Abstract
Virus-induced gene silencing (VIGS) is an RNA-mediated reverse genetics technique that has become an effective tool to investigate gene function in plants. Cotton is one of the most important economic crops globally. In the past decade, VIGS has been successfully applied in cotton functional genomic studies, including those examining abiotic and biotic stress responses and vegetative and reproductive development. This article summarizes the traditional vectors used in the cotton VIGS system, the visible markers used for endogenous gene silencing, the applications of VIGS in cotton functional genomics, and the limitations of VIGS and how they can be addressed in cotton.
1. Introduction
Cotton (Gossypium spp.) is an important economic crop to the textile industry, one of the world’s largest industries [1]. The genus Gossypium contains 52 species. Four of them are cultivated in agricultural production, consisting of two allotetraploids, G. hirsutum and G. barbadense, and two diploids, G. arboretum and G. herbaceum [2,3]. They also serve as ideal models to investigate cell differentiation, cell elongation, cell wall biosynthesis, and polyploidization in plants [1].
The lack of accurate genomic information has always been a restriction in cotton breeding. In recent years, the genome sequences of both diploid and tetraploid cottons have been successfully assembled [4]. Technical advances in high-throughput sequencing and bioinformatics analysis have brought a new epoch for the genomic investigation of cotton. Rich genomic resources will not only contribute to the deep understanding of cotton genome evolution and trait domestication but also accelerate the research on functional genomes in cotton [4]. Understanding the function and regulation of cotton genes is an important step in manipulating these genes in agricultural production. Therefore, the development of a fast and cost-efficient tool is urgently required to fill the gap between cotton genomics and functional genomics. At present, agrobacteria-mediated gene transformation is the main method used to acquire transgenic plants. However, tissue cultures and plant regeneration are time-consuming procedures. Moreover, large genome sizes, gene duplication with functional redundancy, polyploidy, and only a few widely used receptors greatly limit the application of gene transformation in cotton [5]. Consequently, virus-induced gene silencing (VIGS) has become a rapid and effective tool in silencing endogenous genes for cotton functional genomics.
VIGS is an RNA interference-mediated high-throughput reverse genetics technique for functional gene analysis in plants [6]. It knocks down gene expression through posttranscriptional gene silencing (PTGS) [7] (Figure 1). VIGS was first used to describe the recovery of viral symptoms in plants after virus infection [8].
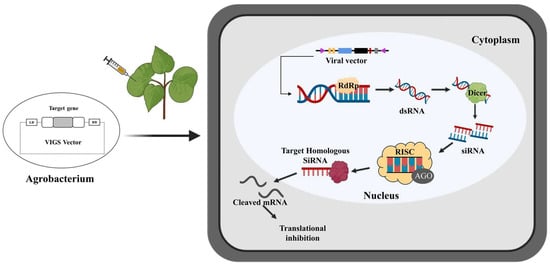
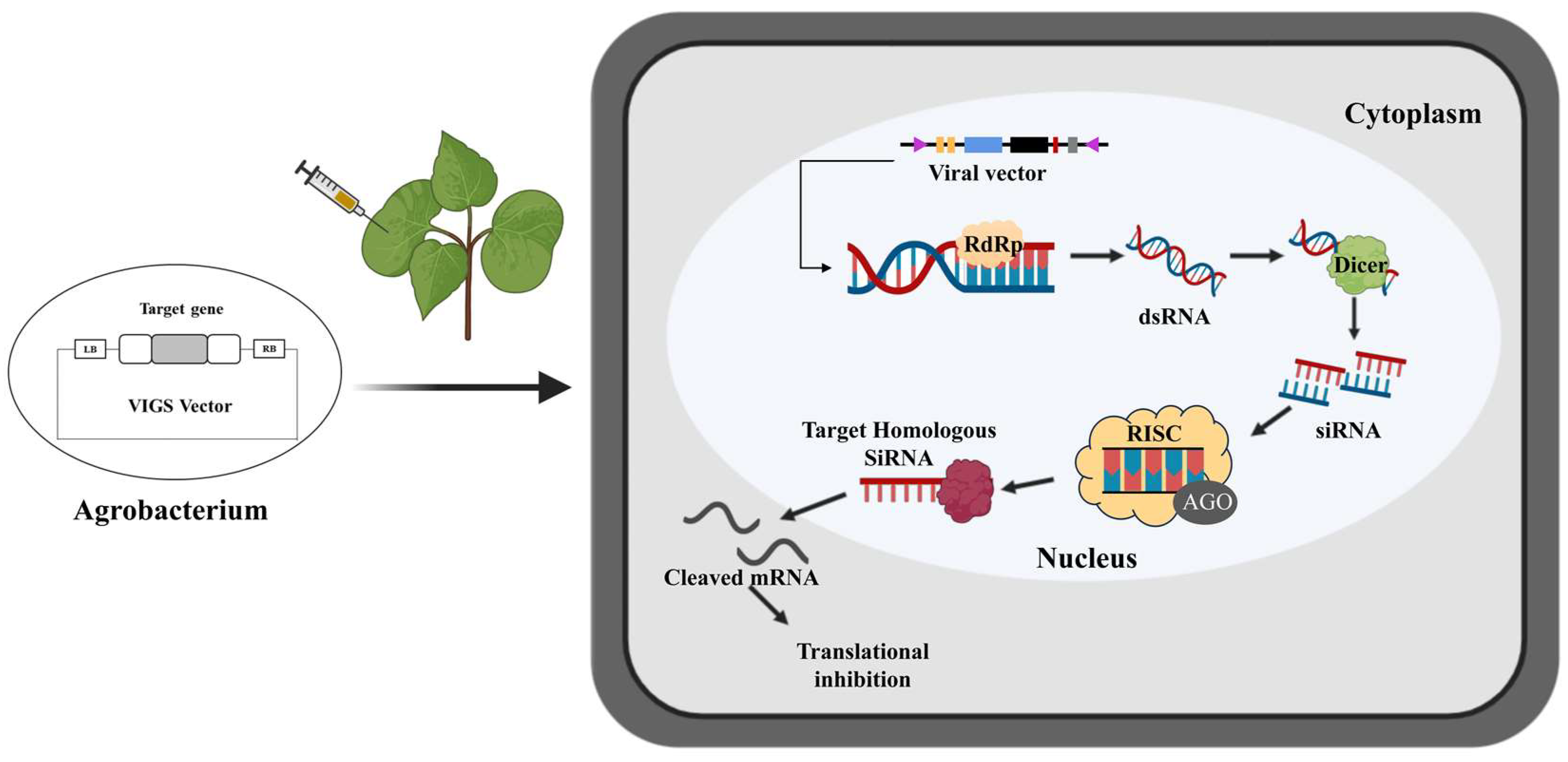
Figure 1.
Molecular mechanism of virus-induced gene silencing viral vectors injected into cotton through an agrobacterium carrying the targeted gene. The target gene is fused into the VIGS vector and transformed into Agrobacterium. The strains enter the cotton leaf through injection. After infection, T-DNA containing the viral genome is transcribed by the cotton RNA polymerase. With the help of RNA-dependent RNA polymerase (RdRP) (yellow), the single-stranded RNA (ssRNA) viral transcripts produce double-stranded RNA (dsRNA). The dsRNAs are further cleaved into small (21–24 nt) short interfering RNAs (siRNAs) by a dicer (green). Then, amplified siRNAs, together with the AGO protein, form an RNA-induced silencing complex (RISC). RISC uses these siRNAs to accurately establish homologous RNAs in cells, which triggers the endo-nucleolytic cleavage and translational inhibition of the cognate target mRNA, thereby producing PTGS. The single-stranded siRNAs are amplified and propagated as a mobile silencing signal throughout the plant, leading to target gene silencing in plant organs far from the infected site.
Researchers then proved that this is a natural defense mechanism in plants induced by virus infection [9]. Therefore, researchers engineered virus genomes (complementary DNA (cDNA)) into recombinant viral vectors containing sequences of homologous host genes, which can trigger homologous endogenous gene silencing in plants. Thus, VIGS has been widely applied for rapid and large-scale analyses of gene functions and functional genomics in many higher plants, such as Arabidopsis [10], tobacco (Nicotiana benthamiana) [11], wheat (Triticum aestivum) [12], alfalfa (Medicago truncatula) [13], tomato (Solanum lycopersicum) [14], and poplar (Populus euphratica) [15].
In this review, we discuss the VIGS method in cotton, including modifications and applications, especially for genes whose function has been described. We also elaborate on the challenges and restrictions in the use of this method and suggest future directions for its improvement.
2. VIGS Vectors for Gene Functional Analysis in Cotton
In the past few decades, various plant viruses have been engineered using the VIGS system for a large number of plant species. As required, many vectors have been developed and applied in the VIGS system. VIGS vectors contain three kinds of vectors: DNA, RNA, and satellite virus vectors. DNA viruses make up only a minority of plant viruses, with large genome structures and limited movement in plants [16]. Single-stranded DNA (ssDNA) viruses belong to the largest known family of plant DNA viruses [16]. Other types of DNA-based viruses, such as African cassava mosaic virus (ACMV) and Cotton leaf crumple virus (CLCrV), have been efficiently converted into VIGS vectors and successfully applied to cassava and cotton plants [17,18]. RNA viruses are the earliest and most widely used viral vector to establish the VIGS system, due to their small molecular weight and high infection efficiency. RNA virus vectors contain Tobacco mosaic virus (TMV) [19], Tobacco rattle virus (TRV) [20], Barley stripe mosaic virus (BSMV) [21], Bean pod mottle virus (BPMV) [22], Potato X virus (PXV) [23], etc. TMV was the earliest vector based on the model RNA virus [19]. A recombinant virus containing the coding sequence of Nicotiana benthamiana phytoene desaturase (NbPDS) was constructed and used in plants to successfully knockdown NbPDS [19]. Among these RNA virus vectors, the TRV-induced gene silencing (TRV-VIGS) system has many advantages, such as a high silencing efficiency, long duration, mild virus symptoms in host plants, and gene silencing in various tissues, resulting in its wide use [7]. Satellite viruses do not induce any plant diseases, and they are usually unrelated to any disease or interference with the true gene-silencing phenotype [24].
