Resistance to Amino Acid Biosynthesis Inhibiting-Herbicides in Amaranthus palmeri Populations from Aragon (Spain)
Abstract
1. Introduction
2. Results and Discussion
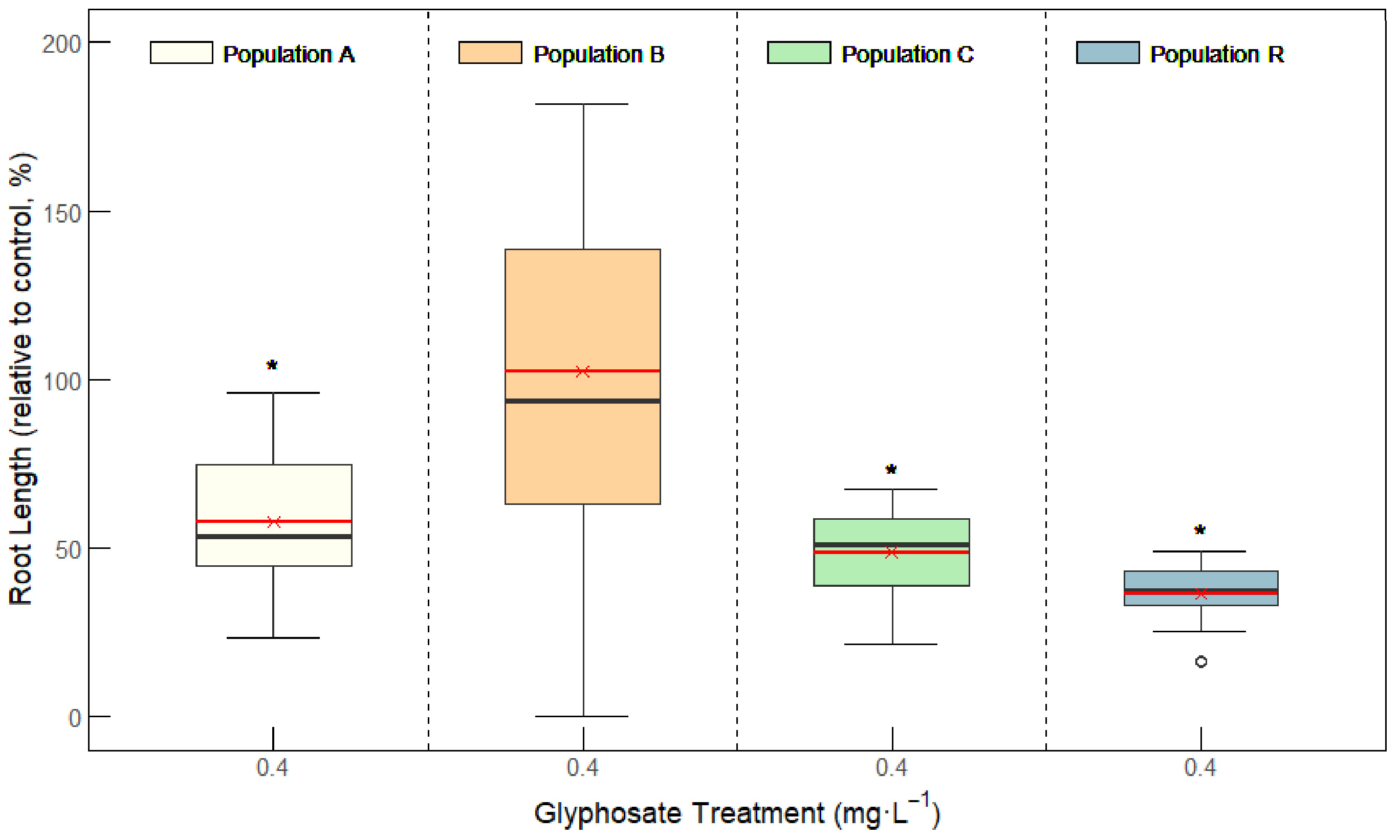
2.1. Resistance to Glyphosate
2.1.1. Glyphosate Dose–Response Bioassays in Vertical Plates
2.1.2. Hydroponic Tanks and Shikimate Content
2.1.3. EPSPS Gene Copy Number
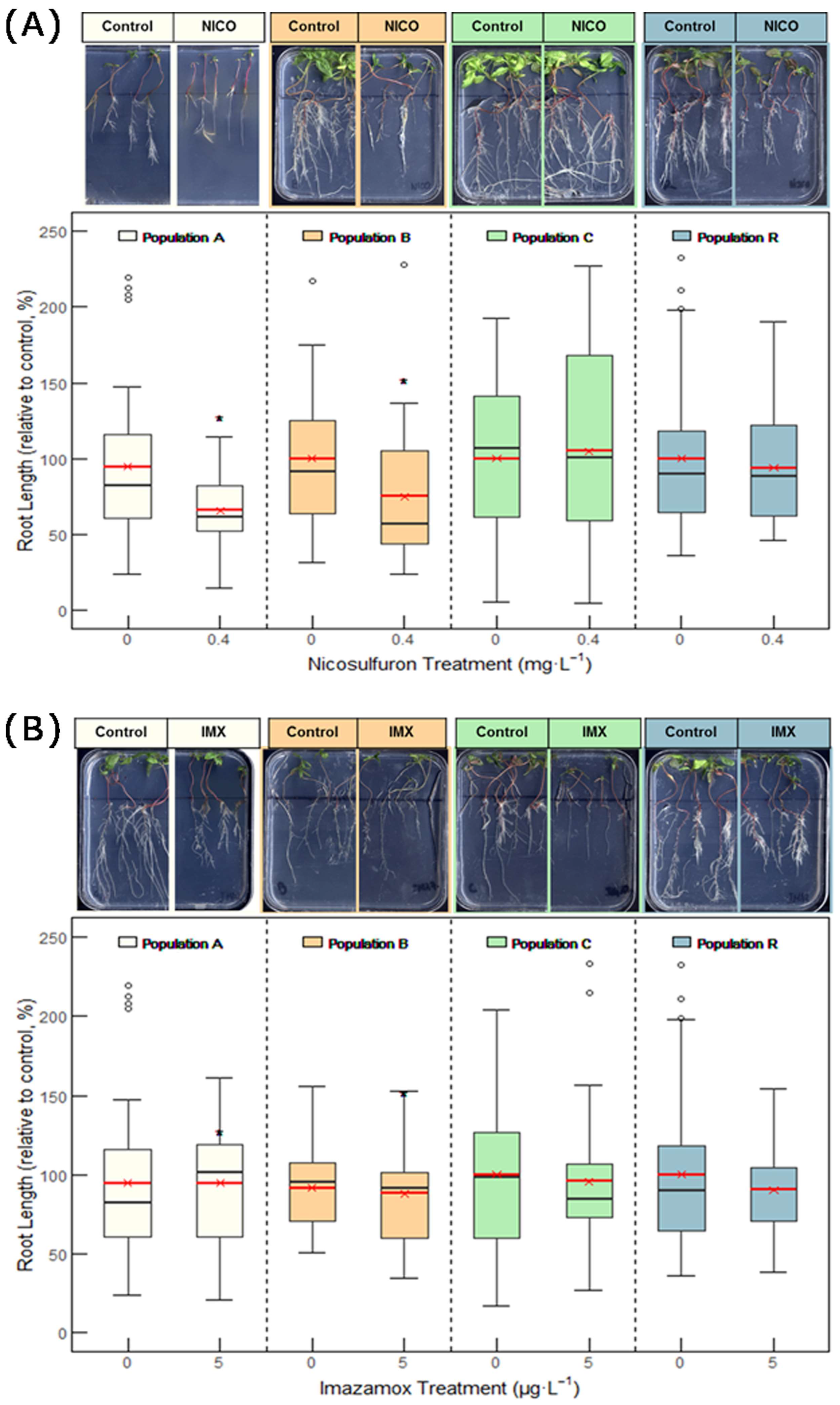
2.2. Resistance to ALS Inhibitors
2.2.1. Dose–Response Bioassays in Vertical Plates
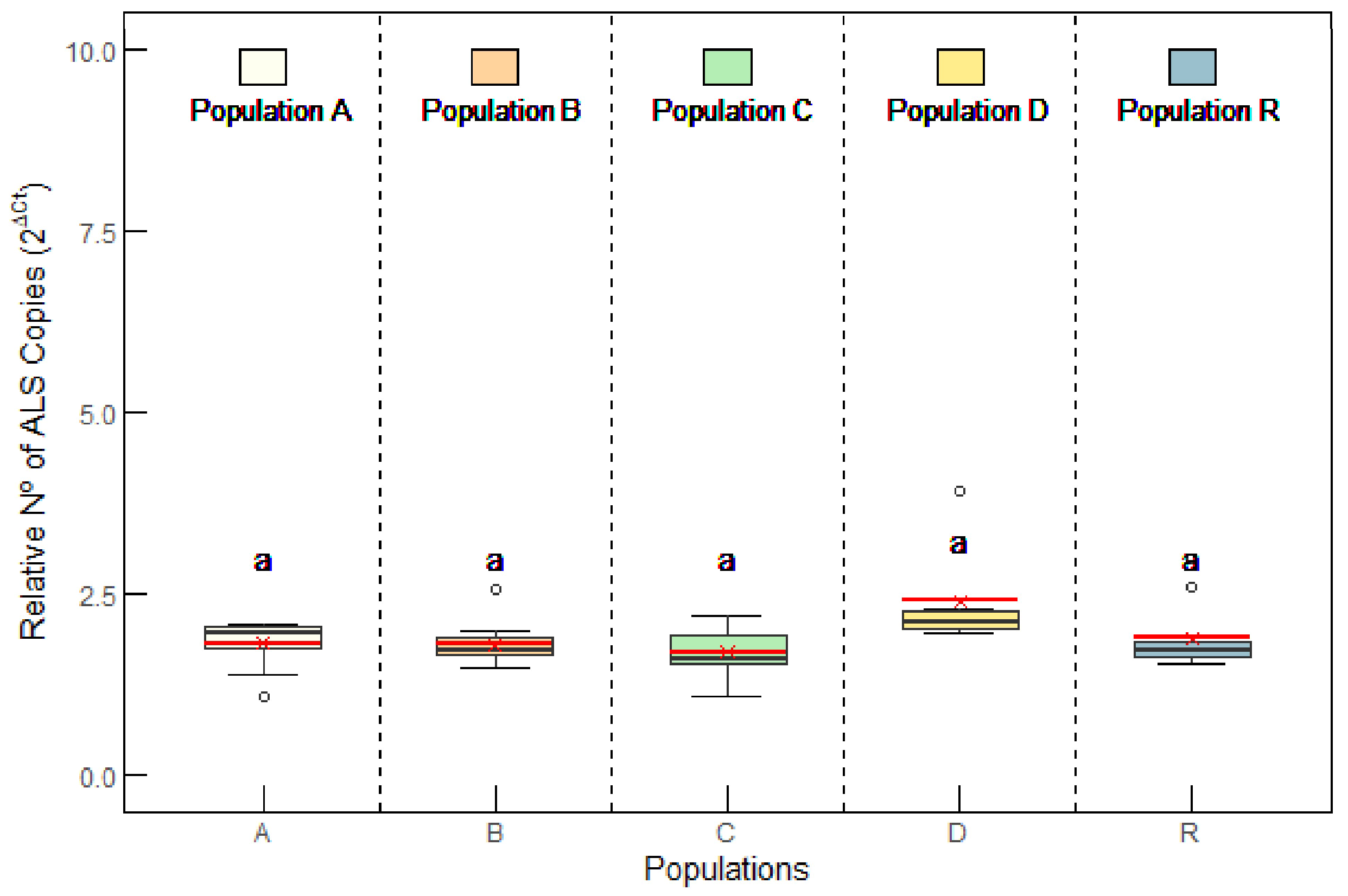
2.2.2. ALS Gene Copy Number
2.2.3. ALS Mutations
3. Materials and Methods
3.1. Plant Material
3.2. Nicosulfuron and Imazamox Dose–Response Bioassay with Vertical Plates
3.3. Plant Growth in Hydroponic Tanks and Glyphosate Application
3.4. Shikimate Content Determination
3.5. Genomic DNA Extraction
3.6. EPSPS and ALS Gene Copy Number
3.7. ALS Gene Sequencing
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Acetolactate synthase |
| CPS | Carbamoylphosphate synthetase |
| EPSPS | 5-Enolpyruvylshikimate-3-phosphate synthase |
| IWM | Integrated Weed Management |
| NTSR | Non-target-site resistance |
| qRT-PCR | Quantitative real-time PCR |
| TSR | Target-site resistance |
References
- Bensch, C.N.; Horak, M.J.; Peterson, D. Interference of Redroot pigweed (Amaranthus retroflexus), Palmer amaranth (A. palmeri), and Common waterhemp (A. rudis) in Soybean. Weed Sci. 2003, 51, 37–43. [Google Scholar] [CrossRef]
- Massinga, R.A.; Currie, R.S.; Horak, M.J.; Boyer, J. Interference of Palmer amaranth in Corn. Weed Sci. 2001, 49, 202–208. [Google Scholar] [CrossRef]
- Moore, J.W.; Murray, D.S.; Westerman, R.B. Palmer amaranth (Amaranthus palmeri) effects on the harvest and yield of grain Sorghum (Sorghum bicolor). Weed Technol. 2004, 18, 23–29. [Google Scholar] [CrossRef]
- Morgan, G.D.; Baumann, P.A.; Chandler, J.M. Competitive impact of Palmer amaranth (Amaranthus palmeri) on Cotton (Gossypium hirsutum) development and yield. Weed Technol. 2001, 15, 408–412. [Google Scholar] [CrossRef]
