A Three-Dimensional Phenotype Extraction Method Based on Point Cloud Segmentation for All-Period Cotton Multiple Organs
Abstract
1. Introduction
- A novel full-period cotton organ segmentation model is proposed. The network architecture is optimized based on DGCNN and integrated with residual modules to enhance feature extraction capabilities. This method significantly improves the segmentation accuracy of cotton organs across all growth periods, achieving a 4.86% increase in mIoU compared to baseline models.
- An improved algorithm for precise segmentation of individual organs based on region growing is developed. By integrating point-to-point distance mapping and curvature-normal features, the method effectively addresses the problem of organ overlap in cotton, enabling precise segmentation of organs, such as leaves, stems, and buds. In the most challenging task of overlapping leaf segmentation, the method achieves an R2 of 0.962 and an RMSE of 2.0. Based on this improved algorithm, the bell drop rate is innovatively calculated, providing a novel technical approach for cotton growth monitoring and yield estimation.
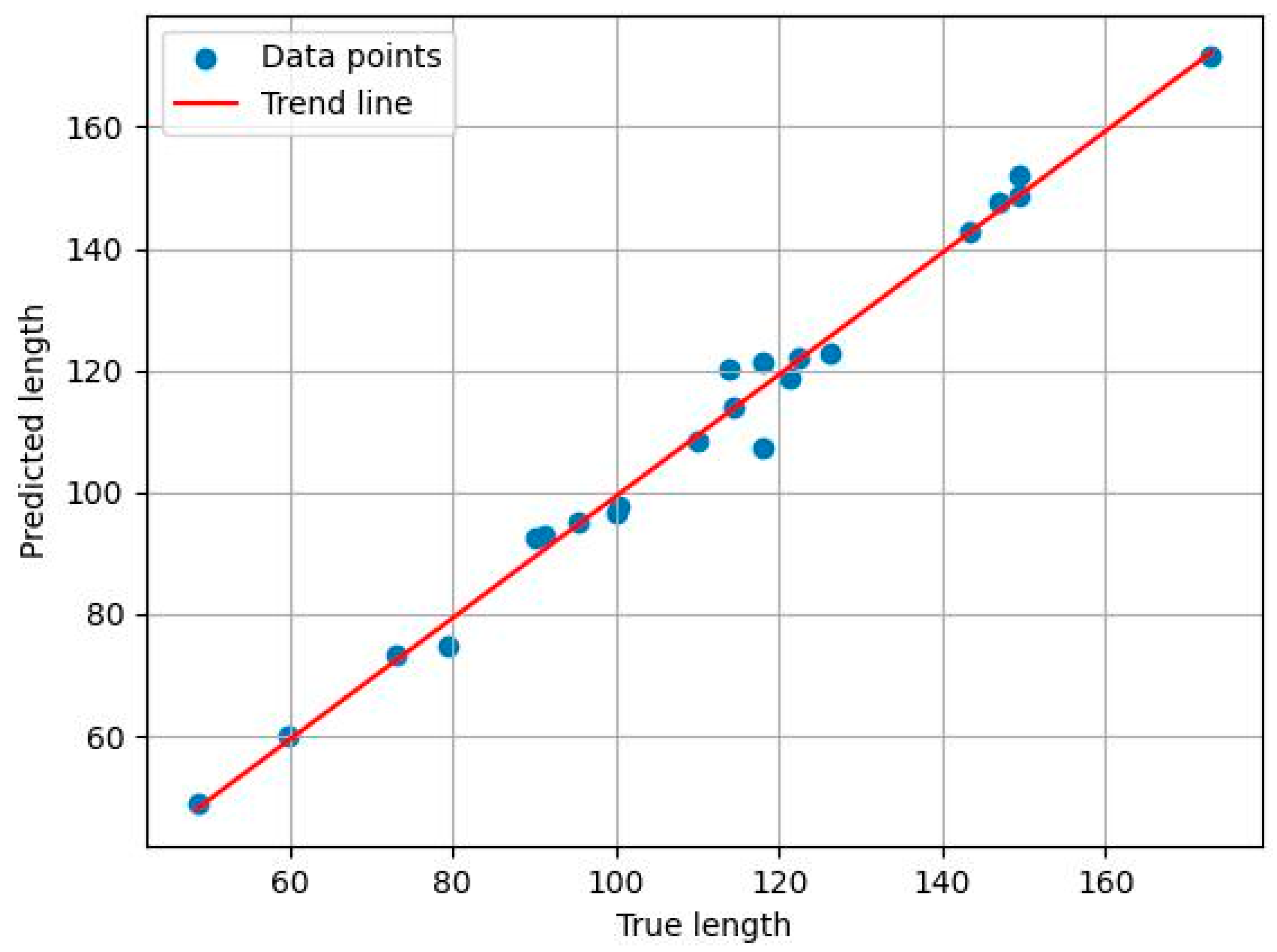
- A phenotypic computation framework applicable to different growth periods is developed. By calculating plant height and stem length and comparing them with ground-truth measurements, the framework achieves a mean relative error of only 0.973, fully demonstrating its effectiveness in extracting key phenotypic parameters. This provides reliable support for precision agriculture and intelligent breeding.
