Genome-Wide Association Analysis of Sweet Pepper (Capsicum annuum) Based on Agronomic Traits Using PepperSNP50K
Abstract
1. Introduction
2. Results
2.1. Descriptive Statistical Analysis of Phenotypes of 217 Sweet Pepper Genotypes
2.2. Population Analysis of 217 Sweet Pepper Germplasm Samples
2.3. Whole-Genome Association Analysis of Sweet Pepper Traits
2.4. Genome-Wide Association Study (GWAS)-Identified Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and DNA Extract
4.2. Agronomic Trait Survey
4.3. Genotyping Using PepperSNP50K Liquid-Phase Breeding Chip
4.4. Phylogenetic and Population Analyses
4.5. Genome-Wide Association Study
4.6. Gene Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hickey, J.M.; Chiurugwi, T.; Mackay, I.; Powell, W.; Implementing Genomic Selection in CGIAR Breeding Programs Workshop Participants. Genomic Prediction Unifies Animal and Plant Breeding Programs to Form Platforms for Biological Discovery. Nat. Genet. 2017, 49, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jia, Z.; Li, J.; Kang, D.; Li, M.; Ma, S.; Cheng, Q.; Shen, H.; Sun, L. Development of a 45K Pepper GBTS Liquid-Phase Gene Chip and Its Application in Genome-Wide Association Studies. Front. Plant Sci. 2024, 15, 1405190. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.S.; Yang, T.J.; Hong, S.Y.; Kwon, Y.S.; Woo, J.G.; Park, H.G. Determination of Cytoplasmic Male Sterile Factors in Onion Plants (Allium cepa L.) Using PCR-RFLP and SNP Markers. Mol. Cells 2006, 21, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Robin, A.H.K.; Natarajan, S.; Jung, H.J.; Nou, I.S. Varietal Identification of Open-Pollinated Onion Cultivars Using a Nanofluidic Array of Single Nucleotide Polymorphism (SNP) Markers. Agronomy 2018, 8, 179. [Google Scholar] [CrossRef]