- Ward, S.M.; Webster, T.M.; Steckel, L.E. Palmer amaranth (Amaranthus palmeri): A review. Weed Technol. 2013, 27, 12–27. [Google Scholar] [CrossRef]
- Steckel, L.E. The dioecious Amaranthus spp.: Here to stay. Weed Technol. 2007, 21, 567–570. [Google Scholar] [CrossRef]
- Heap, I. The International Survey of Herbicide Resistant Weeds. Available online: https://www.weedscience.org/Home.aspx (accessed on 31 March 2025).
- Duke, S.O.; Powles, S.B. Glyphosate: A once-in-a-century herbicide. Pest Manag. Sci. 2008, 64, 319–325. [Google Scholar] [CrossRef]
- Steinrücken, H.C.; Amrhein, N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem. Biophys. Res. Commun. 1980, 94, 1207–1212. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, W.; Zhang, Y.; Liu, K.K. Action mechanisms of acetolactate synthase-inhibiting herbicides. Pestic. Biochem. Physiol. 2007, 89, 89–96. [Google Scholar] [CrossRef]
- Ray, T.B. Site of action of chlorsulfuron: Inhibition of valine and isoleucine biosynthesis in plants. Plant Physiol. 1984, 75, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Gaines, T.A.; Zhang, W.; Wang, D.; Bukun, B.; Chisholm, S.T.; Shaner, D.L.; Nissen, S.J.; Patzoldt, W.L.; Tranel, P.J.; Culpepper, A.S.; et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 2010, 107, 1029–1034. [Google Scholar] [CrossRef]
- Singh, S.; Singh, V.; Lawton-Rauh, A.; Bagavathiannan, M.V.; Roma-Burgos, N. EPSPS gene amplification primarily confers glyphosate resistance among Arkansas Palmer amaranth (Amaranthus palmeri) populations. Weed Sci. 2018, 66, 293–300. [Google Scholar] [CrossRef]
- Chahal, P.S.; Varanasi, V.K.; Jugulam, M.; Jhala, A.J. Glyphosate-resistant Palmer amaranth (Amaranthus palmeri) in Nebraska: Confirmation, EPSPS gene amplification, and response to POST Corn and Soybean herbicides. Weed Technol. 2017, 31, 80–93. [Google Scholar] [CrossRef]
- Lorentz, L.; Gaines, T.A.; Nissen, S.J.; Westra, P.; Strek, H.J.; Dehne, H.W.; Ruiz-Santaella, J.P.; Beffa, R. Characterization of glyphosate resistance in Amaranthus tuberculatus populations. J. Agric. Food Chem. 2014, 62, 8134–8142. [Google Scholar] [CrossRef]