To date, two viral vector systems suitable for cotton gene silencing have been reported. One is based on TRV, and the other on CLCrV [25,26] (Figure 2). The TRV vector contains a bilateral positive-sense single-stranded RNA virus, including the RNA1 and RNA2 genomes. RNA1 encodes two replication enzymes and a protein rich in cysteine, which is important for viral replication and movement. Conversely, RNA2 encodes one coat protein and two nonessential structural proteins, which can be deleted to insert alien sequences [7,9]. CLCrV belongs to the genus Begomovirus, family Geminiviridae [27]. It is known that CLCrV duplicates in the nucleus, producing double-stranded DNA intermediates as templates for additional genome duplication and transcription [28]. CLCrV is a kind of typical bilateral begomovirus consisting of two 2.6 kb circular single-stranded DNA molecules, DNA-A and DNA-B, which share approximately 200 bp of homologous regions known as common regions [18]. The DNA-A component contains four genes that encode the duplication-related protein, transactivator protein, and coat protein. DNA-B contains two proteins, BV1 and BC1. The BV1 protein is a nuclear shuttle protein (NSP), whereas the BC1 protein mediates the intercellular transport of CLCrV DNA via plasmodesmata [29,30]. The CLCrV-based VIGS vector was first reported to silence genes through particle bombardment in cotton [18].
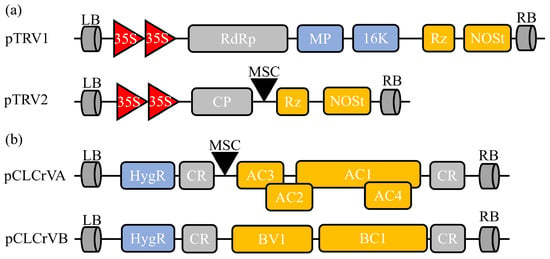
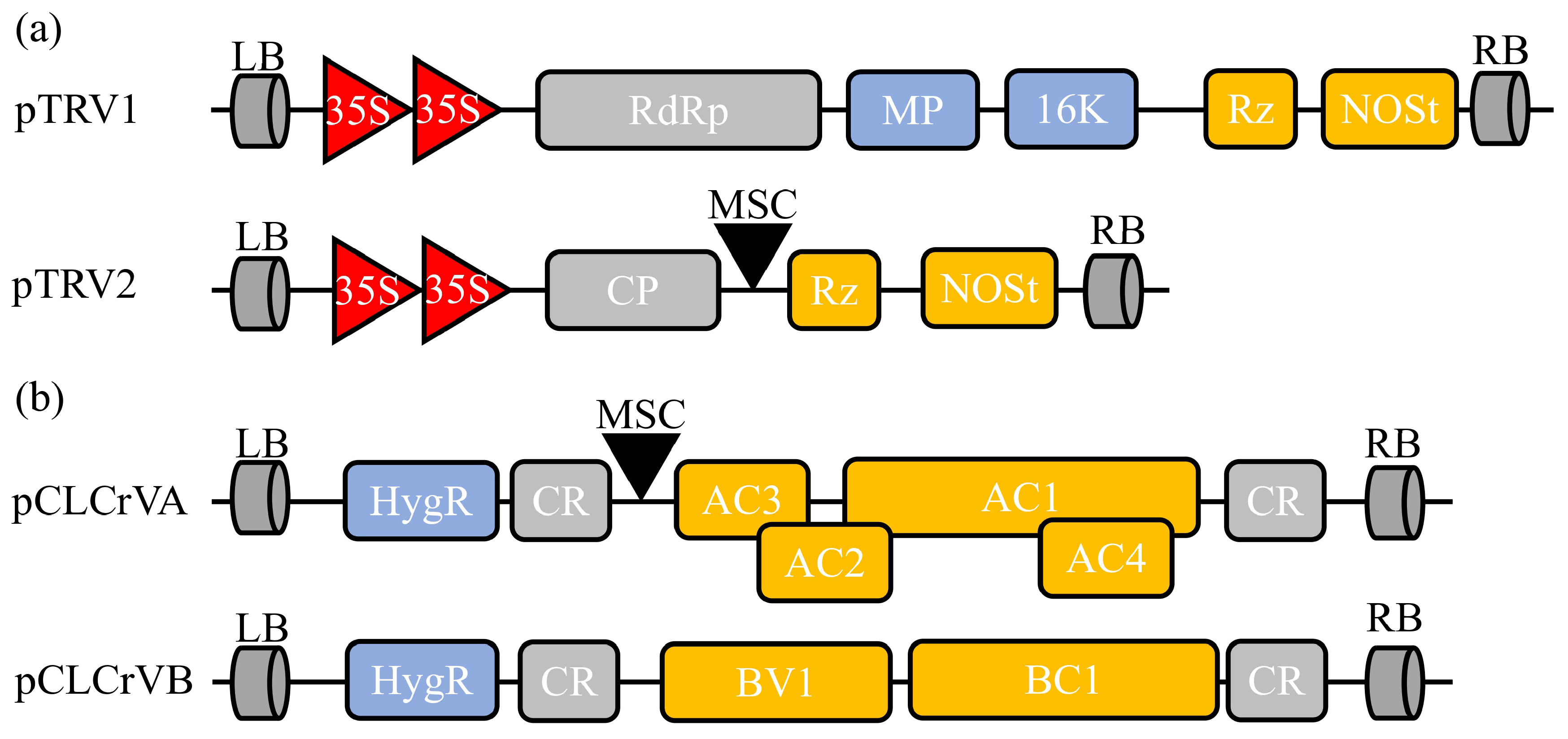
Figure 2.
Schematic diagram of two common VIGS vectors used in cotton. (a) Tobacco rattle virus (TRV)-derived silencing vector contains pTRV1 and pTRV2. LB, left border; RdRp, RNA-dependent RNA polymerase; MP, movement protein; 16 K, 16 Kd protein; Rz, self-cleaving ribozyme; NOSt, NOS terminator; RB, right border; CP, coat protein; MCS, multiple cloning site. (b) Cotton leaf crumple virus (CLCrV)-based silencing vector includes pCLCrVA and pCLCrVB. pCLCrVA consists of four genes (AC1–AC4), and pCLCrVB consists of two genes (BV1 and BC1). The genes are flanked by common regions (CRs), including the origin of replication.
3. Visible Markers for Endogenous Gene Silencing in Cotton
To evaluate and monitor the silencing efficiency of VIGS, marker genes, especially those regulating visible phenotype characteristics, are used, including PDS, chloroplastos alterados 1 gene (CLA1), the green fluorescent protein (GFP), and anthocyanidin synthase (ANS) [19,31,32,33]. The PDS gene is involved in the biosynthesis of carotenoids, and it has been reported in many plant species. It can serve as a visible marker to evaluate the silencing efficiency, as its knockdown in plants can lead to an albino phenotype, due to the lack of chlorophyll [34]. CLA1 encodes the 1-deoxy-Dxylulose 5-phosphate synthase related to chloroplast development, and it is highly conserved in various plants [35]. The silencing of CLA1 results in a bleached phenotype [35]. The GFP signal can be easily observed in plant cells, and the silencing efficiency of a modified TRV–GFP vector has been evaluated in various plant species [36]. It has been shown that the infection efficiency of the vector is equal to that of the original TRV vector [36].
In cotton, CLA1 is widely used as a positive control to determine the silencing efficiency of VIGS [26]. This gene has been silenced in the four cultivated varieties: G. hirsutum, G. barbadense, G. arboretum, and G. herbaceum [37,38]. However, not all G. hirsutum varieties have an equal response to TRV-mediated silencing, which is an important consideration for future research on gene function [38]. Moreover, compared to G. hirsutum, the silencing of CLA1 is more prominent in G. arboretum and G. herbaceum, implying that the efficiency of TRV-mediated VIGS may be influenced by the ploidy level and appears to be higher in diploids [38]. Two cotton phenotypic scorable endogenous genes, PDS and phytoene synthase (PSY), have been tested as positive controls in TRV-based VIGS [39]. The silencing of PDS in both G. hirsutum and G. arboretum showed the photobleaching effect. Meanwhile, knocking down the expression of PSY in red leaf cotton resulted in green cotton leaves with no red color patches. These results suggested that PDS and PSY could be used as positive controls in the VIGS system. However, although the silencing of CLA1 and PDS can lead to visible photobleaching, it can also result in wilting and plant death. Therefore, it cannot be used for gene silencing throughout the entire plant growth period. To address this issue, the cotton pigment gland formation gene (PGF), which has been reported to regulate gland formation, has been developed as a new marker gene to track the efficiency of gene silencing in infected tissues [26]. The silencing of PGF in cotton does not influence normal growth and development, and the number of glands is highly correlated with the expression level of PGF [26,40]; therefore, the PGF gene is a perfect marker for the entire cotton growth period. In a previous study, the proanthocyanidin (PA) metabolic-related genes ANS and anthocyanidin reductase (ANR) were also silenced via VIGS in G. barbadense. Transcripts of these two genes were dramatically reduced. The brownish phenotype of the cotton plants emerged from 8 to 10 days post-vaccination in the ANS-silenced lines, whereas no visible phenotype difference between the ANR-silenced lines and vector control plants was detected [37]. These results indicate that ANS and ANR serve as mild marker genes for endogenous gene silencing in VIGS.