- Varshney, R.; Graner, A.; Sorrells, M. Genomics-Assisted Breeding for Crop Improvement. Trends Plant Sci. 2005, 10, 621–630. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-Wide Genetic Marker Discovery and Genotyping Using next-Generation Sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef]
- Gupta, P.K.; Rustgi, S.; Mir, R.R. Array-Based High-Throughput DNA Markers for Crop Improvement. Heredity 2008, 101, 5–18. [Google Scholar] [CrossRef]
- Haile, M.; Ro, N.; Ko, H.-C.; Oh, H.; Lee, G.-A. A Comprehensive Genome-Wide Association Study of Carotenoid and Capsaicinoid Contents in Capsicum chinense Germplasm. Int. J. Mol. Sci. 2023, 24, 13885. [Google Scholar] [CrossRef]
- Varshney, R.K.; Terauchi, R.; McCouch, S.R. Harvesting the Promising Fruits of Genomics: Applying Genome Sequencing Technologies to Crop Breeding. PLoS Biol. 2014, 12, e1001883. [Google Scholar] [CrossRef]
- Wang, J.; Chu, S.; Zhang, H.; Zhu, Y.; Cheng, H.; Yu, D. Development and Application of a Novel Genome-Wide SNP Array Reveals Domestication History in Soybean. Sci. Rep. 2016, 6, 20728. [Google Scholar] [CrossRef]
- Bevan, M.W.; Uauy, C. Genomics Reveals New Landscapes for Crop Improvement. Genome Biol. 2013, 14, 206. [Google Scholar] [CrossRef] [PubMed]
- Gunderson, K.L.; Steemers, F.J.; Lee, G.; Mendoza, L.G.; Chee, M.S. A Genome-Wide Scalable SNP Genotyping Assay Using Microarray Technology. Nat. Genet. 2005, 37, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Perkel, J. SNP Genotyping: Six Technologies That Keyed a Revolution. Nat. Methods 2008, 5, 447–453. [Google Scholar] [CrossRef]