- Fernández-Escalada, M.; Gil-Monreal, M.; Zabalza, A.; Royuela, M. Characterization of the Amaranthus palmeri physiological response to glyphosate in susceptible and resistant populations. J. Agric. Food Chem. 2016, 64, 95–106. [Google Scholar] [CrossRef]
- Dominguez-Valenzuela, J.A.; Gherekhloo, J.; Fernández-Moreno, P.T.; Cruz-Hipolito, H.E.; Alcántara-de la Cruz, R.; Sánchez-González, E.; De Prado, R. First confirmation and characterization of target and non-target site resistance to glyphosate in Palmer amaranth (Amaranthus palmeri) from Mexico. Plant Physiol. Biochem. 2017, 115, 212–218. [Google Scholar] [CrossRef]
- Palma-Bautista, C.; Torra, J.; Garcia, M.J.; Bracamonte, E.; Rojano-Delgado, A.M.; Alcántara-de la Cruz, R.; De Prado, R. Reduced absorption and impaired translocation endows glyphosate resistance in Amaranthus palmeri harvested in glyphosate-resistant Soybean from Argentina. J. Agric. Food Chem. 2019, 67, 1052–1060. [Google Scholar] [CrossRef]
- Meyer, C.J.; Peter, F.; Norsworthy, J.K.; Beffa, R. Uptake, translocation, and metabolism of glyphosate, glufosinate, and dicamba mixtures in Echinochloa crus-galli and Amaranthus palmeri. Pest Manag. Sci. 2020, 76, 3078–3087. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.; Du, L.; Zhang, X.; Liu, W.; Wang, J. Unravelling the effect of two herbicide resistance mutations on acetolactate synthase kinetics and growth traits. J. Exp. Bot. 2020, 71, 3535–3542. [Google Scholar] [CrossRef]
- Nakka, S.; Thompson, C.R.; Peterson, D.E.; Jugulam, M. Target site–based and non–target site based resistance to ALS inhibitors in Palmer amaranth (Amaranthus palmeri). Weed Sci. 2017, 65, 681–689. [Google Scholar] [CrossRef]
- Küpper, A.; Borgato, E.A.; Patterson, E.L.; Netto, A.G.; Nicolai, M.; De Carvalho, S.J.P.; Nissen, S.J.; Gaines, T.A.; Christoffoleti, P.J. Multiple resistance to glyphosate and acetolactate synthase inhibitors in Palmer amaranth (Amaranthus palmeri) identified in Brazil. Weed Sci. 2017, 65, 317–326. [Google Scholar] [CrossRef]
