BrWRKY22 Protein Sequences Retrieval
4.2. Phylogenetics Analysis of the BrWRKY22 Protein
4.3. BrWRKY22 Gene Structure Analysis
4.4. Identification of Cis-Acting Elements in the Promoter Region of BrWRKY22
4.5. Plant Materials and Growth Condition
4.6. Yeast Two-Hybrid (Y2H) Assay
4.7. Yeast One-Hybrid Assay
4.8. Subcellular Localization of BrWRKY22 in Arabidopsis and Nicotiana benthamiana
4.9. Transformation of the pCBIM-Bra037368-Flag Vector into Arabidopsis thaliana
4.10. Agrobacterium-Mediated Transformation of the OE Vector in Brassica rapa
4.11. SDS-PAGE and Western Blotting
4.12. Real-Time PCR Analysis
4.13. Measurement of Chlorophyll and Hormones
4.14. Dual-Luciferase Assay
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, M.; Wang, H.; Li, X.; Mason, A.S.; Fu, D. Rapeseed as an Ornamental. Horticulturae 2022, 8, 27. [Google Scholar] [CrossRef]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 15–18. [Google Scholar] [CrossRef]
- Jung, C.; Müller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Wingler, A. Interactions between flowering and senescence regulation and the influence of low temperature in Arabidopsis and crop plants Ann. Appl. Biol. 2011, 159, 320–338. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef]
- Hensel, L.L.; Grbic, V.; Baumgarten, D.A.; Bleecker, A.B. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 1993, 5, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ren, G.; Zhang, K.; Li, Z.; Miao, Y.; Guo, H. Leaf senescence: Progression, regulation, and application. Mol. Hort. 2021, 1, 5. [Google Scholar] [CrossRef]
- Daviere, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Zhou, D.; Zhang, Y.; Song, R.; Li, C.; Li, J.; Gao, J. The Roles of Gibberellins in Regulating Leaf Development. Plants 2023, 12, 1243. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Li, Y.; Yao, X.H.; Qiao, K.; Lin, W.; Liu, B.H.; Zhang, D.W.; Lin, H.H. NAP is involved in GA-mediated chlorophyll degradation and leaf senescence by interacting with DELLAs in Arabidopsis. Plant Cell Rep. 2019, 39, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. JIPB 2020, 62, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, M.; Shih, C.F.; Yang, C.H. Expression of an antisense Brassica oleracea GIGANTEA (BoGI) gene in transgenic broccoli causes delayed flowering, leaf senescence, and post-harvest yellowing retardation. Plant Mol. Biol. Rep. 2015, 33, 1499–1509. [Google Scholar] [CrossRef]
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Kracher, B.; Somssich, I.E. Induced genome-wide binding of three arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 2017, 29, 20–38. [Google Scholar] [CrossRef]
- Ullah, A.; Sun, H.; Hakim; Yang, X.; Zhang, X. A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant. 2018, 162, 439–454. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, C.X.; Li, G.X.; Wu, Y.R.; Zheng, S.J. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015, 84, 56–69. [Google Scholar] [CrossRef]
- Xie, W.; Li, X.; Wang, S.; Yuan, M. OsWRKY53 Promotes Abscisic Acid Accumulation to Accelerate Leaf Senescence and Inhibit Seed Germination by Downregulating Abscisic Acid Catabolic Genes in Rice. Front. Plant Sci. 2022, 12, 816156. [Google Scholar] [CrossRef]
- Schluttenhofer, C.; Yuan, L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef]
- Chen, L.; Lei, W.; He, W.; Wang, Y.; Tian, J.; Gong, J.; Hao, B.; Cheng, X.; Shu, Y.; Fan, Z. Mapping of Two Major QTLs Controlling Flowering Time in Brassica napus Using a High-Density Genetic Map. Plants 2022, 11, 2635. [Google Scholar] [CrossRef] [PubMed]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef]