- Li, Y.; Tao, Y.; Bai, A.; Yu, Z.; Yuan, S.; Wang, H.; Liu, T.; Hou, X.; Li, Y. High Expression of Ethylene Response Factor BcERF98 Delays the Flowering Time of Non-Heading Chinese Cabbage. Planta 2024, 260, 50. [Google Scholar] [CrossRef]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop Breeding Chips and Genotyping Platforms: Progress, Challenges, and Perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Yu, H.; Xie, W.; Li, J.; Zhou, F.; Zhang, Q. A Wholegenome SNP Array (RICE6K) for Genomic Breeding in Rice. Plant Biotechnol. J. 2013, 12, 28–37. [Google Scholar] [CrossRef]
- Thomson, M.J. Large-Scale Deployment of a Rice 6 K SNP Array for Genetics and Breeding Applications. Rice 2017, 10, 40. [Google Scholar] [CrossRef]
- McCouch, S.R.; Zhao, K.; Wright, M.; Tung, C.-W.; Ebana, K.; Thomson, M.; Reynolds, A.; Wang, D.; DeClerck, G.; Ali, M.L.; et al. Development of Genome-Wide SNP Assays for Rice. Breed. Sci. 2010, 60, 524–535. [Google Scholar] [CrossRef]
- Chen, H.; Xie, W.; He, H.; Yu, H.; Chen, W.; Li, J.; Yu, R.; Yao, Y.; Zhang, W.; He, Y.; et al. A High-Density SNP Genotyping Array for Rice Biology and Molecular Breeding. Mol. Plant 2014, 7, 541–553. [Google Scholar] [CrossRef]
- Ganal, M.W.; Durstewitz, G.; Polley, A.; Bérard, A.; Buckler, E.S.; Charcosset, A.; Clarke, J.D.; Graner, E.-M.; Hansen, M.; Joets, J.; et al. A Large Maize (Zea Mays L.) SNP Genotyping Array: Development and Germplasm Genotyping, and Genetic Mapping to Compare with the B73 Reference Genome. PLoS ONE 2011, 6, e28334. [Google Scholar] [CrossRef]