- Barco-Antoñanzas, M.; Gil-Monreal, M.; Eceiza, M.V.; Royuela, M.; Zabalza, A. Primary metabolism in an Amaranthus palmeri population with multiple resistance to glyphosate and pyrithiobac herbicides. Plant Sci. 2022, 318, 111212. [Google Scholar] [CrossRef]
- Sauer, J. Recent migration and evolution of the dioecious Amaranths. Evolution 1957, 11, 11–31. [Google Scholar] [CrossRef]
- Matzrafi, M.; Scarabel, L.; Milani, A.; Iamonico, D.; Torra, J.; Recasens, J.; Montull, J.M.; Llenes, J.M.; Gazoulis, I.; Tataridas, A.; et al. Amaranthus palmeri S. Watson: A new threat to agriculture in Europe and the Mediterranean region. Weed Res. 2023, 65, e12596. [Google Scholar] [CrossRef]
- Larran, A.S.; Palmieri, V.E.; Perotti, V.E.; Lieber, L.; Tuesca, D.; Permingeat, H.R. Target-site resistance to acetolactate synthase (ALS)-inhibiting herbicides in Amaranthus palmeri from Argentina. Pest Manag. Sci. 2017, 73, 2578–2584. [Google Scholar] [CrossRef]
- Shimono, A.; Kanbe, H.; Nakamura, S.; Ueno, S.; Yamashita, J.; Asai, M. Initial invasion of glyphosate-resistant Amaranthus palmeri around grain-import ports in Japan. Plants People Planet 2020, 2, 640–648. [Google Scholar] [CrossRef]
- Hamberg, R.C.; Yadav, R.; Hartzler, R.; Owen, M.D. Confirmation of a four-way herbicide-resistant Palmer amaranth (Amaranthus palmeri) population in Iowa. Weed Sci. 2024, 72, 330–338. [Google Scholar] [CrossRef]
- Kaya-Altop, E.; Jabran, K.; Pala, F.; Mennan, H. Multiple resistance to EPSPS and ALS inhibitors in Palmer amaranth (Amaranthus palmeri) identified in Turkey. Weed Res. 2025, 65, e12618. [Google Scholar] [CrossRef]
- Abu-Nassar, J.; Wallach, A.; Winkler, E.; Eizenberg, H.; Matzrafi, M. Occurrence of Amaranthus palmeri in Israeli agriculture: Status of spread and response to glyphosate and trifloxysulfuron. Weed Res. 2025, 65, e12616. [Google Scholar] [CrossRef]