2. Materials and Methods
2.1. Experimental Materials and Data Collection
2.2. Data Composition
2.2.1. Point Cloud Data Acquisition
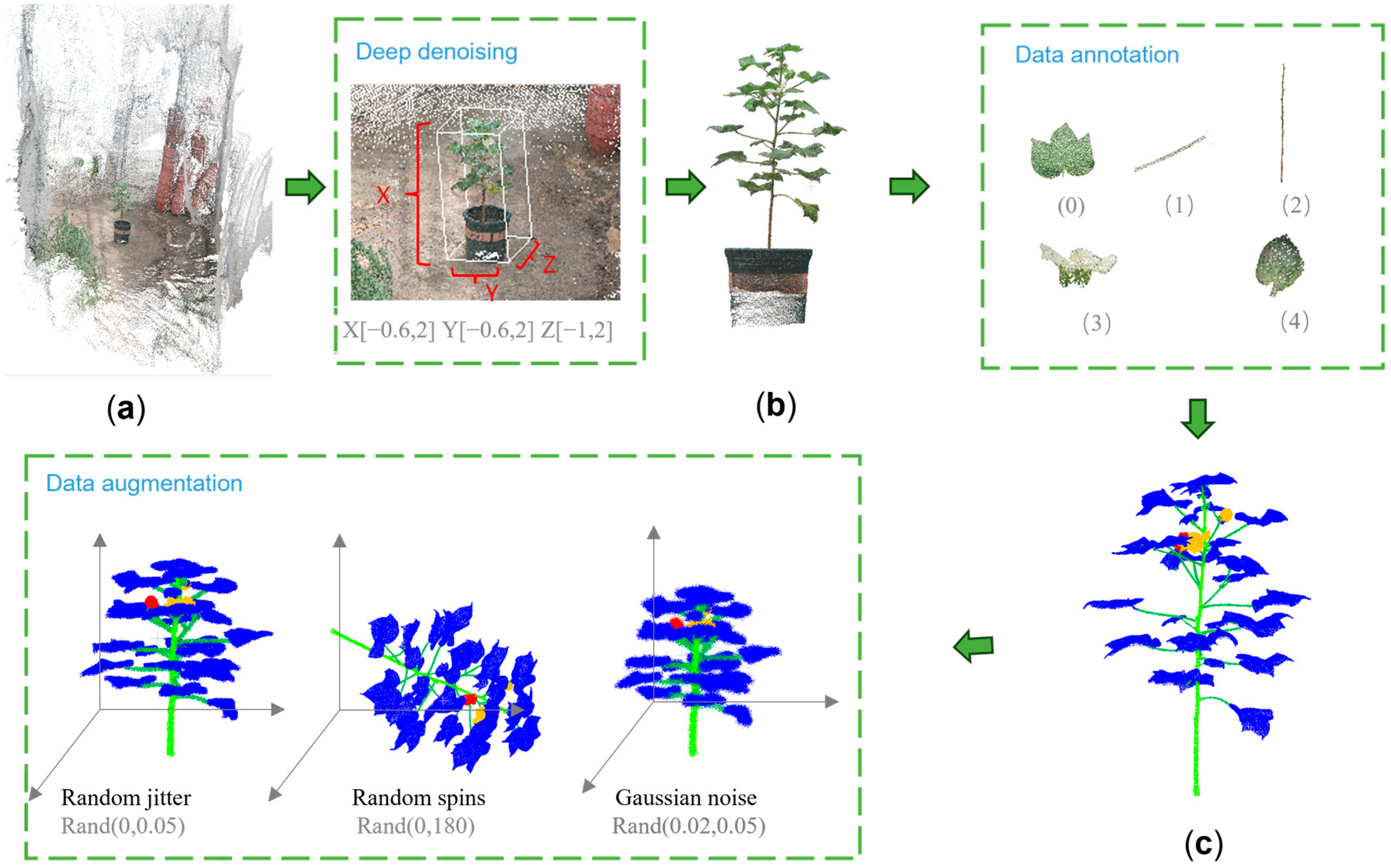
2.2.2. Data Preprocessing
- (1)
- Point cloud denoising
- (2)
- Data classification
- (3)
- Data labeling
- (4)
- Data enhancement
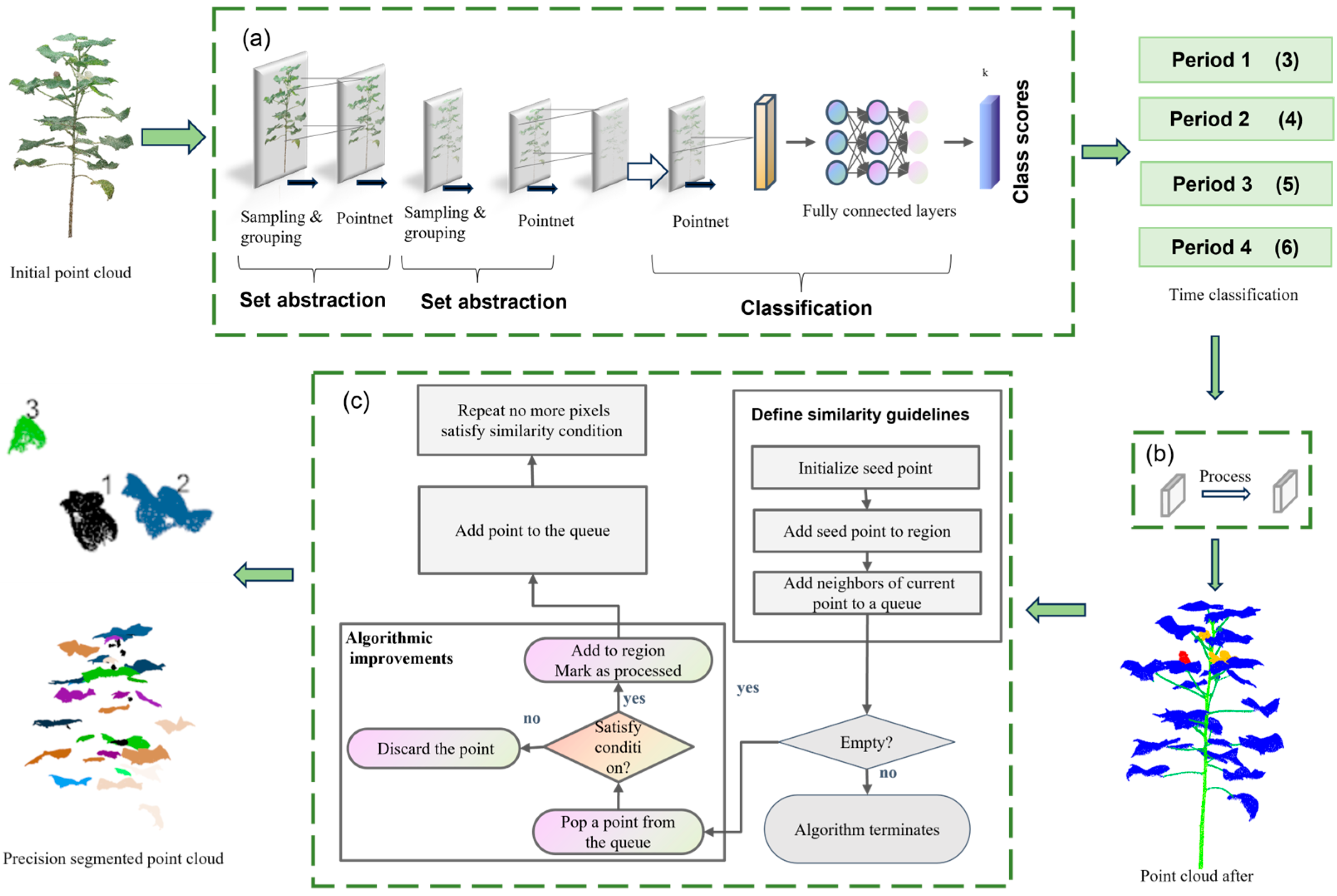
2.3. Point Cloud Segmentation
2.3.1. Cotton Point Cloud Segmentation Process
2.3.2. Cotton Organ Segmentation Architecture
- (1)
- Graph Convolution Module
- (2)
- Residual Convolution Module
2.3.3. Improvement of the Region Growth Algorithm
| Algorithm 1: Improvement of the region growth algorithm |
| Input: P: cotton seedling individual point cloud θ: normal vector threshold k: curvature threshold N: maximal number of regions Output: Segmentation result (leaf or stem regions) |
|
1: Initialize U = 0, U′ = U; // All points are unprocessed 2: Initialize empty regions R[i]; // List to store regions 3: Initialize queue with seed points from P // Step 1: Region growing for each seed point based on normal vector and curvature 4: for (k = 1 to N) do 5: for each point Pi in P: 6: if Pi normal_vector < θ: 7: if Pi curvature < k: 8: Grow point Pi into the nearest region R[min_region_id] 9: Add Pi to region R[min_region_id] 10: else: 11: Add Pi as a new seed point for the next iteration 12: else: 13: Discard point Pi // Do not grow