- Wen, W.; He, Z.; Gao, F.; Liu, J.; Jin, H.; Zhai, S.; Qu, Y.; Xia, X. A High-Density Consensus Map of Common Wheat Integrating Four Mapping Populations Scanned by the 90K SNP Array. Front. Plant Sci. 2017, 8, 1389. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Dong, Z.; Zhao, L.; Ren, Y.; Zhang, N.; Chen, F. The Wheat 660K SNP Array Demonstrates Great Potential for Marker-assisted Selection in Polyploid Wheat. Plant Biotechnol. J. 2020, 18, 1354–1360. [Google Scholar] [CrossRef]
- Tang, B.; Yang, H.; Yin, Q.; Miao, W.; Lei, Y.; Cui, Q.; Cheng, J.; Zhang, X.; Chen, Y.; Du, J.; et al. Fertility Restorer Gene CaRf and PepperSNP50K Provide a Promising Breeding System for Hybrid Pepper. Hortic. Res. 2024, 11, uhae223. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Qin, C.; Tang, X.; Zhou, H.; Hu, Y.; Zhao, Z.; Cui, J.; Li, B.; Wu, Z.; Yu, J.; et al. Development of a SNP Array and Its Application to Genetic Mapping and Diversity Assessment in Pepper (Capsicum spp.). Sci Rep 2016, 6, 33293. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Jang, S.J.; Jeong, H.B.; Lee, S.Y.; Venkatesh, J.; Lee, J.H.; Kwon, J.K.; Kang, B.C. A Mutation in Zeaxanthin Epoxidase Contributes to Orange Coloration and Alters Carotenoid Contents in Pepper Fruit (Capsicum annuum). Plant J. 2021, 106, 1692–1707. [Google Scholar] [CrossRef]
- León-Chan, R.G.; Lightbourn-Rojas, L.A.; López-Meyer, M.; Amarillas, L.; Heredia, J.B.; Martínez-Bastidas, T.F.; Villicaña, C.; León-Félix, J. Differential Gene Expression of Anthocyanin Biosynthetic Genes under Low Temperature and Ultraviolet-B Radiation in Bell Pepper (Capsicum annuum). Int. J. Agric. Biol. 2020, 23, 501–508. [Google Scholar] [CrossRef]