- Recasens, J.; Conesa, J.A. Presencia de la mala hierba Amaranthus palmeni en el NE de la Península Ibérica. Una amenaza como potencial invasora de cultivos extensivos de regadío. Bol. Sanid. Veg. Plagas 2011, 37, 129–132. [Google Scholar]
- Torra, J.; Royo-Esnal, A.; Romano, Y.; Osuna, M.D.; León, R.G.; Recasens, J. Amaranthus palmeri a new invasive weed in Spain with herbicide resistant biotypes. Agronomy 2020, 10, 993. [Google Scholar] [CrossRef]
- Manicardi, A.; Scarabel, L.; Llenes, J.M.; Montull, J.M.; Osuna, M.D.; Torra Farré, J.; Milani, A. Genetic basis and origin of resistance to acetolactate synthase inhibitors in Amaranthus palmeri from Spain and Italy. Pest Manag. Sci. 2023, 79, 4886–4896. [Google Scholar] [CrossRef]
- Manicardi, A.; Milani, A.; Scarabel, L.; Mora, G.; Recasens, J.; Llenes, J.M.; Montull, J.M.; Torra, J. First report of glyphosate resistance in an Amaranthus palmeri population from Europe. Weed Res. 2023, 65, e12579. [Google Scholar] [CrossRef]
- Whitaker, J.R.; Burton, J.D.; York, A.C.; Jordan, D.L.; Chandi, A. Physiology of glyphosate-resistant and glyphosate-susceptible Palmer amaranth (Amaranthus palmeri) biotypes collected from North Carolina. Int. J. Agron. 2013, 2013, 429294. [Google Scholar] [CrossRef]
- Yanniccari, M.; Gaines, T.; Scursoni, J.; Prado, R.D.; Vila-Aiub, M. Global patterns of herbicide resistance evolution in Amaranthus spp.: An analysis comparing species, cropping regions and herbicides. Adv. Weed Sci. 2022, 40, e0202200037. [Google Scholar] [CrossRef]
- Wiersma, A.T.; Gaines, T.A.; Preston, C.; Hamilton, J.P.; Giacomini, D.; Robin Buell, C.; Leach, J.E.; Westra, P. Gene amplification of 5-enol-pyruvylshikimate-3-phosphate synthase in glyphosate-resistant Kochia scoparia. Planta 2015, 241, 463–474. [Google Scholar] [CrossRef]
- Gaines, T.A.; Duke, S.O.; Morran, S.; Rigon, C.A.G.; Tranel, P.J.; Küpper, A.; Dayan, F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020, 295, 10307–10330. [Google Scholar] [CrossRef]