4. VIGS for Studying Abiotic Stress Response in Cotton
Abiotic stresses, such as drought, soil salinity and alkalinity, and extreme temperature stresses, can induce a series of physiological and biochemical reactions and inhibit the normal growth and development of plants [41,42,43]. VIGS has been widely used to study gene function in cotton under abiotic stress (Table 1). The following sections list the application of VIGS to characterize abiotic-stress-related genes in cotton.
4.1. Drought Stress Tolerance
Drought is an important limiting factor in cotton production. Over half of the global cotton supply is grown in areas with severe water shortages. Cotton is recognized as drought-resistant. VIGS is a valuable tool for the functional validation of drought-responsive genes in transcriptional profiles under drought stress in cotton.
By integrating genome-wide RNA sequencing (RNA-Seq) and loss-of-function screening using VIGS with a comprehensive biochemical assay, GhWRKY59 was identified as an important transcription factor that regulates cotton drought stress [44]. Further analysis showed that GhWRKY59 is phosphorylated by a classical mitogen-activated protein kinase (MAPK) cascade composed of GhMAP3K15-GhMKK4-GhMPK6 and that GhWRKY59 positively regulates GhMPK6 activation through the feedback control of GhMAP3K expression. Moreover, GhWRKY59 directly regulates the expression of abscisic acid (ABA)-independent GhDREB2 and drought-responsive genes by binding to their promoters [44]. This study revealed a complete MAP kinase cascade that phosphorylates and activates an important WRKY transcription factor, and it elucidated a regulatory module composed of GhMAP3K15-GhMKK4-GhMPK6-GhWRKY59-GhDREB2, which is involved in regulating the drought response in cotton. Vacuolar HC-ATPase (V-ATPase) is responsible for the deacidification of the cytosol and the excitation of secondary transport in the vacuolar membrane [45]. Various V-ATPase-related genes have been proven to be involved in regulating the plant response to water deficit. However, their roles in the cotton drought response have not been reported. The silencing of GhVHA-A in cotton via VIGS decreased the resistance to the drought response and induced a decrease in chlorophyll content and antioxidant enzyme activity, implying that GhVHA-A is a candidate gene to enhance resistance to drought stress in cotton [45]. The tubby-like protein (TULP), which is involved in the plant response to various stresses, has rarely been reported in cotton. The expression of GhTULP30 was activated by drought stress [46]. The knockdown of the expression of cotton GhTULP30 through VIGS slowed down the rate of stomatal closure under drought stress and reduced the width and length of stomata, which provided a reference and direction for further exploring the role of TULP in cotton [46]. N6-methyladenosine (m6A) is the most common internal modification in mRNA, and it is believed to be involved in a series of developmental and biological processes. However, the function of m6A modification in drought stress is still poorly understood. The silencing of two Ca2+ signal-related genes, GhECA1 and GhCNGC4, via VIGS reduced drought tolerance, resulting in a decrease in m6A enrichment in silenced lines [47]. This finding revealed a novel posttranscriptional modification mechanism that involves the methylation of m6A on the targeted transcripts of the ABA and Ca2+ signaling pathway in regulating the cotton drought response.
4.2. Salt Stress Tolerance
Soil salinization is one of the most urgent issues worldwide, due to its negative impact on agricultural production. It has adversely affected over 800 million hectares of arable land globally, accounting for over 6% of the world’s total agricultural land [48]. An appropriate salt concentration can serve as a nutritional component to promote cotton growth. However, a high salt concentration results in the early maturity or senescence of leaves, leading to a decrease in cotton yield [43]. The usefulness of VIGS in studying cotton salt stress resistance has also been confirmed.
The plant lipoxygenase (LOX) gene is a member of the non-heme iron-containing dioxygenase family, and it catalyzes the oxidation of polyunsaturated fatty acid to multifunctional oxygenase. The roles of LOX genes have been widely investigated in abiotic and biotic stresses; however, their functions in cotton are still poorly understood. The silencing of two cotton LOX genes (GhLOX12 and GhLOX13) through VIGS exhibited significantly higher chlorophyll degradation and a higher accumulation of H2O2, malondialdehyde (MDA), and proline, thus improving sensitivity to salt stress [49]. The salt overly sensitive (SOS) pathway is conserved in plants, and it is mainly responsible for transporting intracellular Na+ out of the cytoplasm to reduce the content of Na+ under salt stress conditions [50]. Knocking down the expression of GhSOS1 via VIGS makes cotton plants more sensitive to salt stress with decreased growth, insufficient root vitality, and increased Na+ content and Na+/K+ ratio in the roots, stems, and leaves [50]. In addition to positive regulatory genes, there are some factors that negatively respond to salt stress in cotton. The drought-induced 19 (Di19) protein is a Cys2/His2 (C2H2)-type zinc finger protein that plays an important role in normal growth and response to abiotic stress in plants. In a previous study, through VIGS, the silencing of GhDi19-3 and GhDi19-4 reduced sensitivity to salt stress, with significantly decreased levels of H2O2, MDA, and peroxidase (POD), while the activity of superoxide dismutase (SOD) dramatically increased sensitivity [51]. Further analysis indicated that the expressions of the Ca2+ signal and ABA-related genes were markedly changed, suggesting that GhDi19-3 and GhDi19-4 respond to salt stress by participating in Ca2+ and ABA signaling. Long noncoding RNAs (lncRNAs) have been proven to be involved in many biological processes and responses to environmental stress. However, the function of the lncRNA response to salt stress in cotton is poorly understood. β-glucosidases (BGLUs) hydrolyze β-D-glycosidic bonds and retain the heteromeric configuration that is involved in plant biotic and abiotic stresses [52]. Knocking down the expression of GhBGLU24-A via VIGS resulted in a salt-resistant phenotype in cotton while also enhancing plant height and fresh weight [53]. Meanwhile, the lncRNA TRABA binds to the promoter of GhBGLU24-A to inhibit its expression. Further analysis showed that the GhBGLU24-A interacted with the RING-type E3 ubiquitin ligase GhRUBL, and, by using VIGS, these two genes were both confirmed to be involved in endoplasmic reticulum (ER) stress. Moreover, the function of GhRUBL under salt stress was also investigated using the VIGS method, and the silencing of cotton GhRUBL increased salt resistance. These findings help to understand the regulatory mechanism of the lncRNA TRABA in regulating cotton salt and ER stresses, enabling potential suggestions regarding the development of more resilient crops.
4.3. Cold and Heat Stress Tolerance
The extreme temperature fluctuations caused by climate change suppress normal plant growth and threaten crop productivity. Due to global warming, plants are increasingly affected by temperature stress, including cold and heat stresses. Plants have developed sophisticated strategies to quickly sense and effectively respond to temperature stress. The activation of temperature-related genes is important in resisting temperature stress in plants. VIGS is an effective tool to investigate functional genes under temperature stress in cotton.
The members of the P4 subfamily of P-type ATPases are associated with lipid asymmetry between the two lipid lobules of the Arabidopsis plasma membrane and are crucial for low-temperature tolerance; however, the functions of P4-ATPases in cotton are still unclear. The silencing of GbPATP through VIGS made cotton plants more sensitive to low temperatures and increased the content of MDA with lower catalase (CAT) activity, implying that GbPATP acts as an important regulatory factor in cotton low-temperature tolerance [54]. One short-chain alcohol dehydrogenase, GhSAD1, which responds to low-temperature tolerance, was identified in a genome-wide association study (GWAS) of 200 cotton materials [55]. Knocking down the expression of a haploid genotype of GhSAD1 (GhSAD1HapB) decreased the tolerance of cotton to low temperatures. Further analysis showed that GhSAD1HapB modulated the activity of the C-repeat binding factor, which regulates the ABA signaling pathway. Moreover, GhSAD1HapB activated the expression of COLD-REGULATED (COR) genes and increased the number of metabolites related to low-temperature tolerance. These findings improve the understanding of the mechanisms underlying the GhSAD1-mediated low-temperature response in cotton. Natural antisense transcripts (NATs) have been known to be important regulatory factors of gene expression under abiotic stresses in model plants. However, their function under low-temperature stress is poorly understood in crops. CAN1 was identified from the NATs of the leaves of G. hirsutum and G. barbadense under low-temperature stress [56]. VIGS experiments indicated that CAN1 significantly improved the cold tolerance of cotton, and further investigations showed that CAN1 regulated the expression of SnRK2.8 in response to low-temperature stress. These findings indicate the potential of NATs for application in breeding cold-resistant cotton. Mitogen-activated protein kinase kinase kinases (MAP3Ks) regulate various plant biological processes, but their functions in cotton are still unclear. At present, there are few studies examining high-temperature stress in cotton. Knocking down the expression of GhMAP3K65 through VIGS improved cotton tolerance to heat stress, which provided further insight into the regulatory mechanisms of a Raf-like MAP3K65 protein in cotton [57].
5. VIGS for Studying Biotic Stress Response in Cotton
In addition to abiotic stress, several biotic stresses, including pest and pathogen stresses, can also affect the growth and development of plants. Wilt diseases, such as Fusarium proliferatum and Verticillium dahliae, are recognized as the most damaging environmental stresses in cotton. These diseases can induce serious growth arrest and a loss of yield in cotton. VIGS has been an effective tool to investigate the function of cotton genes under biotic stress (Table 1).