this point 14: end for 15: Update regions based on the newly classified points: R = R ∪ R′ 16: U = U ∪ U′ // Update the processed points // Step 2: Merge unprocessed points to the nearest region based on Euclidean distance 17: for each unprocessed point Pi in P do 18: Find the nearest region R[min_region_id] using Euclidean distance 19: Add Pi to region R[min_region_id] 20: end for // Step 3: Return all regions after growing and merging 21: Return the segmented regions R[i] |
2.4. Cotton Phenotype Extraction
2.4.1. Calculation of Bell Drop Rate
2.4.2. Calculation of Plant Height and Stem Length
3. Experimental Results and Analysis
3.1. Environment and Setting
3.2. Assessment of Indicators
3.2.1. Model Comparison Experiment Evaluation
3.2.2. Phenotype Extraction Assessment
3.3. Point Cloud Segmentation Results and Analysis
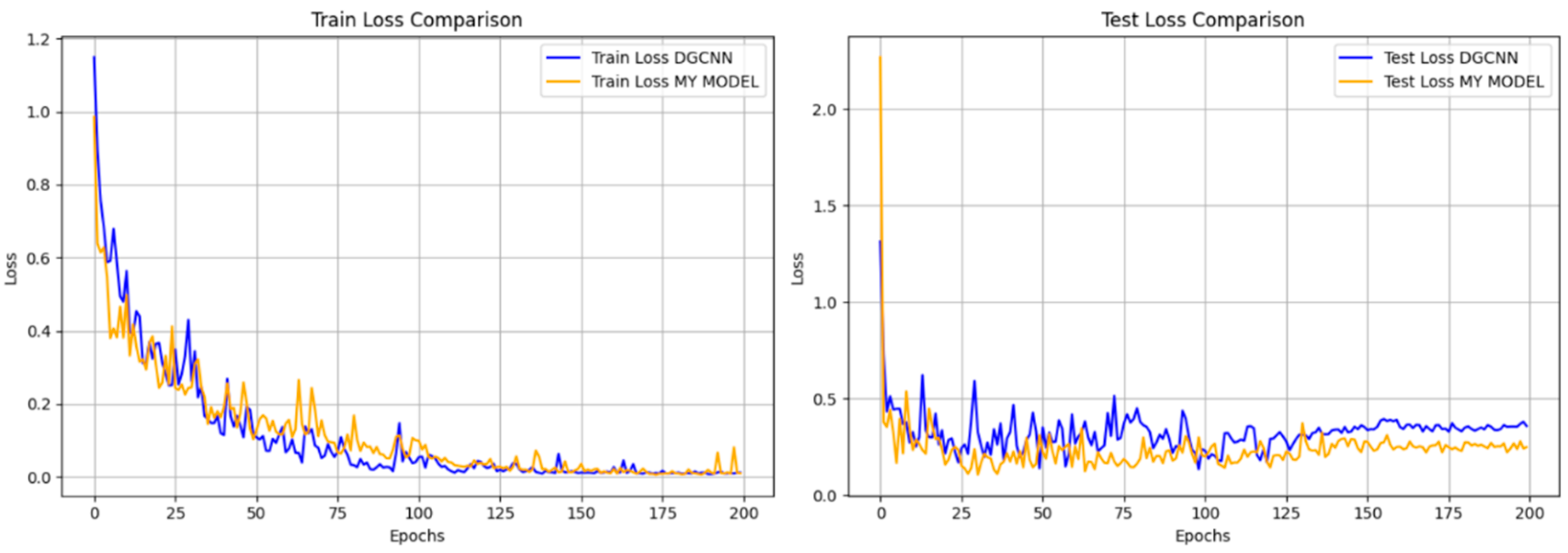
3.3.1. Network Performance Analysis
3.3.2. Results of Comparative Analysis of Models for Cotton Organ Segmentation
3.3.3. Results of the Analysis of Cotton Organ Segmentation at Various Periods of Time
3.3.4. Cotton Organ Segmentation Results of Different Organ Analysis
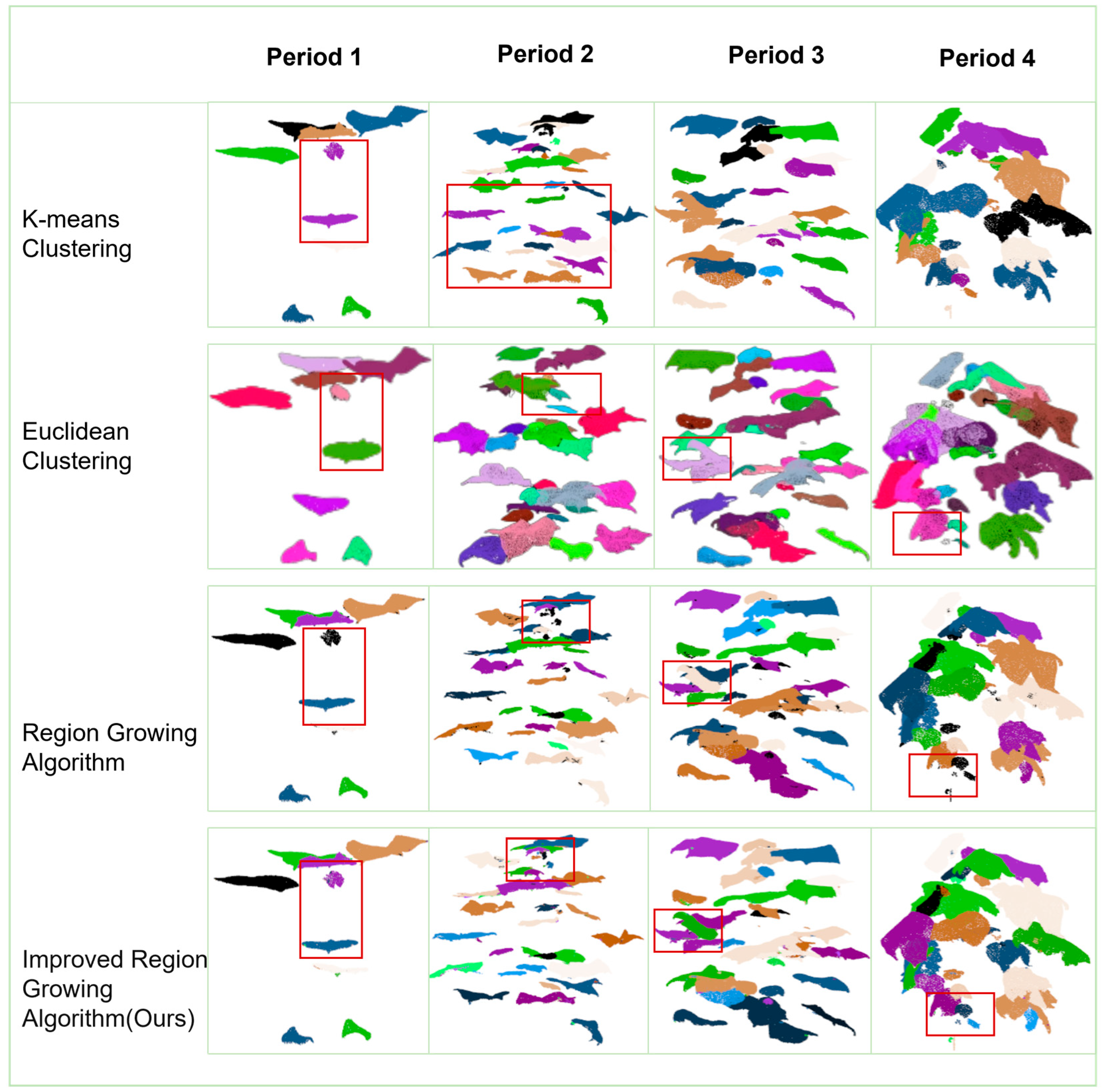
3.3.5. Results of Precise Segmentation of Individual Organs in Cotton
3.4. Results of Phenotypic Parameter Extraction
4. Discussion
- (1)
- For the effect of external perturbations on the 3D reconstruction of plants, this paper adopts the 3D reconstruction method based on the neural vector field to process the video data captured by cell phone. The method has the advantages of low cost [43] and high reconstruction accuracy [44], and the resulting point cloud data can better meet the subsequent experimental needs of organ segmentation and phenotypic parameter extraction. In order to better observe the morphological characteristics of cotton at various growth stages, most of the data acquisition work was carried out in a greenhouse environment. However, some cotton plants were also moved outdoors at specific time periods for photographing. Comparative analysis showed that the data collected outside the greenhouse did not differ much from the data inside the greenhouse in terms of reconstruction effect under small external stimulus conditions, and all of them were able to capture the details of the plants better. Based on this finding, a further attempt was made to perform video acquisition and 3D reconstruction in the experimental field. The results showed that compared with the acquisition method under greenhouse conditions, the video captured directly in the natural environment had a poorer reconstruction effect with significantly more background noise, and the reconstruction effect of the data captured in the experimental field is shown in Figure 15.