- Singh, S.; Singh, V.; Salas-Perez, R.A.; Bagavathiannan, M.V.; Lawton-Rauh, A.; Roma-Burgos, N. Target-site mutation accumulation among ALS inhibitor-resistant Palmer amaranth. Pest Manag. Sci. 2018, 75, 1131–1139. [Google Scholar] [CrossRef]
- Iwakami, S.; Shimono, Y.; Manabe, Y.; Endo, M.; Shibaike, H.; Uchino, A.; Tominaga, T. Copy number variation in acetolactate synthase genes of thifensulfuron-methyl resistant Alopecurus aequalis (Shortawn Foxtail) accessions in Japan. Front. Plant Sci. 2017, 8, 254. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.P.; Tranel, P.J. Target-site mutations conferring herbicide resistance. Plants 2019, 8, 382. [Google Scholar] [CrossRef]
- Yu, Q.; Powles, S.B. Resistance to AHAS inhibitor herbicides: Current understanding. Pest Manag. Sci. 2014, 70, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- Molin, W.T.; Nandula, V.K.; Wright, A.A.; Bond, J.A. Transfer and expression of ALS inhibitor resistance from Palmer amaranth (Amaranthus palmeri) to an A. spinosus × A. palmeri hybrid. Weed Sci. 2016, 64, 240–247. [Google Scholar] [CrossRef]
- Whaley, C.M.; Wilson, H.P.; Westwood, J.H. A new mutation in plant ALS confers resistance to five classes of ALS-inhibiting herbicides. Weed Sci. 2007, 55, 83–90. [Google Scholar] [CrossRef]
- Sibony, M.; Michel, A.; Haas, H.U.; Rubin, B.; Hurle, K. Sulfometuron-resistant Amaranthus retroflexus: Cross-resistance and molecular basis for resistance to acetolactate synthase-inhibiting herbicides. Weed Res. 2001, 41, 509–522. [Google Scholar] [CrossRef]
- Ji, M.; Yu, H.; Cui, H.; Chen, J.; Yu, J.; Li, X. A new Pro-197-Ile mutation in Amaranthus palmeri associated with acetolactate synthase-inhibiting herbicide resistance. Plants 2025, 14, 525. [Google Scholar] [CrossRef]
- Rangani, G.; Noguera, M.; Salas-Perez, R.; Benedetti, L.; Roma-Burgos, N. Mechanism of resistance to S-metolachlor in Palmer amaranth. Front. Plant Sci. 2021, 12, 652581. [Google Scholar] [CrossRef] [PubMed]
- Brabham, C.; Norsworthy, J.K.; Houston, M.M.; Varanasi, V.K.; Barber, T. Confirmation of S-metolachlor resistance in Palmer amaranth (Amaranthus palmeri). Weed Technol. 2019, 33, 720–726. [Google Scholar] [CrossRef]