5.1. Verticillium Stress Tolerance
Verticillium wilt is a serious disease caused by Verticillium dahliae wilt. Once infected with V. dahliae, the root xylem vessels become blocked, resulting in a decrease in yield and fiber quality [58]. Many functional genes that respond to Verticillium stress have been investigated to improve the understanding of the molecular mechanism underlying cotton resistance to Verticillium.
Pectin is the main component of the primary cell wall in plants and serves as the main barrier against pathogens. Pectin methylesterases (PMEs) catalyze the demethylation of the pectin galacturonic domain in plant cell walls. Their activity is regulated by PME inhibitors (PMEIs). The silencing of GhPMEI3 in cotton via VIGS enhanced susceptibility to V. dahliae [59]. Glutathione S-transferases (GSTs) have been classified into soluble and microsomal proteins, and they play crucial roles in detoxifying antioxidants and exogenous substances. GSTs protect cells from abiotic or biotic stresses by catalyzing the reduction in tripeptide glutathione (GSH) to bind to various exogenous or endogenous substrates in vivo [60]. The virus-induced silencing of the GST gene (Gh_A09G1509) made the resistant cotton cultivar Nongda601 dramatically susceptible to V. dahliae [60]. Further analysis revealed that the GST gene was crucial for achieving a subtle balance between the production and scavenging of H2O2 under V. dahliae stress [60]. Suberin acts as a stress-induced resistance barrier in the root cell wall. CYP86A1 encodes the cytochrome P450 fatty acid ω-hydroxylase, which has been reported to be an important enzyme in the biosynthesis of suberin. However, the function of CYP86A1 in responding to fungi and the mechanisms associated with immune responses are poorly understood. GbCYP86A1-1 was identified as a disease-resistance-related gene, and the knocking down of GbCYP86A1-1 through VIGS particularly increased susceptibility to V. dahliae in cotton [61]. These results emphasize the function of GbCYP86A1-1 in defense against fungi and its potential molecular immune mechanisms during this process [61]. Changes in the active structure of the actin cytoskeleton are a common host response to pathogen attack. The role of the cotton actin-binding protein VILLIN2 (GhVLN2) in host defense against soilborne fungus V. dahliae wilt was investigated, and the silencing of GhVLN2 via VIGS reduced the degree of actin filament bundles and interfered with cotton plant growth, leading to the formation of twisted organs and brittle stems, as well as a decrease in cellulose content in cell walls [62]. These results revealed that the regulatory expression and functional transfer of GhVLN2 promote the regulation of the dynamic remodeling of the actin cytoskeleton in the host immune response to V. dahliae. The role of lncRNA in the cotton V. dahliae stress response is still unclear. Comprehensive lncRNA profiles of the cotton disease response to defend against V. dahliae were constructed, and the virus-induced silencing of two key lncRNAs, GhlncNAT-ANX2 and GhlncNAT-RLP7, in cotton improved tolerance to V. dahliae and Botrytis cinerea, which may be related to the enhanced expression of LOX1 and LOX2 [63]. This is the first description of lncRNA in responding to fungal disease, and it provides new clues for elucidating the mechanism underlying the cotton disease response.
5.2. Bemisia Tabaci (Whitefly) Stress Tolerance
Whitefly has caused significant damage to global cotton production. Information on how cotton plants perceive and defend against this destructive pest is limited. RNA-seq was employed to compare two cotton cultivars exhibiting strong resistance and sensitivity to the whitefly. Analysis revealed that the transcriptional response of cotton to whitefly invasion involves transcription factors, genes encoding protein kinases, phytohormone signals, and metabolite synthesis. The virus-induced silencing of GhMPK3 in cotton led to the inhibition of the MPK–WRKY–jasmonic acid (JA) and ethylene (ET) pathways, and it increased susceptibility to the whitefly [64].
6. VIGS for Studying Vegetative Development in Cotton
Cotton vegetative development includes seed germination, root elongation, seedling development, and plant growth. VIGS has been widely used to investigate the functional genes related to vegetative development in cotton (Table 1).
6.1. VIGS for Studying Seed Germination in Cotton
Seed germination is a critical stage in the crop life cycle, and it is closely associated with seedling survival rate and crop yield. A low germination rate, emergence rate, and vigor of seeds lead to a decrease in cotton production. Thus, it is of significance to investigate the function and regulation of genes during seed germination in cotton. Recently, an improved VIGS system was developed by simplifying the seed imbibition (Si-VIGS) of the widely used TRV to expand the study of functional genes during germination [65]. Through the functional screening of the cDNA library of germinated cotton seeds, an important regulation gene, galactose synthase 2 (GhGOLS2), for cotton seed germination was discovered. The silencing of GhGOLS2 in cotton through the Si-VIGS system dramatically suppressed the seed germination process.
6.2. VIGS for Studying Root Development in Cotton
Roots absorb water and nutrients supporting plants to respond to environmental stress and significantly affecting many important agronomic traits. However, cotton’s root structure is poorly understood. A GWAS of cotton root traits from 200 upland cotton accessions revealed two candidate genes, GhTRL1-A05 and GhPIN8-D04 [66]. The virus-induced silencing of these two genes in cotton resulted in a reduced root length and surface area.
6.3. VIGS for Studying Stem Development in Cotton
Stems ultimately originate from the shoot apical meristem (SAM), which is the main organ responsible for transporting molecules from underground roots to the aboveground parts of plants. The virus-induced gene silencing of major DELLA genes particularly increased secondary cell wall (SCW) formation in the cotton stem xylem and phloem [67]. The DELLA protein in cotton has been found to interact with the SCW-related NAC protein, and the silencing of NAC genes via VIGS suppressed the development of SCW by downregulating lignin biosynthesis and deposition [67]. Through the map-based cloning strategy, one key repressor of the formation of the cotton stem trichome GoSTR was identified based on an F2 segregating population that originated from a cross between TM-1 and J220 [68]. Knocking down the expression of GoSTR in J220 and Hai7124 through VIGS led to pubescent stems but no apparent leaf trichome changes.
6.4. VIGS for Studying Leaf Development in Cotton
Leaves are derived from the peripheral zone of SAM. The shapes of cotton leaves vary greatly, including okra, Sea-Island, super-okra, and broad leaf shapes, and this is regulated by a multiple allele locus, L2 [69]. Understanding the genetic basis of cotton leaf morphological variation is crucial for improving agricultural production. The L2 loci was identified as a homeodomain leucine zipper (HD-Zip) transcription factor homologous to the LATE MERISTEM IDENTITY1 (LMI1) gene (GhOKRA) [69,70]. The silencing of GhOKRA through VIGS resulted in a change in phenotype from okra to broad leaf. This study provides a theoretical direction for breeders to generate a superior cotton ideotype. Ethylene accumulates with the senescence process, and it is the main accelerant of leaf senescence. ETHYLENE INSENSITIVE3 (EIN3), as the core transcriptional enhancer, activates the expression of extensive downstream genes during leaf senescence. Cotton LINT YIELD INCREASING (GhLYI) belongs to the EIN3-LIKE 1 (EIL1) gene, which acts as an ethylene signal response factor and a positive regulator of senescence [71]. GhLYI directly binds to the promoter of SENESCENCE-ASSOCIATED GENE 20 (SAG20) to activate its expression. The silencing of GhSAG20 via VIGS in cotton can delay leaf senescence [71].
7. VIGS for Studying Flowering in Cotton
The role of genes that regulate the development of anther and fertility is still yet to be determined. The silencing of acyl-CoA N-acyltransferase (GhACNAT) through VIGS in cotton led to indehiscent anthers, reduced filaments and stamens, and plant sterility [72]. Further analysis revealed that GhACNAT played a critical role in controlling fertility by regulating lipid biosynthesis and the JA biogenesis pathways. Glycerol-3-phosphate acyltransferases (GPATs) are crucial for various biological processes, such as male fertility, and they have been widely investigated. However, their exact role and potential regulatory mechanisms in cotton anther development are poorly understood. GhGPAT12/25 has been confirmed to regulate tapetum degradation, anther cuticle formation, and pollen wall development [73]. Further investigation showed that the expression of GhGPAT12/25 might be activated by GhMYB80s in the regulation of male fertility. The virus-induced gene silencing of GhMYB80s in cotton exhibited a dramatic reduction in male fertility [73]. These findings provide insights into the regulatory mechanism of cotton anther development. The use of male sterility plays an important role in improving cotton yield and fiber quality. A complete male sterile line (ms5ms6) is widely used in the development of hybrid cotton globally. Duplicate mutations of GhCYP450 genes, which encode the cytochrome P450 protein, are responsible for causing male sterility in the cotton ms5ms6 accession [74]. The suppression of GhCYP450 in cotton via VIGS resulted in shorter filaments and no mature pollen grains.