- (2)
- The impact of the number of training points on the effect of point cloud segmentation. In the prediction of the model segmentation results, this paper found the number of samples has a great impact on the model segmentation accuracy (Table 5). In periods 1–4 of cotton training using DGCNN training, for example, the model trained using 2048 points shows poorer results in the prediction of other points. In further research, it may be considered to increase the number of trained plant point clouds, thereby achieving better segmentation results for plants [45].
- (3)
- Multiscale feature-guided optimization of regional growth segmentation aims at the problem that different cotton organs have local overlapping and fuzzy boundaries in space, which makes it difficult to segment them accurately. In this paper, based on the seven-dimensional feature information (x, y, z, Nx, Ny, Nz, labels) contained in the point cloud dataset, we combine the point cloud coordinate features and normal vector features for the region growth determination and expansion. Two optimization strategies, point distance mapping and curvature normal vector, are introduced to design and implement an improved region-growing algorithm. The algorithm can realize fine-grained precise segmentation of multiple individual organs (including leaves, stalks, flower buds, etc.) of cotton, so as to distinguish organ boundaries more effectively and inhibit the erroneous fusion of overlapping regions.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aslam, S.; Hussain, S.B.; Baber, M.; Shaheen, S.; Aslam, S.; Waheed, R.; Seo, H.; Azhar, M.T. Estimation of drought tolerance indices in upland cotton under water deficit conditions. Agronomy 2023, 13, 984. [Google Scholar] [CrossRef]
- Pabuayon, I.L.B.; Sun, Y.; Guo, W.; Ritchie, G.L. High-throughput phenotyping in cotton: A review. J. Cotton Res. 2019, 2, 1–9. [Google Scholar] [CrossRef]
- Taqdeer, G.; Gill, S.K.; Saini, D.K.; Chopra, Y.; de Koff, J.P.; Sandhu, K.S. A Comprehensive Review of High Throughput Phenotyping and Machine Learning for Plant Stress Phenotyping. Phenomics 2022, 2, 156–183. [Google Scholar]
- Hao, H.; Wu, S.; Li, Y.; Wen, W.; Fan, J.; Zhang, Y.; Zhuang, L.; Xu, L.; Li, H.; Guo, X.; et al. Automatic acquisition, analysis and wilting measurement of cotton 3D phenotype based on point cloud. Biosyst. Eng. 2024, 239, 173–189. [Google Scholar] [CrossRef]
- Gupta, P.K.; Rustgi, S.; Kulwal, P.L. Linkage disequilibrium and association studies in higher plants: Present status and future prospects. Plant Mol. Biol. 2005, 57, 461–485. [Google Scholar] [CrossRef]
- Guo, Q.; Jin, S.; Li, M.; Yang, Q.; Xu, K.; Ju, Y.; Zhang, J.; Xuan, J.; Liu, J.; Su, Y.; et al. Application of deep learning in ecological resource research: Theories, methods, and challenges. Sci. China Earth Sci. 2020, 63, 1457–1474. [Google Scholar] [CrossRef]
- Eltner, A.; Sofia, G. Structure from motion photogrammetric technique. In Developments in Earth Surface Processes; Elsevier: Amsterdam, The Netherlands, 2020; Volume 23, pp. 1–24. [Google Scholar]
- Deng, Q.; Zhao, J.; Li, R.; Liu, G.; Hu, Y.; Ye, Z.; Zhou, G. A precise segmentation algorithm of pumpkin seedling point cloud stem based on CPHNet. Plants 2024, 13, 2300. [Google Scholar] [CrossRef]
- Shen, J.; Wu, T.; Zhao, J.; Wu, Z.; Huang, Y.; Gao, P.; Zhang, L. Organ segmentation and phenotypic trait extraction of cotton seedling point clouds based on a 3D lightweight network. Agronomy 2024, 14, 1083. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, Y.; Zhou, J.; Xu, X.; Miao, M. Cucumber seedling segmentation network based on a multiview geometric graph encoder from 3D point clouds. Plant Phenomics 2024, 6, 0254. [Google Scholar] [CrossRef]
- Yan, J.; Tan, F.; Li, C.; Jin, S.; Zhang, C.; Gao, P.; Xu, W. Stem–Leaf segmentation and phenotypic trait extraction of individual plant using a precise and efficient point cloud segmentation network. Comput. Electron. Agric. 2024, 220, 108839. [Google Scholar] [CrossRef]