- Gil-Monreal, M.; Zabalza, A.; Missihoun, T.D.; Dörmann, P.; Bartels, D.; Royuela, M. Induction of the PDH bypass and upregulation of the ALDH7B4 in plants treated with herbicides inhibiting amino acid biosynthesis. Plant Sci. 2017, 264, 16–28. [Google Scholar] [CrossRef]
- Labhilili, M.; Joudrier, P.; Gautier, M.F. Characterization of cDNAs encoding Triticum durum dehydrins and their expression patterns in cultivars that differ in drought tolerance. Plant Sci. 1995, 112, 219–230. [Google Scholar] [CrossRef]
- Eceiza, M.V.; Gil-Monreal, M.; Barco-Antoñanzas, M.; Zabalza, A.; Royuela, M. The moderate oxidative stress induced by glyphosate is not detected in Amaranthus palmeri plants overexpressing EPSPS. J. Plant Physiol. 2022, 274, 153720. [Google Scholar] [CrossRef]
- Culpepper, A.S.; Grey, T.L.; Vencill, W.K.; Kichler, J.M.; Webster, T.M.; Brown, S.M.; York, A.C.; Davis, J.W.; Hanna, W.W. Glyphosate-resistant Palmer amaranth (Amaranthus palmeri) confirmed in Georgia. Weed Sci. 2006, 54, 620–626. [Google Scholar] [CrossRef]
- Hess, M.; Barralis, G.; Bleiholder, H.; Buhr, L.; Eggers, T.H.; Hack, H.; Stauss, R. Use of the extended BBCH scale—General for the descriptions of the growth stages of mono; and dicotyledonous weed species. Weed Res. 1997, 37, 433–441. [Google Scholar] [CrossRef]
- Ma, R.; Kaundun, S.S.; Tranel, P.J.; Riggins, C.W.; McGinness, D.L.; Hager, A.G.; Hawkes, T.; McIndoe, E.; Riechers, D.E. Distinct detoxification mechanisms confer resistance to mesotrione and atrazine in a population of Waterhemp. Plant Physiol. 2013, 163, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| CAD Domain | BE Domain | |||||
|---|---|---|---|---|---|---|
| Ala122 | Pro197 | Ala205 | Trp574 | Ser653 | Gly654 | |
| Population B | ||||||
| Non-mutated | 100% | 100% | 100% | 100% | 100% | 100% |
| Population R | ||||||
| Non-mutated | 100% | 100% | 100% | 100% | 100% | 100% |
| Population A | ||||||