- Watanabe, K.; Homayouni, A.; Gu, L.; David Ho, T.-H.; Zhang, L.; Ringler, P.; Smith, S.; Rushton, P.J.; Shen, Q.J. Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Elsevier 2017, 236, 214–222. [Google Scholar] [CrossRef]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef] [PubMed]
- Kiranmai, K.; Lokanadha Rao, G.; Pandurangaiah, M.; Nareshkumar, A.; Amaranatha Reddy, V.; Lokesh, U.; Venkatesh, B.; Anthony Johnson, A.M.; Sudhakar, C. A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 2018, 9, 346. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Sun, Y.; Wang, B.; Yu, S.; Dai, H.; Li, H.; Zhang, Z.; Zhang, J. Woodland strawberry WRKY71 acts as a promoter of flowering via a transcriptional regulatory cascade. Hortic. Res. 2020, 7, 137. [Google Scholar] [CrossRef]
- Li, W.; Wang, H.; Yu, D. Arabidopsis WRKY Transcription Factors WRKY12 and WRKY13 Oppositely Regulate Flowering under Short-Day Conditions. Mol. Plant 2016, 9, 1492–1503. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, L.; Yu, D. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 2018, 176, 790–803. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, S.; Li, X.; Chen, Y.; Li, J.; Wang, Y.; Bian, R.; Jin, Y.; Zhu, X.; Zhang, K. Arabidopsis WRKY1 promotes monocarpic senescence by integrative regulation of flowering, leaf senescence, and nitrogen remobilization. Mol. Plant 2024, 17, 1289–1306. [Google Scholar] [CrossRef]
- Zhou, X.; Jiang, Y.; Yu, D. WRKY22 Transcription Factor Mediates Dark-Induced Leaf Senescence in Arabidopsis. Mol. Cells 2011, 31, 303–313. [Google Scholar] [CrossRef]
- Kayum, M.A.; Jung, H.J.; Park, J.I.; Ahmed, N.U.; Saha, G.; Yang, T.J.; Nou, I.S. Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa. Mol. Genet. Genom. 2015, 290, 79–95. [Google Scholar] [CrossRef]
- Yao, Q.Y.; Xia, E.H.; Liu, F.H.; Gao, L.Z. Genome-wide identification and comparative expression analysis reveal a rapid expansion and functional divergence of duplicated genes in the WRKY gene family of cabbage, Brassica oleracea var. capitata. Gene 2015, 557, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Xiang, Y.; Xu, E.; Ge, X.; Li, Z. Major co-localized QTL for plant height, branch initiation height, stem diameter, and flowering time in an alien introgression derived Brassica napus DH population. Front. Plant Sci. 2018, 9, 390. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.Q.; Wei, W.; Tan, X.L.; Shan, W.; Kuang, J.F.; Lu, W.J.; Su, X.G.; Lakshmanan, P.; Lin, H.T.; Chen, J.Y. A NAC transcription factor BrNAC087 is involved in gibberellin-delayed leaf senescence in Chinese flowering cabbage. Postharvest Biol. Technol. 2021, 181, 111673. [Google Scholar] [CrossRef]
- Kaur, S.; Atri, C.; Akhatar, J.; Mittal, M.; Kaur, R.; Banga, S.S. Genetics of days to flowering, maturity and plant height in natural and derived forms of Brassica rapa L. Theor. Appl. Genet. 2021, 134, 473–487. [Google Scholar] [CrossRef]
- Shang, M.; Wang, X.; Zhang, J.; Qi, X.; Ping, A.; Hou, L.; Xing, G.; Li, G.; Li, M. Genetic Regulation of GA Metabolism during Vernalization, Floral Bud Initiation and Development in Pak Choi (Brassica rapa ssp. Chinensis Makino). Front. Plant Sci. 2017, 8, 1533. [Google Scholar] [CrossRef]