8. VIGS for Studying Fiber Development in Cotton
The process of cotton fiber development takes a long time; VIGS could also function in this process. The silencing of the KATANIN gene through VIGS exhibited shorter cotton fibers and an increased weight ratio of seed oil to endosperm [75]. On the contrary, the suppression of WRINKLED1 expression led to an increase in fiber length but a decrease in the content of oilseed, indicating the potential of increasing fiber length by redistributing carbon flow. These findings provide evidence that the TRV-VIGS system can be used for rapid functional analysis of genes associated with cotton fiber development. A membrane lipid analysis was employed in wild-type cotton and fuzzless–lintless mutant fiber cells and ovules, and the content of phosphatidylinositol (PI) was found to be significantly higher in fiber cells. The genes encoding fatty acid desaturases (Δ15GhFAD), PI synthase (PIS), and PI kinase (PIK) were expressed in a fiber-preferential manner [76]. The silencing of GhD15FAD, GhPIS, or GhPIK via VIGS resulted in shorter cotton fibers. This study provides a foundation for a deeper analysis of the roles of PI and PIP in regulating cotton fiber development. The MYBMIXTA-like (MML) transcription factors GhMML3_A12 and GhMML4_D12 were identified using the map-based cloning strategy; it is interesting that GhMML3_A12 is arranged in tandem with GhMML4_D12 [77,78]. The virus-induced gene silencing of these two genes can inhibit cotton fiber initiation, implying that they play crucial roles in fiber development in cotton. Phytohormones play vital roles in plant growth and development. However, the function and regulatory mechanisms of phytohormones in controlling cotton fiber secondary cell wall formation are largely unknown. The silencing of the auxin response factors GhARF7-1 and GhARF7-2 through VIGS in cotton inhibited fiber elongation, and the length of mature fibers in silenced cotton plants was shorter than in controls; additionally, mature fiber cell wall thickness was apparently less than that of controls [79].

Table 1.
Descriptions of functional genes proved through VIGS in cotton.
Table 1.
Descriptions of functional genes proved through VIGS in cotton.
| Gene | Category | Function | Reference |
|---|---|---|---|
| GhMAP3K15 | Response to drought stress | Positive regulator of drought stress | [44] |
| GhMKK4 | Positive regulator of drought stress | [44] | |
| GhMPK6 | Positive regulator of drought stress | [44] | |
| GhWRKY59 | Positive regulator of drought stress | [44] | |
| GhDREB2 | Positive regulator of drought stress | [44] | |
| GhVHA-A | Positive regulator of drought stress | [45] | |
| GhTULP30 | Positive regulator of drought stress | [46] | |
| GhECA1 | Positive regulator of drought stress | [47] | |
| GhCNGC4 | Positive regulator of drought stress | [47] | |
| GhLOX12 | Response to salt stress | Positive regulator of salt stress | [49] |
| GhLOX13 | Positive regulator of salt stress | [49] | |
| GhSOS1 | Positive regulator of salt stress | [50] | |
| GhDi19-3 | Negative regulator of salt stress | [51] | |
| GhDi19-4 | Negative regulator of salt stress | [51] | |
| GhBGLU24-A | Negative regulator of salt stress | [53] | |
| GhRUBL | Negative regulator of salt stress | [53] | |
| GbPATP | Response to temperature stress | Positive regulator of low-temperature stress | [54] |
| GhSAD1 | Positive regulator of low-temperature stress | [55] | |
| CAN1 | Negative regulator of low-temperature stress | [56] | |
| SnRK2.8 | Positive regulator of low-temperature stress | [56] | |
| GhMAP3K65 | Negative regulator of high temperature stress | [57] | |
| GhPMEI3 | Response to verticillium stress | Positive regulator of verticillium stress | [59] |
| GhGST | Positive regulator of verticillium stress | [60] | |
| GbCYP86A1-1 | Positive regulator of verticillium stress | [61] | |
| GhVLN2 | Negative regulator of verticillium stress | [62] | |
| GhlncNAT-ANX2 | Positive regulator of verticillium stress | [63] | |
| GhlncNAT-RLP7 | Positive regulator of verticillium stress | [63] | |
| GhMPK3 | Response to whitefly stress | Positive regulator of whitefly stress | [64] |
| GhGOLS2 | Seed germination | Positive regulator of seed germination | [65] |
| GhTRL1-A05 | Root development | Positive regulator of root length and surface area | [66] |
| GhPIN8-D04 | Positive regulator of root length and surface area | [66] | |
| DELLA | Stem development | Negative regulator of stem secondary cell wall formation | [67] |
| NAC | Positive regulator of stem secondary cell wall formation | [67] | |
| GoSTR | Negative regulator of stem trichome formation | [68] | |
| GhOKRA | Leave development | Regulator of leaf shape | [69,70] |
| GhLYI | Positive regulator of leaf senescence | [71] | |
| GhSAG20 | Positive regulator of leaf senescence | [71] | |
| GhACNAT | Flowering | Positive regulator of fertility | [72] |
| GhMYB80s | Positive regulator of male fertility | [73] | |
| GhCYP450 | Positive regulator of male fertility | [74] | |
| KATANIN | Fiber development | Positive regulator of fiber length | [75] |
| WRINKLED1 | Negative regulator of fiber length | [75] | |
| GhD15FAD | Positive regulator of fiber length | [76] | |
| GhPIS | Positive regulator of fiber length | [76] | |
| GhPIK | Positive regulator of fiber length | [76] | |
| GhMML3_A12 | Positive regulator of fiber initiation | [77] | |
| GhMML4_D12 | Positive regulator of fiber initiation | [78] | |
| GhARF7-1 | Positive regulator of fiber secondary cell wall formation | [79] | |
| GhARF7-2 | Positive regulator of fiber secondary cell wall formation | [79] |
9. Limitations of VIGS and How They Can Be Addressed in Cotton
VIGS is one of the most promising and effective tools to investigate functional genes in cotton. Its main advantage is the ability to generate rapid phenotypes without the need for stable plant transformation. VIGS is inexpensive compared with other tools, such as T-DNA, transposon insertion technology, and genome editing methods like the CRISPR–Cas system. However, the VIGS method still has several limitations.
Firstly, the gene silencing duration of VIGS is generally species-specific. Although the VIGS method system has a high silencing efficiency during the cotton seedling stage, its efficiency dramatically decreases after flowering. Therefore, in order to make this method suitable for gene function research during the flowering or fiber elongation stage, a friction inoculation method was employed to prolong the period of target gene silencing [26]. Moreover, the improved system can prolong the period of VIGS in other plants [80,81]. The CLA1 gene was used as a positive control in the improved system in cotton. First, the Agrobacterium strain carrying the CLA1 silencing vector was injected into Nicotiana tabacum, and then newly grown tobacco leaves exhibited a macular phenotype two weeks later [26]. Next, about 1–3 g of the macular phenotype tobacco leaves was collected and placed in a mortar, followed by a small amount of phosphate-buffered solution (PBS), and ground into juice. The cotton leaves were inoculated and stained with 500-mesh emery paper. After dipping a grinding rod in the juice and gently rubbing it on the cotton leaves, the leaves displayed slight wounds, but epidermal cells remained undamaged. The newly grown cotton leaves exhibited whitening after two weeks in a greenhouse [26]. A real-time fluorescence quantitative PCR analysis revealed that CLA1 expression was dramatically reduced. These findings indicate that the improved friction inoculation method in cotton can extend the silencing period of target genes.
Secondly, the differential penetrance of the phenotype in vegetative and reproductive tissues is another challenge of the VIGS method, which requires the phenotype screening of a large number of plants. To date, research has revealed that the silencing effect is usually regional, dividing the entire plant or being limited to the gene-silencing regions of plants with few successive nodes [80,82]. Therefore, an appropriate selection of positive controls is essential for applying the VIGS system in various tissues during the cotton development stage. As an internal reference gene, PGF could reflect the silencing efficiency of the target gene in real time based on changes in the number of glands in the newly grown tissue of cotton plants, including leaves, stems, buds, sepals, and bolls [26]. Moreover, the PGF gene is one of the best visible markers for tracing the entire growth period of cotton. Therefore, PGF is the perfect candidate gene for evaluating the silencing efficiency in both vegetative and reproductive tissues throughout cotton growth.
Furthermore, changes in environmental conditions, including lighting, temperature, and humidity, can influence the silencing efficiency in the process of abiotic stress treatment [83,84]. The silencing efficiency is decreased at a high temperature due to reduced virus reproduction [85]. This can be resolved by pre-validating virus proliferation and keeping plants inoculated with VIGS vectors under optimal environmental conditions until silencing after abiotic stress treatment.
10. Conclusions and Future Prospects
As is well known, the transgenic expression of target genes is the most popular and effective way to investigate functional genes. However, due to the genotypic dependence of cotton genetic transformation, it is a time-consuming and laborious task that is sometimes difficult to employ. VIGS is a feasible method for quickly verifying gene function, including abiotic and biotic stress responses, and vegetative and reproductive development in cotton (Figure 3).