| Non-mutated | 100% | 100% | 100% | 50% | 100% | 100% |
| Mutated | 50% (Leu) | |||||
| Population C | ||||||
| Non-mutated | 100% | 43% | 100% | 43% | 100% | 100% |
| Mutated | 28.5% Thr 28.5% (Pro/Thr) | 28.5% (Leu) 28.5% (Trp/Leu) | ||||
| Population D | ||||||
| Non-mutated | 100% | 100% | 100% | 34% | 100% | 100% |
| Mutated | 66% (Trp/Leu) | |||||
| Population Code | Location | Coordinates | Collection Year | Site Background |
|---|---|---|---|---|
| A | Albelda (HU) | 41.841621, 0.496103 | 2020 | Crop field |
| B | Bujaraloz (ZA) | 41.483769, 0.150913 | 2020 | Roadside |
| C | Caspe (ZA) | 41.235783, 0.054056 | 2020 | Roadside |
| D | Tamarite de Litera (HU) | 41.766663, 0.344205 | 2022 | Crop field |
| R | Altorricon (HU) | 41.771093, 0.376554 | 2021 | Roadside |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trebol-Aizpurua, E.; Eceiza, M.V.; Jimenez-Martinez, C.; Marí, A.I.; Royuela, M.; Zabalza, A.; Gil-Monreal, M. Resistance to Amino Acid Biosynthesis Inhibiting-Herbicides in Amaranthus palmeri Populations from Aragon (Spain). Plants 2025, 14, 1505. https://doi.org/10.3390/plants14101505
Trebol-Aizpurua E, Eceiza MV, Jimenez-Martinez C, Marí AI, Royuela M, Zabalza A, Gil-Monreal M. Resistance to Amino Acid Biosynthesis Inhibiting-Herbicides in Amaranthus palmeri Populations from Aragon (Spain). Plants. 2025; 14(10):1505. https://doi.org/10.3390/plants14101505
Chicago/Turabian StyleTrebol-Aizpurua, Eneko, Mikel V. Eceiza, Clara Jimenez-Martinez, Ana I. Marí, Mercedes Royuela, Ana Zabalza, and Miriam Gil-Monreal. 2025. "Resistance to Amino Acid Biosynthesis Inhibiting-Herbicides in Amaranthus palmeri Populations from Aragon (Spain)" Plants 14, no. 10: 1505. https://doi.org/10.3390/plants14101505
APA StyleTrebol-Aizpurua, E., Eceiza, M. V., Jimenez-Martinez, C., Marí, A. I., Royuela, M., Zabalza, A., & Gil-Monreal, M. (2025). Resistance to Amino Acid Biosynthesis Inhibiting-Herbicides in Amaranthus palmeri Populations from Aragon (Spain). Plants, 14(10), 1505. https://doi.org/10.3390/plants14101505