- Lee, H.-Y. Uncovering Candidate Genes Controlling Major Fruit-Related Traits in Pepper via Genotype-by-Sequencing Based QTL Mapping and Genome-Wide Association Study. Front. Plant Sci. 2020, 11, 1100. [Google Scholar] [CrossRef]
- Wu, L.; Wang, P.; Wang, Y.; Cheng, Q.; Lu, Q.; Liu, J.; Li, T.; Ai, Y.; Yang, W.; Sun, L.; et al. Genome-Wide Correlation of 36 Agronomic Traits in the 287 Pepper (Capsicum) Accessions Obtained from the SLAF-Seq-Based GWAS. Int. J. Mol. Sci. 2019, 20, 5675. [Google Scholar] [CrossRef]
- Bode, A.M.; Dong, Z. The Two Faces of Capsaicin. Cancer Res. 2011, 71, 2809–2814. [Google Scholar] [CrossRef]
- Hernández-Pérez, T.; Gómez-García, M.D.R.; Valverde, M.E.; Paredes-López, O. Capsicum annuum (Hot Pepper): An Ancient Latin-American Crop with Outstanding Bioactive Compounds and Nutraceutical Potential. A Review. Comp. Rev. Food Sci. Food Safe 2020, 19, 2972–2993. [Google Scholar] [CrossRef]
- Ochoa-Alejo, N.; Reyes-Valdés, M.H.; Martínez, O. Estimating Transcriptome Diversity and Specialization in Capsicum annuum L. Plants 2024, 13, 983. [Google Scholar] [CrossRef] [PubMed]
- Flores-Díaz, A.; Escoto-Sandoval, C.; Cervantes-Hernández, F.; Ordaz-Ortiz, J.J.; Hayano-Kanashiro, C.; Reyes-Valdés, H.; Garcés-Claver, A.; Ochoa-Alejo, N.; Martínez, O. Gene Functional Networks from Time Expression Profiles: A Constructive Approach Demonstrated in Chili Pepper (Capsicum annuum L.). Plants 2023, 12, 1148. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhao, J.; Sun, H.; Xiong, C.; Sun, X.; Wang, X.; Wang, Z.; Jarret, R.; Wang, J.; Tang, B.; et al. Genomes of Cultivated and Wild Capsicum Species Provide Insights into Pepper Domestication and Population Differentiation. Nat. Commun. 2023, 14, 5487. [Google Scholar] [CrossRef] [PubMed]