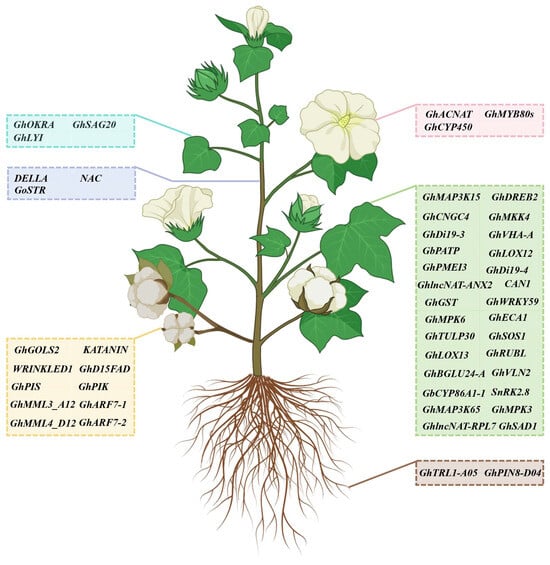
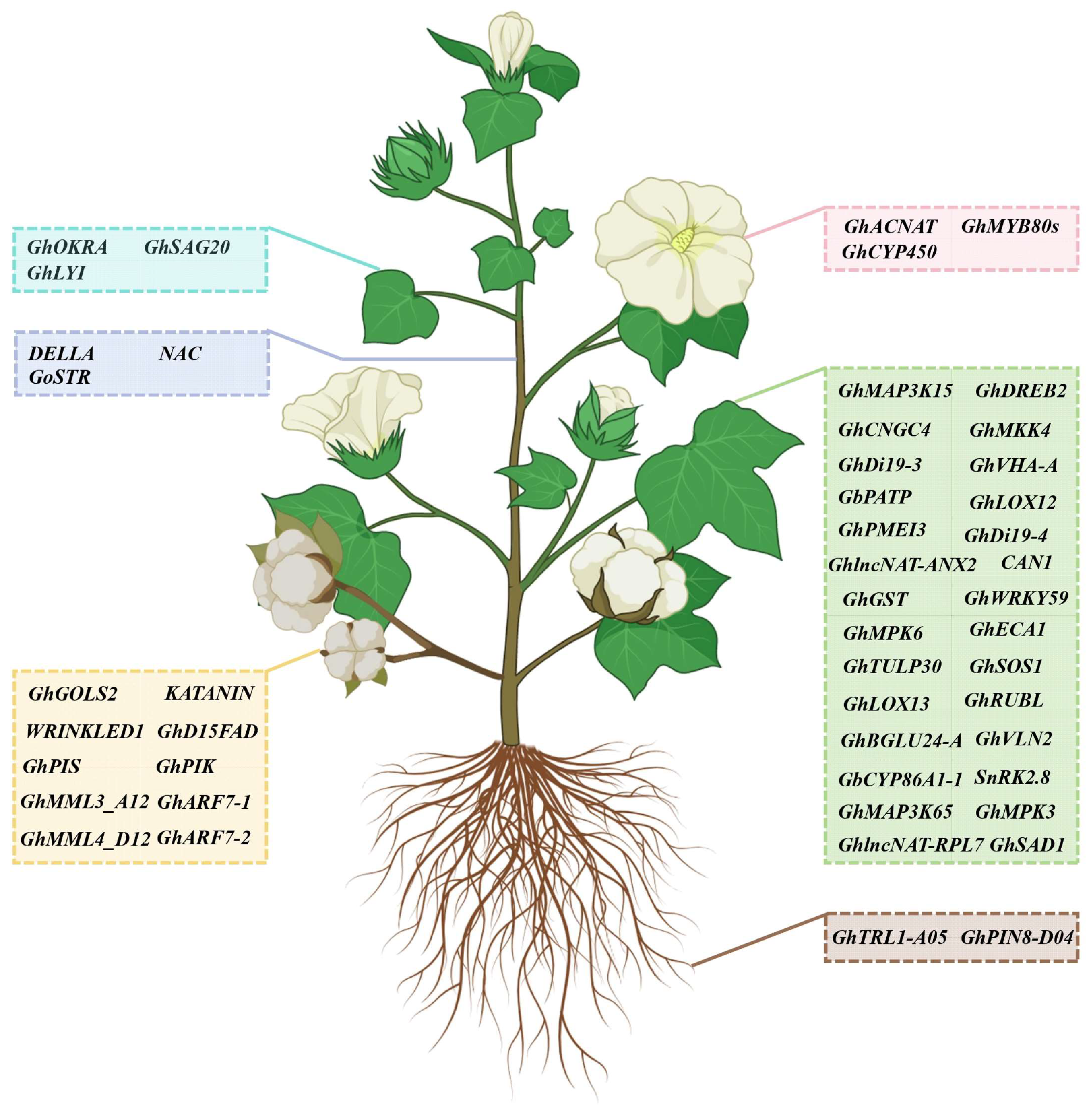
Figure 3.
A representation of functional genes validated via VIGS in cotton. The blue box represents genes that function in the regulation of leaf development; the purple box indicates genes that function in the regulation of stem development; the pink box represents genes that function in the regulation of flower development; the yellow box indicates genes that function in the regulation of fiber and seed development; the brown box represents genes that function in the regulation of root development; the green box indicates genes that respond to abiotic and biotic stresses.
Improvement in the inoculation method could expand the application of VIGS in cotton. A leaf injection is traditionally used. However, this restricts the application to the early stage of cotton development, which is important for seedling survival and crop yield. Recently, through the simplified seed imbibition of TRV, an improved VIGS system was developed to expand functional gene investigations during the germination of cotton seeds [65]. This system has been validated by suppressing the seed-germination-related gene GhGOLS2 in cotton. Moreover, the efficient duration of virus-induced silencing and applicability in several cotton varieties has been successfully estimated. This finding allowed for the establishment of a novel VIGS system to study functional genes during the cotton seed germination and early seedling stages, expanding the application scope of VIGS and promoting research on cotton functional genomics. In addition, the findings and applicability of the PGF gene demonstrate its potential as a positive control for evaluating and tracking the efficiency of VIGS in cotton [26,40]. The silencing of PGF in cotton has no effect on normal growth and development. Moreover, a change in the gland number is highly associated with the expression level of the PGF gene. Therefore, it is a perfect indicator to trace the silencing efficiency through the entire growth period in cotton. Additionally, the method of friction inoculation has been developed and employed to prolong the silencing efficiency of VIGS in cotton [26], aiding in the study of functional genes during both the vegetative stage and reproductive stage. The improved VIGS method has become a powerful tool for the rapid analysis and investigation of unknown gene functions in cotton.
Collectively, these studies serve as crucial references in the improvement and application of VIGS in cotton. However, due to the transient effect and non-inheritable nature, the VIGS method cannot be used for breeding purposes. Recently, it has been proposed that high-pressure double-stranded RNA (dsRNA) spraying directed at the nucleus can produce stable RNA-directed DNA methylation (RdDM) [86,87]. These new findings provide enormous potential for applications of VIGS-based technology in cotton.
Author Contributions
Conceptualization, Z.Z. and K.Z.; writing–original draft preparation: Z.Z., K.Z., Y.F., Y.Y., X.C. and M.H.; writing–review and editing: X.C. and Y.T.; supervision: Y.T.; project management: Y.T.; funding acquisition: Y.T. All authors have read and agreed to the published version of the manuscript.
Funding
The work was financially supported by the National Natural Science Foundation of China (32200284) and the Natural Science Foundation of Jiangsu Province (BK20210877).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhai, Z.Y.; Zhang, K.X.; Fang, Y.; Yang, Y.J.; Cao, X.; Liu, L.; Tian, Y. Systematically and comprehensively understanding the regulation of cotton fiber initiation: A review. Plants 2023, 12, 3771. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Flagel, L.E.; Wendel, J.F.; Udall, J.A. Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Genom. 2012, 13, 302. [Google Scholar] [CrossRef]
- Wen, X.; Chen, Z.; Yang, Z.; Wang, M.; Jin, S.; Wang, G.; Zhang, L.; Wang, L.; Li, J.; Saeed, S.; et al. A comprehensive overview of cotton genomics, biotechnology and molecular biological studies. Sci. China Life Sci. 2023, 66, 2214–2256. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Jones, D.C.; Liu, F.; Zhang, B. From sequencing to genome editing for cotton improvement. Trends Biotechnol. 2021, 39, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Dommes, A.B.; Gross, T.; Herbert, D.B.; Kivivirta, K.I.; Becker, A. Virus-induced gene silencing: Empowering genetics in non-model organisms. J. Exp. Bot. 2019, 70, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Hao, M.; Tian, B.; Cao, G.; Wei, F.; Xie, Z. A methodological advance of tobacco rattle virus-induced gene silencing for functional genomics in plants. Front. Plant Sci. 2021, 12, 671091. [Google Scholar] [CrossRef] [PubMed]
- Kammen, A.V. Virus-induced gene silencing in infected and transgenic plants. Trends Plant Sci. 1997, 2, 409–411. [Google Scholar] [CrossRef]
- 9 Jagram, N.; Dasgupta, I. Principles and practice of virus induced gene silencing for functional genomics in plants. Virus Genes 2023, 59, 173–187. [Google Scholar] [CrossRef]
- Choi, I.; Jeon, Y.; Pai, H.S. Brix protein APPAN plays a role in ribosomal RNA processing in Arabidopsis. Plant Sci. 2023, 333, 111721. [Google Scholar] [CrossRef]
- Chen, L.J.; Zou, W.S.; Wu, G.; Lin, H.H.; Xi, D.H. Tobacco alpha-expansion EXPA4 plays a role in Nicotiana benthamiana defence against Tobacco mosaic virus. Planta 2018, 247, 355–368. [Google Scholar] [CrossRef]
- Zhang, J.Q.; Shi, X.T.; Liu, W.D. Targeting wheat fusarium head blight with mycovirus-mediated VIGS. Trends Microbiol. 2023, 31, 1197–1198. [Google Scholar] [CrossRef] [PubMed]
- Serwatowska, J.; Lund, O.S.; Johansen, I.E. Transient posttranscriptional gene silencing in Medicago truncatula: Virus-induced gene silencing (VIGS). Methods Mol. Biol. 2018, 1822, 115–122. [Google Scholar] [PubMed]
- Xu, A.; Wei, L.; Ke, J.J.; Peng, C.F.; Li, P.Y.; Fan, C.Q.; Yu, X.; Li, B. ETI signaling nodes are involved in resistance of Hawaii 7996 to Ralstonia solanacearum-induced bacterial wilt disease in tomato. Plant Signal. Behav. 2023, 18, 2194747. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Sun, J.; Yao, J.; Wang, S.; Ding, M.; Zhang, H.; Qian, Z.; Zhao, N.; Sa, G.; Zhao, R.; et al. High rates of virus-induced gene silencing by tobacco rattle virus in Populus. Tree Physiol. 2015, 35, 1016–1029. [Google Scholar] [CrossRef]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A.; Ictv Report, C. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef]
- Aimone, C.D.; De Leon, L.; Dallas, M.M.; Ndunguru, J.; Ascencio-Ibanez, J.T.; Hanley-Bowdoin, L. A new type of satellite associated with cassava mosaic begomoviruses. J. Virol. 2021, 95, e0043221. [Google Scholar] [CrossRef]
- Lei, J.F.; Li, Y.; Dai, P.H.; Liu, C.; Zhao, Y.; You, Y.Z.; Qu, Y.Y.; Chen, Q.J.; Liu, X.D. Efficient virus-mediated genome editing in cotton using the CRISPR/Cas9 system. Front. Plant Sci. 2022, 13, 1032799. [Google Scholar] [CrossRef]
- Kumagai, M.H.; Donson, J.; Cioppa, G.D.; Harvey, D.; Hanley, H.; Grill, L.K. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar] [CrossRef]
- Singh, A.K.; Ghosh, D.; Chakraborty, S. Optimization of Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) in tomato. Methods Mol. Biol. 2022, 2408, 133–145. [Google Scholar]