- Sun, T.P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arab. Book 2008, 6, e0103. [Google Scholar] [CrossRef]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Qurban, A.Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [Google Scholar] [CrossRef]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Miao, Y.; Laun, T.; Zimmerman, P.; Zentgraf, U. Targets of AtWRKY 53 transcription factor and its role during the senescence of Arabidopsis leaf. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Sun, X.; Liu, B.; Zhu, D.; Bai, X.; Cai, H.; Ji, W.; Cao, L.; Wu, J.; Wang, M.; et al. Ectopic Expression of a WRKY Homolog from Glycine soja Alters Flowering Time in Arabidopsis. PLoS ONE 2013, 8, e73295. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Z.; Wang, Z.Q.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.Q.; Li, G.X.; Wu, Y.R.; Yang, J.L.; Ma, J.F.; et al. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 2018, 219, 149–162. [Google Scholar] [CrossRef]
- Wu, Z.; Li, T.; Cao, X.; Zhang, D.; Teng, N. LilyWRKY factor LlWRKY22 promotes thermotolerance through autoactivation and activation of LlDREB2B. Hortic. Res. 2022, 9, uhac186. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wei, X.; Cui, X.; Wang, J.; Wang, Y.; Sun, M.; Zhao, P.; Yang, B.; Wang, Q.; Jiang, Y.Q. The transcription factor WRKY22 modulates ethylene biosynthesis and root development through transactivating the transcription of ACS5 and ACO5 in Arabidopsis. Physiol. Plant 2024, 176, e14371. [Google Scholar] [CrossRef]
- Long, Q.; Du, M.; Long, J.; Xie, Y.; Zhang, J.; Xu, L.; He, Y.; Li, Q.; Chen, S.; Zou, X. Transcription factor WRKY22 regulates canker susceptibility in sweet orange (Citrus sinensis Osbeck) by enhancing cell enlargement and CsLOB1 expression. Hortic. Res. 2021, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Tan, L.; Guo, X.; Lou, S.; Dan, X.; Han, Y.; Zeng, C.; Zhang, H.; Yang, K.; Chen, L.; et al. Allelic variation in the promoter of WRKY22 enhances humid adaptation of Arabidopsis thaliana. Mol. Plant 2025, 18, 42–58. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Kracher, B.; Ross, A.; Kramer, K.; Finkemeier, I.; Somssich, I.E. Principles and characteristics of the Arabidopsis WRKY regulatory network during early MAMP-triggered immunity. Plant J. 2018, 96, 487–502. [Google Scholar] [CrossRef]
- Li, S.; Khoso, M.A.; Xu, H.; Zhang, C.; Liu, Z.; Wagan, S.; Dinislam, K.; Liu, L. WRKY Transcription Factors (TFs) as Key Regulators of Plant Resilience to Environmental Stresses: Current Perspective. Agronomy 2024, 14, 2421. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef]
- Teotia, S.; Tang, G. To bloom or not to bloom: Role of microRNAs in plant flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef]
- Wilson, R.N.; Heckman, J.W.; Somerville, C.R. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 1992, 100, 403–408. [Google Scholar] [CrossRef]
- Xu, Y.L.; Gage, D.A.; Zeevaart, J.A. Gibberellins and stem growth in Arabidopsis thaliana. Effects of photoperiod on expression of the GA4 and GA5 loci. Plant Physiol. 1997, 114, 1471–1476. [Google Scholar] [CrossRef]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef]
- Moon, J.; Suh, S.S.; Lee, H.; Choi, K.R.; Hong, C.B.; Paek, N.C.; Kim, S.G.; Lee, I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003, 35, 613–623. [Google Scholar] [CrossRef]
- Tanaka, R.; Oster, U.; Kruse, E.; Rudiger, W.; Grimm, B. Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 1999, 120, 695–704. [Google Scholar] [CrossRef]
- Tamary, E.; Nevo, R.; Naveh, L.; Zaidman, S.; Kiss, V.; Savidor, A.; Levin, Y.; Eyal, Y.; Reich, Z.; Adam, Z. Chlorophyll catabolism precedes changes in chloroplast structure and proteome during leaf senescence. Plant Direct. 2019, 3, e00127. [Google Scholar] [CrossRef]
- Fryer, M. The antioxidant effects of thylakoid Vitamin E (α-tocopherol). Plant Cell Environ. 2006, 15, 381–392. [Google Scholar] [CrossRef]