- Liu, J.J.; Liu, Y.H.; Doonan, J. Point cloud based iterative segmentation technique for 3d plant phenotyping. In Proceedings of the 2018 IEEE International Conference on Information and Automation (ICIA), Wuyi Mountains, China, 11–13 August 2018; IEEE: New York, NY, USA, 2018; pp. 1072–1077. [Google Scholar]
- Lin, C.; Han, J.; Xie, L.; Hu, F. Cylinder space segmentation method for field crop population using 3D point cloud. Trans. Chin. Soc. Agric. Eng. 2021, 37, 175–182. [Google Scholar]
- Peng, C.; Li, S.; Miao, Y.; Zhang, Z.; Zhang, M.; Han, M. Stem-leaf segmentation and phenotypic trait extraction of tomatoes using three-dimensional point cloud. Trans. Chin. Soc. Agric. Eng. 2022, 38, 187–194. [Google Scholar]
- Chen, R.; Han, S.; Xu, J.; Su, H. Point-based multi-view stereo network. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 2 November–27 October 2019; pp. 1538–1547. [Google Scholar]
- Zhou, Y.; Qi, Y.; Xiang, L. Automatic Extraction Method of Phenotypic Parameters for Phoebe zhennan Seedlings Based on 3D Point Cloud. Agriculture 2025, 15, 834. [Google Scholar] [CrossRef]
- Sun, B.; Zain, M.; Zhang, L.; Han, D.; Sun, C. Stem-Leaf Segmentation and Morphological Traits Extraction in Rapeseed Seedlings Using a Three-Dimensional Point Cloud. Agronomy 2025, 15, 276. [Google Scholar] [CrossRef]
- Ibrahim, M.; Wang, H.; Iqbal, I.A.; Miao, Y.; Albaqami, H.; Blom, H.; Mian, A. Forest Stem Extraction and Modeling (FoSEM): A LiDAR-Based Framework for Accurate Tree Stem Extraction and Modeling in Radiata Pine Plantations. Remote Sens. 2025, 17, 445. [Google Scholar] [CrossRef]
- Mu, S.; Liu, J.; Zhang, P.; Yuan, J.; Liu, X. YS3AM: Adaptive 3D Reconstruction and Harvesting Target Detection for Clustered Green Asparagus. Agriculture 2025, 15, 407. [Google Scholar] [CrossRef]
- Mildenhall, B.; Srinivasan, P.P.; Tancik, M.; Barron, J.T.; Ramamoorthi, R.; Ng, R. Nerf: Representing scenes as neural radiance fields for view synthesis. Commun. ACM 2021, 65, 99–106. [Google Scholar] [CrossRef]
- Arshad, M.A.; Jubery, T.; Afful, J.; Jignasu, A.; Balu, A.; Ganapathysubramanian, B.; Sarkar, S.; Krishnamurthy, A. Evaluating Neural Radiance Fields for 3D Plant Geometry Reconstruction in Field Conditions. Plant Phenomics 2024, 6, 0235. [Google Scholar] [CrossRef]
- Zhu, X.; Huang, Z.; Li, B. Three-dimensional phenotyping pipeline of potted plants based on neural radiation fields and path segmentation. Plants 2024, 13, 3368. [Google Scholar] [CrossRef]
- Korycki, A.; Yeaton, C.; Gilbert, G.S.; Josephson, C.; McGuire, S. NeRF-Accelerated Ecological Monitoring in Mixed-Evergreen Redwood Forest. arXiv 2024, arXiv:2410.07418. [Google Scholar] [CrossRef]
- Wang, Y.; Wen, W.; Wu, S.; Wang, C.; Yu, Z.; Guo, X.; Zhao, C. Maize plant phenotyping: Comparing 3D laser scanning, multi-view stereo reconstruction, and 3D digitizing estimates. Remote Sens. 2018, 11, 63. [Google Scholar] [CrossRef]
- Zhu, Q.; Fan, L.; Weng, N. Advancements in point cloud data augmentation for deep learning: A survey. Pattern Recognit. 2024, 153, 110532. [Google Scholar] [CrossRef]
- Miao, Y.; Peng, C.; Wang, L.; Qiu, R.; Li, H.; Zhang, M. Measurement method of maize morphological parameters based on point cloud image conversion. Comput. Electron. Agric. 2022, 199, 107174. [Google Scholar] [CrossRef]
- Liu, H.; Zhong, H.; Xie, G.; Zhang, P. Tree Species Classification Based on Point Cloud Completion. Forests 2025, 16, 280. [Google Scholar] [CrossRef]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. Pointnet++: Deep hierarchical feature learning on point sets in a metric space. Adv. Neural Inf. Process. Syst. 2017, 30, 5105–5114. [Google Scholar]
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. Pointnet: Deep learning on point sets for 3d classification and segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 652–660. [Google Scholar]
- Wang, Y.; Sun, Y.; Liu, Z.; Sarma, S.E.; Bronstein, M.M.; Solomon, J.M. Dynamic graph cnn for learning on point clouds. ACM Trans. Graph. 2019, 38, 1–12. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Cunningham, P.; Delany, S.J. k-Nearest neighbour classifiers: (with Python examples). arXiv 2020, arXiv:2004.04523. [Google Scholar]