- Nimmakayala, P.; Abburi, V.L.; Abburi, L.; Alaparthi, S.B.; Cantrell, R.; Park, M.; Choi, D.; Hankins, G.; Malkaram, S.; Reddy, U.K. Linkage Disequilibrium and Population-Structure Analysis among Capsicum annuum L. Cultivars for Use in Association Mapping. Mol. Genet. Genom. 2014, 289, 513–521. [Google Scholar] [CrossRef]
- Efrati, A.; Eyal, Y.; Paran, I. Molecular Mapping of the Chlorophyll Retainer (Cl) Mutation in Pepper (Capsicum spp.) and Screening for Candidate Genes Using Tomato ESTs Homologous to Structural Genes of the Chlorophyll Catabolism Pathway. Genome 2005, 48, 347–351. [Google Scholar] [CrossRef]
- Lightbourn, G.J.; Griesbach, R.J.; Novotny, J.A.; Clevidence, B.A.; Rao, D.D.; Stommel, J.R. Effects of Anthocyanin and Carotenoid Combinations on Foliage and Immature Fruit Color of Capsicum annuum L. J. Hered. 2008, 99, 105–111. [Google Scholar] [CrossRef]
- Thorup, T.A.; Tanyolac, B.; Livingstone, K.D.; Popovsky, S.; Paran, I.; Jahn, M. Candidate Gene Analysis of Organ Pigmentation Loci in the Solanaceae. Proc. Natl. Acad. Sci. USA 2000, 97, 11192. [Google Scholar]
- Hurtado-Hemandez, H.; Smith, P.G. Inheritance of Mature Fruit Color in Capsicum armuum L. Mater. Methods 1985, 76, 211–213. [Google Scholar]
- Kumar, P.; Lal, S.; Thakur, P.; Sharma, S. Inheritance of Mature Fruit Colour in Capsicum. Indian J. Plant Genet. Resour. 2009, 22, 36–40. [Google Scholar]