- Barja, M.V.; Ezquerro, M.; Beretta, S.; Diretto, G.; Florez-Sarasa, I.; Feixes, E.; Fiore, A.; Karlova, R.; Fernie, A.R.; Beekwilder, J.; et al. Several geranylgeranyl diphosphate synthase isoforms supply metabolic substrates for carotenoid biosynthesis in tomato. New Phytol. 2021, 231, 255–272. [Google Scholar] [CrossRef]
- Keller, Y.; Bouvier, F.; d’Harlingue, A.; Camara, B. Metabolic compartmentation of plastid prenyllipid biosynthesis: Evidence for the involvement of a multifunctional CHLP. Eur. J. Biochem. 1998, 251, 413–417. [Google Scholar] [CrossRef]
- Beale, S.I. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef]
- Nagata, N.; Tanaka, R.; Satoh, S.; Tanaka, A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell. 2005, 17, 233–240. [Google Scholar] [CrossRef]
- Tommaso, P.D.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.M.; Taly, J.F.; Notredame, C. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011, 39, 13–17. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Zheng, X.; Jehanzeb, M.; Habiba; Zhang, Y.; Li, L.; Miao, Y. Characterization of S40-like proteins and their roles in response to environmental cues and leaf senescence in rice. BMC Plant Biol. 2019, 19, 174. [Google Scholar] [CrossRef]
- Norkunas, K.; Harding, R.; Dale, J.; Dugdale, B. Improving agroinfiltration-based transient gene expression in Nicotiana benthamiana. Plant Methods 2018, 14, 71. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Ren, Y.; Yang, J.; Luo, B.; Jiang, Y.; Chen, H.; Hong, Y.; Wu, B.; Miao, Y. Structure of Pigment Metabolic Pathways and Their Contributions to White Tepal Color Formation of Chinese Narcissus tazetta var. chinensis cv Jinzhanyintai. Int. J. Mol. Sci. 2017, 18, 1923. [Google Scholar] [CrossRef]
- Yang, J.; Ren, Y.; Zhang, D.; Chen, X.; Huang, J.; Xu, Y.; Aucapiña, C.B.; Zhang, Y.; Miao, Y. Transcriptome-based WGCNA analysis reveals regulated metabolite fluxes between floral color and scent in Narcissus tazetta flower. Int. J. Mol. Sci. 2021, 22, 8249. [Google Scholar] [CrossRef]
- Rodrigquez, C.E.; Shinyashiki, M.; Froines, J.; Yu, R.C.; Fukuto, J.M.; Cho, A.K. An examination of quinone toxicity using the yeast Saccharomyces cerevisiae model system. Toxicology 2004, 201, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Yu, J.; Cai, Q.; Chen, Y.; Li, Y.; Ren, Y.; Miao, Y. Triple-localized WHIRLY2 Influences Leaf Senescence and Silique Development via Carbon Allocation. Plant Physiol. 2020, 184, 1348–1362. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, J.B.G.; Zhuo, T.; Li, X.; Wu, X.; Wu, Z.; Habiba; Ren, Y.; Miao, Y. Overexpressing BrWRKY22 Delays Flowering and Leaf Senescence via Inhibition of GA Biosynthesis in Brassica rapa. Plants 2025, 14, 1658. https://doi.org/10.3390/plants14111658
Uddin JBG, Zhuo T, Li X, Wu X, Wu Z, Habiba, Ren Y, Miao Y. Overexpressing BrWRKY22 Delays Flowering and Leaf Senescence via Inhibition of GA Biosynthesis in Brassica rapa. Plants. 2025; 14(11):1658. https://doi.org/10.3390/plants14111658
Chicago/Turabian StyleUddin, Junaite Bin Gias, Tingzhen Zhuo, Xiaojie Li, Xuan Wu, Zhuoyu Wu, Habiba, Yujun Ren, and Ying Miao. 2025. "Overexpressing BrWRKY22 Delays Flowering and Leaf Senescence via Inhibition of GA Biosynthesis in Brassica rapa" Plants 14, no. 11: 1658. https://doi.org/10.3390/plants14111658
APA StyleUddin, J. B. G., Zhuo, T., Li, X., Wu, X., Wu, Z., Habiba, Ren, Y., & Miao, Y. (2025). Overexpressing BrWRKY22 Delays Flowering and Leaf Senescence via Inhibition of GA Biosynthesis in Brassica rapa. Plants, 14(11), 1658. https://doi.org/10.3390/plants14111658