- Song, X.; Cui, T.; Zhang, D.; Yang, L.; He, X.; Zhang, K. Reconstruction and spatial distribution analysis of maize seedlings based on multiple clustering of point clouds. Comput. Electron. Agric. 2025, 235, 110196. [Google Scholar] [CrossRef]
- Miao, Y.; Li, S.; Wang, L.; Li, H.; Qiu, R.; Zhang, M. A single plant segmentation method of maize point cloud based on Euclidean clustering and K-means clustering. Comput. Electron. Agric. 2023, 210, 107951. [Google Scholar] [CrossRef]
- Ge, Y.; Tang, H.; Xia, D.; Wang, L.; Zhao, B.; Teaway, J.W.; Chen, H.; Zhou, T. Automated measurements of discontinuity geometric properties from a 3D-point cloud based on a modified region growing algorithm. Eng. Geol. 2018, 242, 44–54. [Google Scholar] [CrossRef]
- Feng, J.; Ma, X.; Guan, H.; Zhu, K.; Yu, S. Calculation method of soybean plant height based on depth information. Acta Opt. Sin. 2019, 39, 258–268. [Google Scholar]
- Zhu, C.; Miao, T.; Xu, T.; Li, N.; Deng, H.; Zhou, Y. Segmentation and phenotypic trait extraction of maize point cloud stem-leaf based on skeleton and optimal transportation distances. Trans. Chin. Soc. Agric. Eng. 2021, 37, 188–198. [Google Scholar]
- Zhou, D.; Fang, J.; Song, X.; Guan, X.; Yin, J.; Dai, Y.; Yang, R. Iou loss for 2d/3d object detection. In Proceedings of the 2019 International Conference on 3D Vision (3DV), Québec City, QC, Canada, 16–19 September 2019; IEEE: New York, NY, USA, 2019; pp. 85–94. [Google Scholar]
- Li, G.; Muller, M.; Thabet, A.; Ghanem, B. DeepGCNs: Can GCNs go as deep as CNNs? In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 2 November–27 October 2019; pp. 9267–9276. [Google Scholar]
- Qian, G.; Li, Y.; Peng, H.; Mai, J.; Hammoud, H.; Elhoseiny, M.; Ghanem, B. Pointnext: Revisiting pointnet++ with improved training and scaling strategies. Adv. Neural Inf. Process. Syst. 2022, 35, 23192–23204. [Google Scholar]
- Deng, X.; Zhang, W.; Ding, Q.; Zhang, X. Pointvector: A vector representation in point cloud analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 9455–9465. [Google Scholar]
- Qian, G.; Hamdi, A.; Zhang, X.; Ghanem, B. Pix4Point: Image pretrained standard transformers for 3d point cloud understanding. In Proceedings of the 2024 International Conference on 3D Vision (3DV), Davos, Switzerland, 18–21 March 2024; IEEE: New York, NY, USA, 2024; pp. 1280–1290. [Google Scholar]
- Petrovska, I.; Jutzi, B. Vision through Obstacles—3D Geometric Reconstruction and Evaluation of Neural Radiance Fields (NeRFs). Remote Sens. 2024, 16, 1188. [Google Scholar] [CrossRef]
- Jia, Z.; Wang, B.; Chen, C. Drone-NeRF: Efficient NeRF based 3D scene reconstruction for large-scale drone survey. Image Vis. Comput. 2024, 143, 104920. [Google Scholar] [CrossRef]
- Qiu, R.; He, Y.; Zhang, M. Automatic detection and counting of wheat spikelet using semi-automatic labeling and deep learning. Front. Plant Sci. 2022, 13, 872555. [Google Scholar] [CrossRef]
- Xu, G.; Li, X.; Lei, B.; Lv, K. Unsupervised color image segmentation with color-alone feature using region growing pulse coupled neural network. Neurocomputing 2018, 306, 1–16. [Google Scholar] [CrossRef]
- Couder-Castañeda, C.; Orozco-del-Castillo, M.; Padilla-Perez, D.; Medina, I. A parallel texture-based region-growing algorithm implemented in OpenMP. Sci. Rep. 2025, 15, 5563. [Google Scholar] [CrossRef]
- Chen, X.; Mao, J.; Zhao, B.; Wu, C.; Qin, M. Facet-Segmentation of Point Cloud Based on Multiscale Hypervoxel Region Growing. J. Indian Soc. Remote Sens. 2025, 1–22, prepublish. [Google Scholar] [CrossRef]
- Yu, Q.; Clausi, D.A. SAR sea-ice image analysis based on iterative region growing using semantics. IEEE Trans. Geosci. Remote Sens. 2007, 45, 3919–3931. [Google Scholar] [CrossRef]










| Model | mIoU (%) | mP (%) | mR (%) | mF1 (%) |
|---|---|---|---|---|
| DGCNN (Baseline) | 62.69 | 67.43 | 71.52 | 69.42 |
| DeepGCNs | 52.78 | 59.63 | 66.68 | 62.97 |
| Pointnext | 61.90 | 66.98 | 72.35 | 69.57 |
| Pointvector | 60.02 | 66.21 | 73.15 | 69.52 |
| Pix4Point | 53.79 | 60.74 | 68.94 | 64.59 |
| Pointnet | 53.49 | 67.50 | 71.53 | 69.46 |
| Pointnet++ | 61.43 | 67.81 | 72.24 | 69.96 |
| Pointnet++ (MSG) | 63.54 | 68.51 | 73.20 | 70.78 |
| ResDGCNN (Ours) | 67.55 | 71.76 | 77.37 | 74.46 |
| Model | mIoU (%) | |||
|---|---|---|---|---|