- Jarugula, S.; Willie, K.; Stewart, L.R. Barley stripe mosaic virus (BSMV) as a virus-induced gene silencing vector in maize seedlings. Virus Genes 2018, 54, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Meziadi, C.; Lintz, J.; Naderpour, M.; Gautier, C.; Blanchet, S.; Noly, A.; Gratias-Weill, A.; Geffroy, V.; Pflieger, S. R-BPMV-mediated resistance to Bean pod mottle virus in Phaseolus vulgaris L. is heat-stable but elevated temperatures boost viral infection in susceptible genotypes. Viruses 2021, 13, 1239. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Gibbs, A.J.; Hajizadeh, M.; Perez, A.; Adams, I.P.; Fribourg, C.E.; Kreuze, J.; Fox, A.; Boonham, N.; Jones, R.A.C. The phylogeography of Potato virus X shows the fingerprints of its human vector. Viruses 2021, 13, 644. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P. Satellite RNAs and satellite viruses. Mol. Plant Microbe Interact. 2016, 29, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Huang, C.; Li, F.; Zhou, X. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 2014, 12, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Si, Z.; Wu, H.; Tian, Y.; Zhang, Z.; Zhang, T.; Hu, Y. Visible gland constantly traces virus-induced gene silencing in cotton. Front. Plant Sci. 2022, 13, 1020841. [Google Scholar] [CrossRef]
- Idris, A.M.; Brown, J.K. Cotton leaf crumple virus is a distinct western hemisphere begomovirus species with complex evolutionary relationships indicative of recombination and reassortment. Phytopathology 2004, 94, 1068–1074. [Google Scholar] [CrossRef]
- Tuttle, J.R.; Idris, A.M.; Brown, J.K.; Haigler, C.H.; Robertson, D. Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 2008, 148, 41–50. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Briddon, R.W.; Brown, J.K.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X. Geminivirus strain demarcation and nomenclature. Arch. Virol. 2008, 153, 783–821. [Google Scholar] [CrossRef]
- Fondong, V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013, 14, 635–649. [Google Scholar] [CrossRef]
- Cheng, G.; Shu, X.; Wang, Z.; Wang, N.; Zhang, F. Establishing a virus-induced gene silencing system in Lycoris chinensis. Plants 2023, 12, 2458. [Google Scholar] [CrossRef] [PubMed]
- Bennypaul, H.S.; Mutti, J.S.; Rustgi, S.; Kumar, N.; Okubara, P.A.; Gill, K.S. Virus-induced gene silencing (VIGS) of genes expressed in root, leaf, and meiotic tissues of wheat. Funct. Integr. Genom. 2012, 12, 143–156. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, Y.; Zhang, L.; Wang, B.; Zhao, Y.; Irfan, M.; Chen, L.; Feng, Y. Regulation of MYB transcription factors of anthocyanin synthesis in lily flowers. Front. Plant Sci. 2021, 12, 761668. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Mo, Q.; Li, J.; Zi, Z.; Xu, M.; Yue, S.; Zhao, H.; Zhu, H.; Wang, G. Establishment of virus-induced gene silencing (VIGS) system in Luffa acutangula using phytoene desaturase (PDS) and tendril synthesis related gene (TEN). Plant Methods 2023, 19, 94. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, F.; Zhao, J.; Xie, K.; Hong, Y.; Liu, Y. Virus-based microRNA expression for gene functional analysis in plants. Plant Physiol. 2010, 153, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Pei, H.; Zhang, S.; Chen, J.; Chen, W.; Yang, R.; Meng, Y.; You, J.; Gao, J.; Ma, N. TRV-GFP: A modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J. Exp. Bot. 2014, 65, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Zhu, Y.; Li, Q.; Liu, J.; Tian, Y.; Liu, Y.; Wu, J. Development of Agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS ONE 2013, 8, e73211. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, R.; Shafiq, M.; Mansoor, S.; Briddon, R.W.; Scheffler, B.E.; Scheffler, J.; Amin, I. Virus-induced gene silencing in cultivated cotton (Gossypium spp.) using Tobacco rattle virus. Mol. Biotechnol. 2015, 58, 65–72. [Google Scholar] [CrossRef]
- Khan, A.H.; Akram, A.; Saeed, M.; ur Rahman, M.; ur Rehman, A.; Mansoor, S.; Amin, I. Establishment of transcriptional gene silencing targeting the promoter regions of GFP, PDS, and PSY genes in cotton using virus-induced gene silencing. Mol. Biotechnol. 2022, 65, 1052–1061. [Google Scholar] [CrossRef]
- Ma, D.; Hu, Y.; Yang, C.; Liu, B.; Fang, L.; Wan, Q.; Liang, W.; Mei, G.; Wang, L.; Wang, H.; et al. Genetic basis for glandular trichome formation in cotton. Nat. Commun. 2016, 7, 10456. [Google Scholar] [CrossRef]
- Zulfiqar, S.; Farooq, M.A.; Zhao, T.; Wang, P.; Tabusam, J.; Wang, Y.; Xuan, S.; Zhao, J.; Chen, X.; Shen, S.; et al. Virus-induced gene silencing (VIGS): A powerful tool for crop improvement and its advancement towards epigenetics. Int. J. Mol. Sci. 2023, 24, 5608. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Saud, S.; Wang, L. Mechanism of cotton resistance to abiotic stress, and recent research advances in the osmoregulation related genes. Front. Plant Sci. 2022, 13, 972635. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, M.; Wang, P.; Cox, K.L.; Duan, L.; Dever, J.K.; Shan, L.; Li, Z.; He, P. Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol. 2017, 215, 1462–1475. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Ni, Z.; Zhang, H.; Chen, Q.; Gao, W.; Cai, Y.; Li, M.; Sun, G.; Qu, Y.Y. The gene encoding subunit A of the vacuolar H+-ATPase from cotton plays an important role in conferring tolerance to water deficit. Front. Plant Sci. 2018, 9, 758. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, J.; Kuang, M.; Zhang, C.; Ma, Q.; Huang, L.; Wang, H.; Fan, S.; Peng, J. Improvement of plant tolerance to drought stress by cotton tubby-like protein 30 through stomatal movement regulation. J. Adv. Res. 2022, 42, 55–67. [Google Scholar] [CrossRef]
- Li, B.; Zhang, M.; Sun, W.; Yue, D.; Ma, Y.; Zhang, B.; Duan, L.; Wang, M.; Lindsey, K.; Nie, X.; et al. N6-methyladenosine RNA modification regulates cotton drought response in a Ca2+ and ABA-dependent manner. Plant Biotechnol. J. 2023, 21, 1270–1285. [Google Scholar] [CrossRef]
- Maryum, Z.; Luqman, T.; Nadeem, S.; Khan, S.; Wang, B.; Ditta, A.; Khan, M.K.R. An overview of salinity stress, mechanism of salinity tolerance and strategies for its management in cotton. Front. Plant Sci. 2022, 13, 907937. [Google Scholar] [CrossRef]
- Shaban, M.; Ahmed, M.M.; Sun, H.; Ullah, A.; Zhu, L. Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genom. 2018, 19, 599. [Google Scholar] [CrossRef]
- Che, B.; Cheng, C.; Fang, J.; Liu, Y.; Jiang, L.; Yu, B. The recretohalophyte tamarix TrSOS1 gene confers enhanced salt tolerance to transgenic hairy root composite cotton seedlings exhibiting virus-induced gene silencing of GhSOS1. Int. J. Mol. Sci. 2019, 20, 2930. [Google Scholar] [CrossRef]
- Zhao, L.; Li, Y.; Li, Y.; Chen, W.; Yao, J.; Fang, S.; Lv, Y.; Zhang, Y.; Zhu, S. Systematical characterization of the sotton Di19 gene family and the role of GhDi19-3 and GhDi19-4 as two negative regulators in response to salt stress. Antioxidants 2022, 11, 2225. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ma, L.; Jiang, W.; Yao, Y.; Tang, Y.; Pang, Y. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula. Plant Physiol. Biochem. 2021, 158, 21–33. [Google Scholar] [CrossRef]
- Cui, C.; Wan, H.; Li, Z.; Ai, N.; Zhou, B. Long noncoding RNA TRABA suppresses beta-glucosidase-encoding BGLU24 to promote salt tolerance in cotton. Plant Physiol. 2023, 6, kiad530. [Google Scholar] [CrossRef]
- Liu, T.; Guo, S.; Lian, Z.; Chen, F.; Yang, Y.; Chen, T.; Ling, X.; Liu, A.; Wang, R.; Zhang, B. A P4-ATPase gene GbPATP of cotton confers chilling tolerance in plants. Plant Cell Physiol. 2015, 56, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Wang, L.; Yang, Y.; Liu, R.; Liu, S.; Chen, J.; Shen, Q.; Ma, H.; Li, Y.; Zhang, S.; et al. Genome-wide association study identifies variants of GhSAD1 conferring cold tolerance in cotton. J. Exp. Bot. 2022, 73, 2222–2237. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Long, X.; Gao, M.; Zhao, Y.; Guan, X. Global identification of natural antisense transcripts in Gossypium hirsutum and Gossypium barbadense under chilling stress. iScience 2023, 26, 107362. [Google Scholar] [CrossRef]