- Tian, S.-L.; Li, L.; Chai, W.-G.; Shah, S.N.M.; Gong, Z.-H. Effects of Silencing Key Genes in the Capsanthin Biosynthetic Pathway on Fruit Color of Detached Pepper Fruits. BMC Plant Biol. 2014, 14, 314. [Google Scholar] [CrossRef]
- Blas, A.L.; Ming, R.; Liu, Z.; Veatch, O.J.; Paull, R.E.; Moore, P.H.; Yu, Q. Cloning of the Papaya Chromoplast-Specific Lycopene β-Cyclase, CpCYC-b, Controlling Fruit Flesh Color Reveals Conserved Microsynteny and a Recombination Hot Spot. Plant Physiol. 2010, 152, 2013–2022. [Google Scholar] [CrossRef]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA Spacer-Length Polymorphisms in Barley: Mendelian Inheritance, Chromosomal Location, and Population Dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Gorsi, B.; Su, Y.-C.; Yost, H.J. MMAPPR: Mutation Mapping Analysis Pipeline for Pooled RNA-Seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A Fast and Effective Tool for Linkage Disequilibrium Decay Analysis Based on Variant Call Format Files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Efficient Multivariate Linear Mixed Model Algorithms for Genome-Wide Association Studies. Nat. Methods 2014, 11, 407–409. [Google Scholar] [CrossRef]
- Duggal, P.; Gillanders, E.M.; Holmes, T.N.; Bailey-Wilson, J.E. Establishing an Adjusted P-Value Threshold to Control the Family-Wide Type 1 Error in Genome Wide Association Studies. BMC Genom. 2008, 9, 516. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
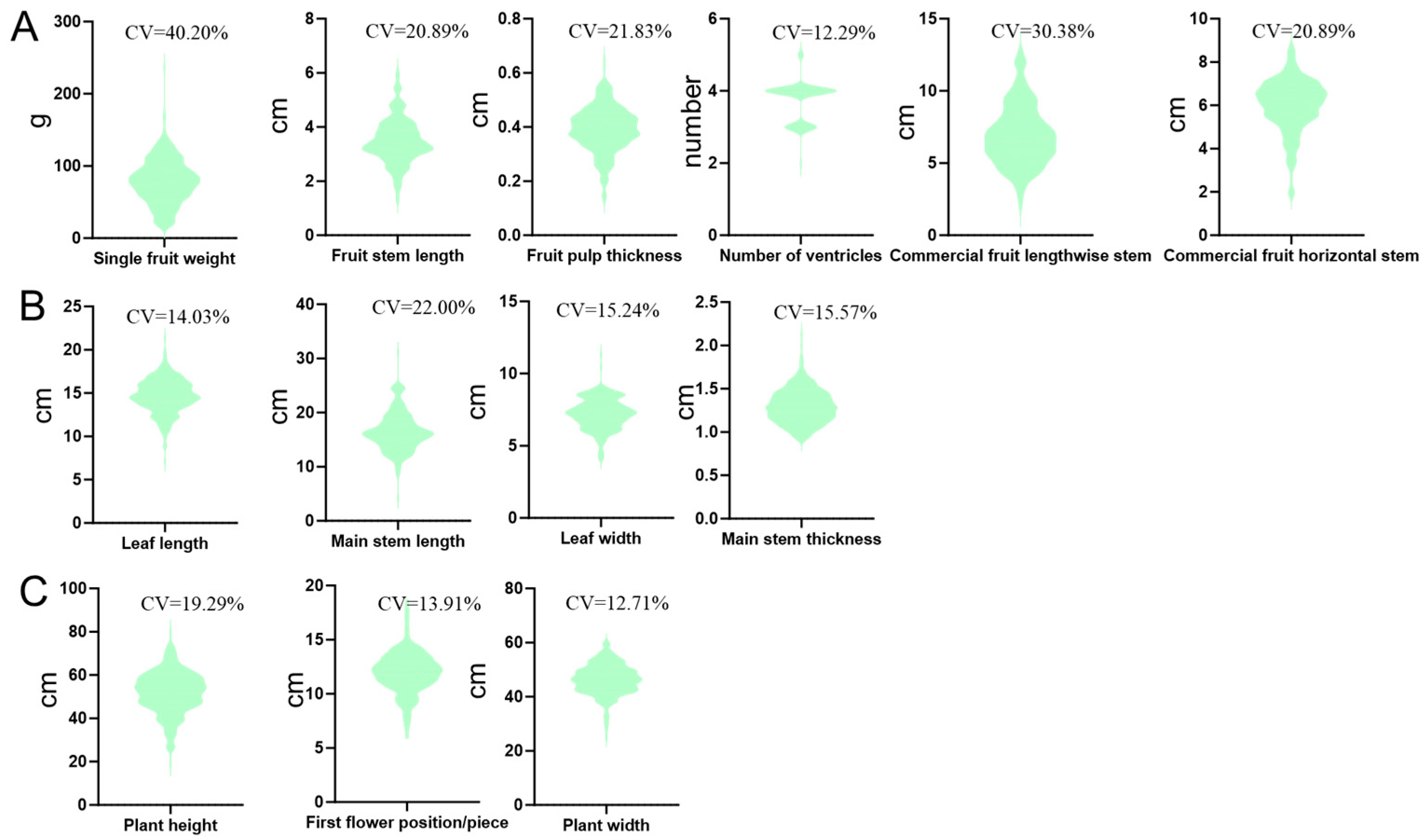
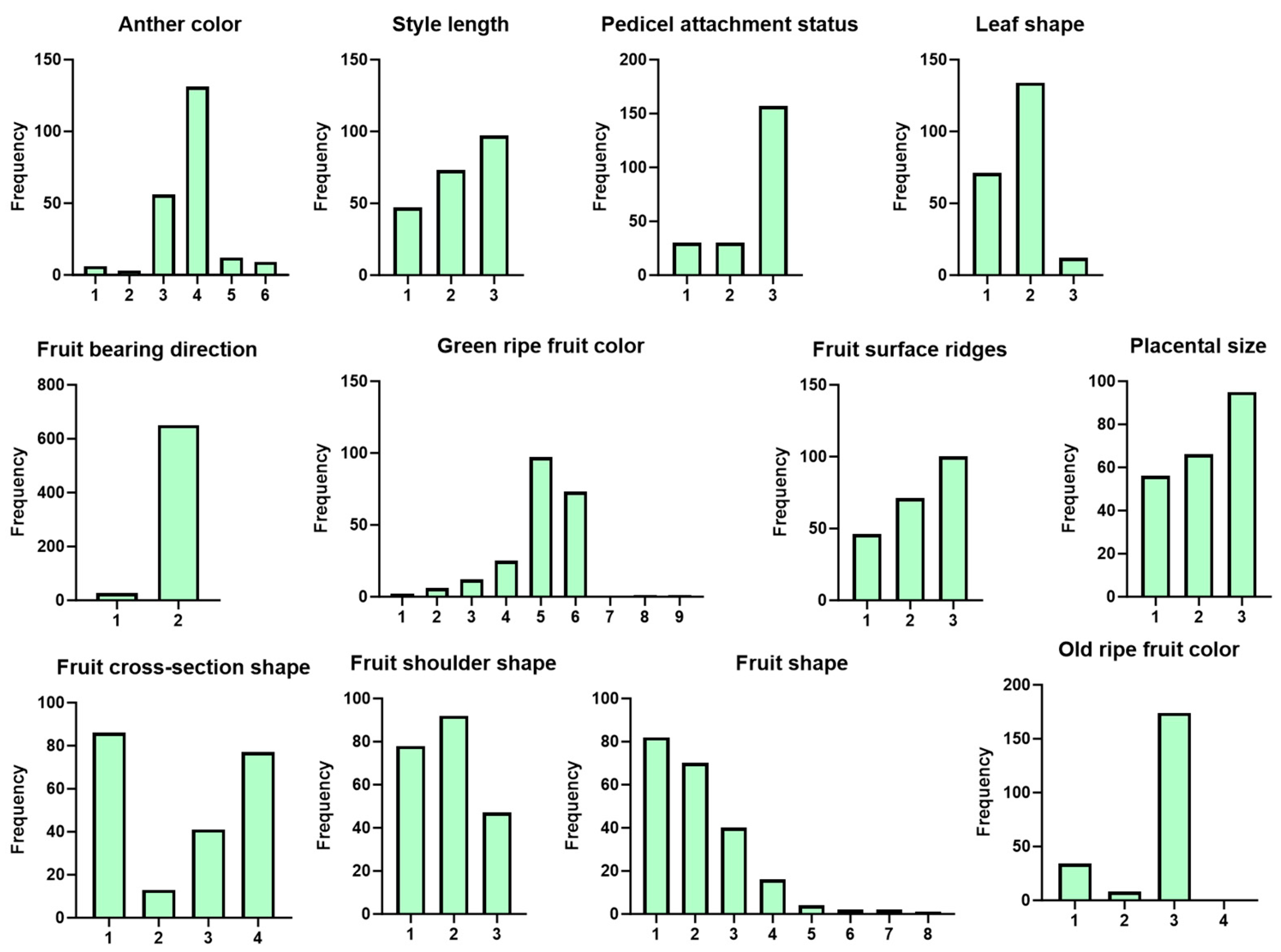
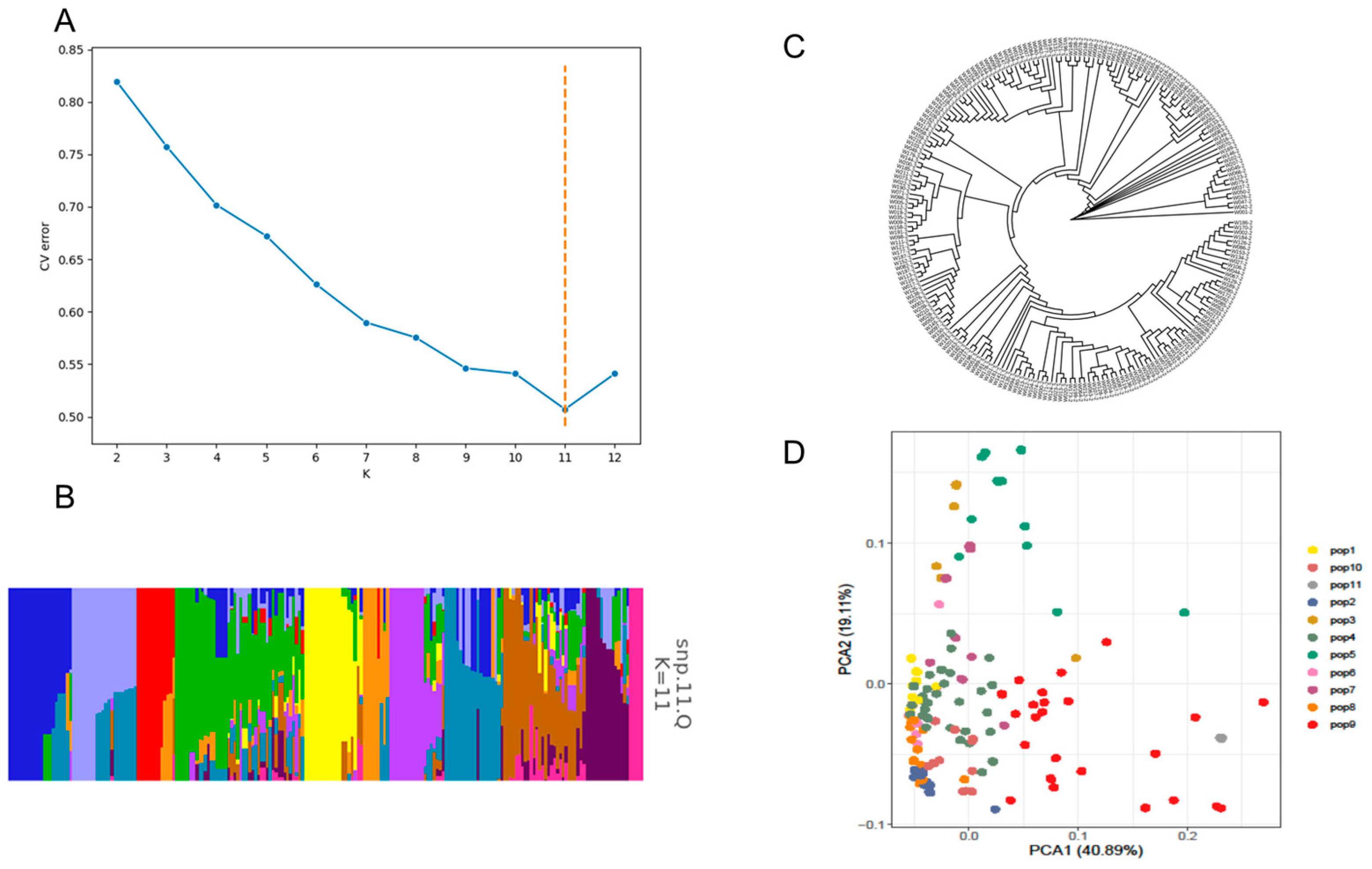
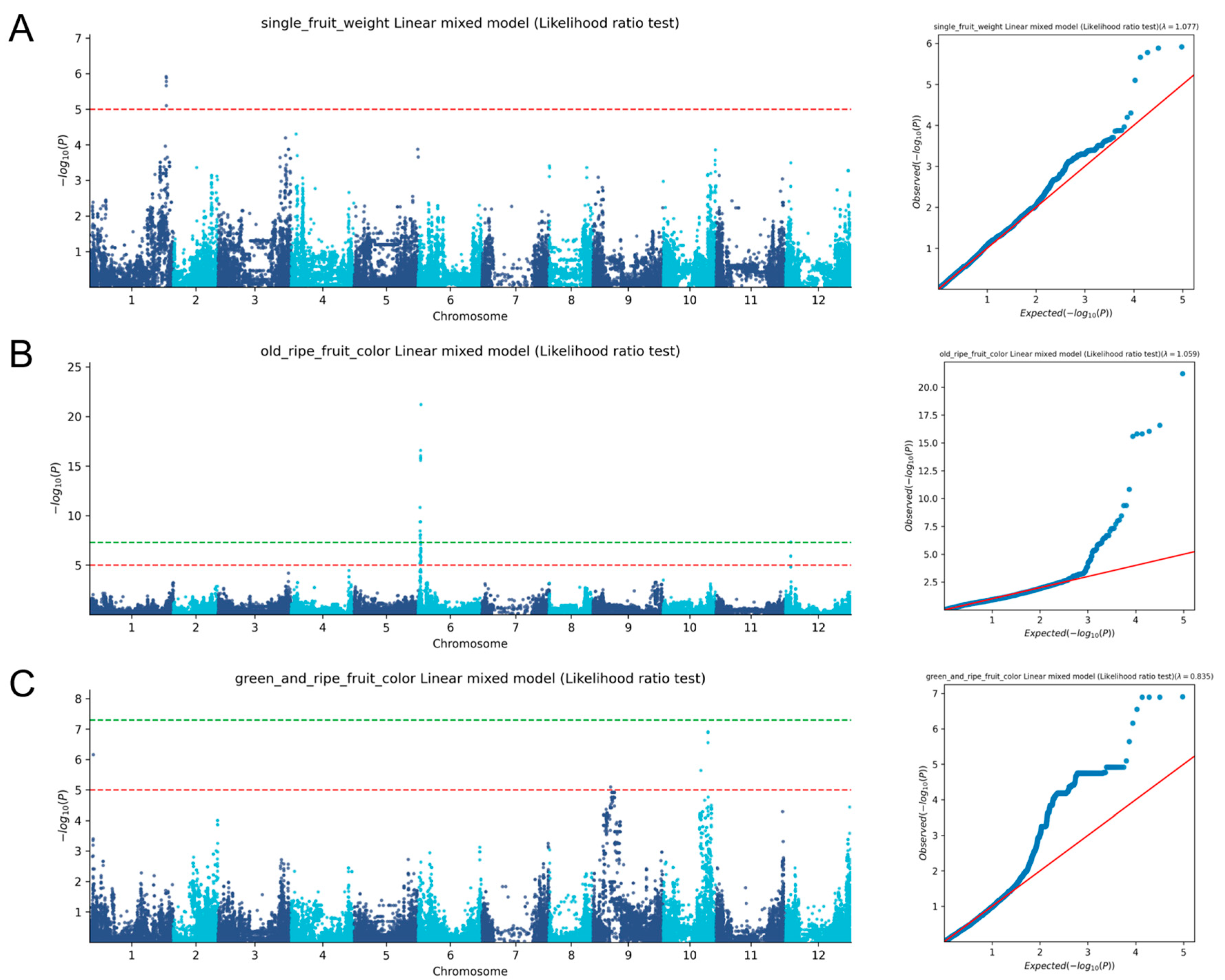
| Trait | Number of Significantly Associated SNPs | Associated Chromosomes |