| Period 1 | Period 2 | Period 3 | Period 4 | |
| DGCNN (Baseline) | 69.39 | 46.01 | 48.04 | 61.34 |
| DeepGCNs | 57.50 | 46.31 | 17.30 | 57.38 |
| Pointnext | 76.44 | 43.59 | 42.13 | 50.66 |
| Pointvector | 67.87 | 47.17 | 44.88 | 53.40 |
| Pix4Point | 64.24 | 38.63 | 17.30 | 52.46 |
| Pointnet | 62.80 | 43.41 | 32.25 | 48.23 |
| Pointnet++ | 73.77 | 43.58 | 35.26 | 55.16 |
| Pointnet++ (MSG) | 72.22 | 51.85 | 39.88 | 60.96 |
| ResDGCNN (Ours) | 77.77 | 53.33 | 54.18 | 66.45 |
| Period | Model | mIoU (%) | ||||
|---|---|---|---|---|---|---|
| Leaf | Stem | Mainstem | Flower | Peach | ||
| Period 1 | DGCNN (Baseline) | 93.60 | 45.35 | 69.24 | - | - |
| DeepGCNs | 93.47 | 26.84 | 52.21 | - | - | |
| Pointnext | 97.44 | 52.53 | 79.37 | - | - | |
| Pointvector | 99.79 | 45.09 | 64.55 | - | - | |
| Pix4Point | 95.23 | 25.30 | 72.20 | - | - | |
| Pointnet | 97.60 | 43.83 | 66.22 | - | - | |
| Pointnet++ | 97.93 | 54.75 | 76.65 | - | - | |
| Pointnet++ (MSG) | 96.84 | 47.51 | 88.90 | - | - | |
| ResDGCNN (Ours) | 98.42 | 71.63 | 87.89 | - | - | |
| Period 2 | DGCNN (Baseline) | 91.24 | 37.06 | 51.92 | 3.82 | - |
| DeepGCNs | 93.80 | 40.24 | 51.22 | 0.00 | - | |
| Pointnext | 92.37 | 23.36 | 58.65 | 0.00 | - | |
| Pointvector | 94.91 | 48.91 | 44.88 | 0.00 | - | |
| Pix4Point | 91.36 | 17.09 | 46.09 | 0.00 | - | |
| Pointnet | 94.62 | 19.1 | 46.4 | 0.23 | - | |
| Pointnet++ | 96.48 | 24.32 | 43.62 | 9.75 | - | |
| Pointnet++ (MSG) | 96.34 | 18.33 | 41.92 | 26.65 | - | |
| ResDGCNN (Ours) | 92.51 | 40.17 | 54.02 | 31.11 | - | |
| Period 3 | DGCNN (Baseline) | 92.74 | 41.96 | 61.10 | 23.06 | 21.37 |
| DeepGCNs | 86.55 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Pointnext | 94.52 | 43.95 | 68.90 | 0.00 | 3.28 | |
| Pointvector | 93.77 | 43.32 | 66.26 | 0.00 | 0.00 | |
| Pix4Point | 86.55 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Pointnet | 93.34 | 24.62 | 7.72 | 3.41 | 3.33 | |
| Pointnet++ | 95.24 | 34.38 | 60.37 | 0.00 | 5.61 | |
| Pointnet++ (MSG) | 94.00 | 33.95 | 78.42 | 56.55 | 9.10 | |
| ResDGCNN (Ours) | 94.53 | 46.62 | 83.26 | 28.88 | 38.69 | |
| Period 4 | DGCNN (Baseline) | 91.85 | 43.41 | 63.03 | - | 47.10 |
| DeepGCNs | 96.22, | 38.62 | 64.37 | - | 30.34 | |
| Pointnext | 93.66 | 18.58 | 61.34 | - | 29.09 | |
| Pointvector | 93.81 | 24.71 | 55.37 | - | 39.74 | |
| Pix4Point | 94.80 | 35.56 | 69.67 | - | 9.80 | |
| Pointnet | 92.72 | 23.12 | 55.33 | - | 1.92 | |
| Pointnet++ | 94.55 | 32.22 | 64.36 | - | 6.31 | |
| Pointnet++ (MSG) | 92.12 | 40.13 | 60.71 | - | 50.90 | |
| ResDGCNN (Ours) | 96.80 | 43.21 | 69.43 | - | 38.38 | |
| Plant | Cotton1 | Cotton2 | Cotton3 | Cotton4 | Cotton5 | Cotton6 |
|---|---|---|---|---|---|---|
| Bell drops rate (%) | 0% | 33% | 57% | 25% | 66% | 25% |
| Spits (pcs) | 1 | 2 | 3 | 3 | 1 | 3 |
| Buds (pcs) | 1 | 3 | 7 | 4 | 3 | 4 |
| Point Number | mIoU (%) | ||
|---|---|---|---|
| Leaf | Stem | Mainstem | |
| 1024 | 94.17 | 47.20 | 83.3 |
| 2048 | 97.30 | 58.33 | 75.52 |
| 3072 | 97.99 | 45.75 | 79.41 |
| 4096 | 97.72 | 42.91 | 77.93 |
| 5120 | 97.52 | 38.15 | 76.16 |
| 10240 | 94.45 | 4.50 | 72.60 |
| 20480 | 92.99 | 1.16 | 71.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, P.; Han, B.; Guo, Q.; Wan, Y.; Zhang, J. A Three-Dimensional Phenotype Extraction Method Based on Point Cloud Segmentation for All-Period Cotton Multiple Organs. Plants 2025, 14, 1578. https://doi.org/10.3390/plants14111578
Chu P, Han B, Guo Q, Wan Y, Zhang J. A Three-Dimensional Phenotype Extraction Method Based on Point Cloud Segmentation for All-Period Cotton Multiple Organs. Plants. 2025; 14(11):1578. https://doi.org/10.3390/plants14111578
Chicago/Turabian StyleChu, Pengyu, Bo Han, Qiang Guo, Yiping Wan, and Jingjing Zhang. 2025. "A Three-Dimensional Phenotype Extraction Method Based on Point Cloud Segmentation for All-Period Cotton Multiple Organs" Plants 14, no. 11: 1578. https://doi.org/10.3390/plants14111578
APA StyleChu, P., Han, B., Guo, Q., Wan, Y., & Zhang, J. (2025). A Three-Dimensional Phenotype Extraction Method Based on Point Cloud Segmentation for All-Period Cotton Multiple Organs. Plants, 14(11), 1578. https://doi.org/10.3390/plants14111578