- Zhai, N.; Jia, H.; Liu, D.; Liu, S.; Ma, M.; Guo, X.; Li, H. GhMAP3K65, a cotton Raf-like MAP3K gene, enhances susceptibility to pathogen infection and heat stress by negatively modulating growth and development in transgenic Nicotiana benthamiana. Int. J. Mol. Sci. 2017, 18, 2462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.F.; Ding, Z.G.; Ma, Q.; Zhang, G.R.; Zhang, S.L.; Li, Z.K.; Wu, L.Q.; Zhang, G.Y.; Ma, Z.Y. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genom. 2013, 14, 637. [Google Scholar] [CrossRef]
- Liu, N.; Sun, Y.; Pei, Y.; Zhang, X.; Wang, P.; Li, X.; Li, F.; Hou, Y. A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 2018, 176, 2202–2220. [Google Scholar] [CrossRef]
- Li, Z.K.; Chen, B.; Li, X.X.; Wang, J.P.; Zhang, Y.; Wang, X.F.; Yan, Y.Y.; Ke, H.F.; Yang, J.; Wu, J.H.; et al. A newly identified cluster of glutathione S-transferase genes provides Verticillium wilt resistance in cotton. Plant J. 2019, 98, 213–227. [Google Scholar] [CrossRef]
- Wang, G.; Xu, J.; Li, L.; Guo, Z.; Si, Q.; Zhu, G.; Wang, X.; Guo, W. GbCYP86A1-1 from Gossypium barbadense positively regulates defence against Verticillium dahliae by cell wall modification and activation of immune pathways. Plant Biotechnol. J. 2020, 18, 222–238. [Google Scholar] [CrossRef] [PubMed]
- Li, W.B.; Song, S.W.; Zhong, M.M.; Liu, L.G.; Su, L.; Han, L.B.; Xia, G.X.; Sun, Y.D.; Wang, H.Y. VILLIN2 regulates cotton defense against Verticillium dahliae by modulating actin cytoskeleton remodeling. Plant Physiol. 2023, 192, 666–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, M.; Li, N.; Wang, H.; Qiu, P.; Pei, L.; Xu, Z.; Wang, T.; Gao, E.; Liu, J.; et al. Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol. J. 2018, 16, 1172–1185. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, L.; Hull, J.J.; Daniell, H.; Jin, S.; Zhang, X. Transcriptome analysis reveals a comprehensive insect resistance response mechanism in cotton to infestation by the phloem feeding insect Bemisia tabaci (whitefly). Plant Biotechnol. J. 2016, 14, 1956–1975. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Hao, M.; Tian, B.; Cao, G.; Chen, W.; Zhang, Q.; Zhang, Y.; Ling, H.; Li, J.; Xie, Z.; et al. A newly established virus-induced gene silencing method via seed imbibition for functional genomics at early germination stages in cotton. Ind. Crops Prod. 2021, 172, 114040. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, S.; Ge, C.; Shen, Q.; Zhang, S.; Ma, H.; Liu, R.; Zhao, X.; Liu, R.; Li, P.; et al. Genome-wide association study reveals that GhTRL1 and GhPIN8 affect cotton root development. Theor. Appl. Genet. 2022, 135, 3161–3176. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, W.; Ran, L.; Chen, Z.; Wang, C.; Dou, Y.; Qin, Y.; Suo, Q.; Li, Y.; Zeng, J.; et al. DELLA-NAC interactions mediate GA signaling to promote secondary cell wall formation in cotton stem. Front. Plant Sci. 2021, 12, 655127. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Zhang, J.; Shi, Y.; Dai, F.; Jiang, T.; Xuan, L.; He, Y.; Zhang, Z.; Deng, J.; Zhang, T.; et al. GoSTR, a negative modulator of stem trichome formation in cotton. Plant J. 2023, 116, 389–403. [Google Scholar] [CrossRef]
- Chang, L.; Fang, L.; Zhu, Y.; Wu, H.; Zhang, Z.; Liu, C.; Li, X.; Zhang, T. Insights into interspecific hybridization events in allotetraploid cotton formation from characterization of a gene-regulating leaf shape. Genetics 2016, 204, 799–806. [Google Scholar] [CrossRef]
- Andres, R.J.; Coneva, V.; Frank, M.H.; Tuttle, J.R.; Samayoa, L.F.; Han, S.W.; Kaur, B.; Zhu, L.; Fang, H.; Bowman, D.T.; et al. Modifications to a LATE MERISTEM IDENTITY1 gene are responsible for the major leaf shapes of upland cotton (Gossypium hirsutum L.). Proc. Natl. Acad. Sci. USA 2016, 114, E57–E66. [Google Scholar] [CrossRef]
- Zhang, Y.; Zang, Y.; Chen, J.; Feng, S.; Zhang, Z.; Hu, Y.; Zhang, T. A truncated ETHYLENE INSENSITIVE3-like protein, GhLYI, regulates senescence in cotton. Plant Physiol. 2023, 193, 1177–1196. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Shen, Y.; Hao, J.; Wu, J.; Ke, L.; Wu, C.; Huang, K.; Luo, B.; Xu, M.; Cheng, X.; et al. Acyl-CoA N-acyltransferase influences fertility by regulating lipid metabolism and jasmonic acid biogenesis in cotton. Sci. Rep. 2015, 5, 11790. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wei, H.; Hao, P.; Wu, A.; Ma, Q.; Zhang, J.; Wang, H.; Fu, X.; Ma, L.; Lu, J.; et al. GhGPAT12/25 are essential for the formation of anther cuticle and pollen exine in cotton (Gossypium hirsutum L.). Front. Plant Sci. 2021, 12, 667739. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Dai, F.; Si, Z.; Fang, L.; Zhang, T. Duplicate mutations of GhCYP450 lead to the production of ms5m6 male sterile line in cotton. Theor. Appl. Genet. 2023, 136, 2. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Ye, J.; Geng, Y.F.; Sun, Y.W.; Gao, S.Q.; Zhang, B.P.; Chen, W.; Chua, N.H. Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol. 2012, 160, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.J.; Xiao, G.H.; Liu, N.J.; Liu, D.; Chen, P.S.; Qin, Y.M.; Zhu, Y.X. Targeted lipidomics studies reveal that linolenic acid promotes cotton fiber elongation by activating phosphatidylinositol and phosphatidylinositol monophosphate biosynthesis. Mol. Plant 2015, 8, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Guan, X.; Yang, N.; Wu, H.; Pan, M.; Liu, B.; Fang, L.; Yang, S.; Hu, Y.; Ye, W.; et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol. 2016, 210, 1298–1310. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Tian, Y.; Wan, Q.; Fang, L.; Guan, X.; Chen, J.; Hu, Y.; Ye, W.; Zhang, H.; Guo, W.; et al. Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytol. 2017, 217, 883–895. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; He, S.P.; Xu, S.W.; Li, L.; Zheng, Y.; Li, X.B. The transcription factor ERF108 interacts with auxin response factors to mediate cotton fiber secondary cell wall biosynthesis. Plant Cell 2023, 35, 4133–4154. [Google Scholar] [CrossRef]
- Becker, A.; Lange, M. VIGS—Genomics goes functional. Trends Plant Sci. 2010, 15, 1–4. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K.S. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011, 16, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Wege, S.; Scholz, A.; Gleissberg, S.; Becker, A. Highly efficient virus-induced gene silencing (VIGS) in california poppy (Eschscholzia californica): An evaluation of VIGS as a strategy to obtain functional data from non-model plants. Ann. Bot. 2007, 100, 641–649. [Google Scholar] [CrossRef]
- Fu, D.Q.; Zhu, B.Z.; Zhu, H.L.; Zhang, H.X.; Xie, Y.H.; Jiang, W.B.; Zhao, X.D.; Luo, Y.B. Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Mol. Cells 2006, 21, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Kotakis, C.; Vrettos, N.; Kotsis, D.; Tsagris, M.; Kotzabasis, K.; Kalantidis, K. Light intensity affects RNA silencing of a transgene in Nicotiana benthamiana plants. BMC Plant Biol. 2010, 10, 220. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, P.; Vanitharani, R.; Ogbe, F.; Fauquet, C.M. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 2005, 138, 1828–1841. [Google Scholar] [CrossRef]
- Dalakouras, A.; Papadopoulou, K.K. Epigenetic Modifications: An unexplored facet of exogenous RNA application in plants. Plants 2020, 9, 673. [Google Scholar] [CrossRef]
- Dalakouras, A.; Vlachostergios, D.; Manavella, P. Epigenetic approaches to crop breeding: Current status and perspectives. J. Exp. Bot. 2021, 72, 5356–5371. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).