|---|---|---|
| Anther color | 1 | 2 |
| Commercial fruit horizontal | 6 | 2, 3, 9, 11 |
| Commercial fruit lengthwise | 1 | 4 |
| Fruit shape | 1 | 1 |
| Fruit surface ridges | 2 | 3, 5 |
| Green and ripe fruit color gene | 4 | 1, 10 |
| Leaf length | 1 | 9 |
| Number of ventricles | 1 | 6 |
| Old ripe fruit color gene | 12 | 6 |
| Pedicel attachment status | 12 | 1, 3, 6, 8, 12 |
| Single fruit weight | 4 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, E.; Lu, J.; Wang, J.; Zang, Q.; Liang, Y.; Tian, R.; Zhang, C.; Jiang, F.; Cheng, Y. Genome-Wide Association Analysis of Sweet Pepper (Capsicum annuum) Based on Agronomic Traits Using PepperSNP50K. Plants 2025, 14, 1506. https://doi.org/10.3390/plants14101506
Wang Y, Li E, Lu J, Wang J, Zang Q, Liang Y, Tian R, Zhang C, Jiang F, Cheng Y. Genome-Wide Association Analysis of Sweet Pepper (Capsicum annuum) Based on Agronomic Traits Using PepperSNP50K. Plants. 2025; 14(10):1506. https://doi.org/10.3390/plants14101506
Chicago/Turabian StyleWang, Yaolong, Entong Li, Jiawei Lu, Jing Wang, Qiaolu Zang, Yanping Liang, Ruxia Tian, Changwei Zhang, Fangling Jiang, and Yan Cheng. 2025. "Genome-Wide Association Analysis of Sweet Pepper (Capsicum annuum) Based on Agronomic Traits Using PepperSNP50K" Plants 14, no. 10: 1506. https://doi.org/10.3390/plants14101506
APA StyleWang, Y., Li, E., Lu, J., Wang, J., Zang, Q., Liang, Y., Tian, R., Zhang, C., Jiang, F., & Cheng, Y. (2025). Genome-Wide Association Analysis of Sweet Pepper (Capsicum annuum) Based on Agronomic Traits Using PepperSNP50K. Plants, 14(10), 1506. https://doi.org/10.3390/plants14101506









