Zinc Finger Protein8 (GhZFP8) Regulates the Initiation of Trichomes in Arabidopsis and the Development of Fiber in Cotton
Abstract
1. Introduction
2. Results
2.1. The Expression Pattern of GhZFP8 in Different Organs
2.2. Correlation of GhZFP8 to the Parameters of Cotton Fiber
2.3. Subcellular Localization of GhZFP8 in Tobacco Leaves
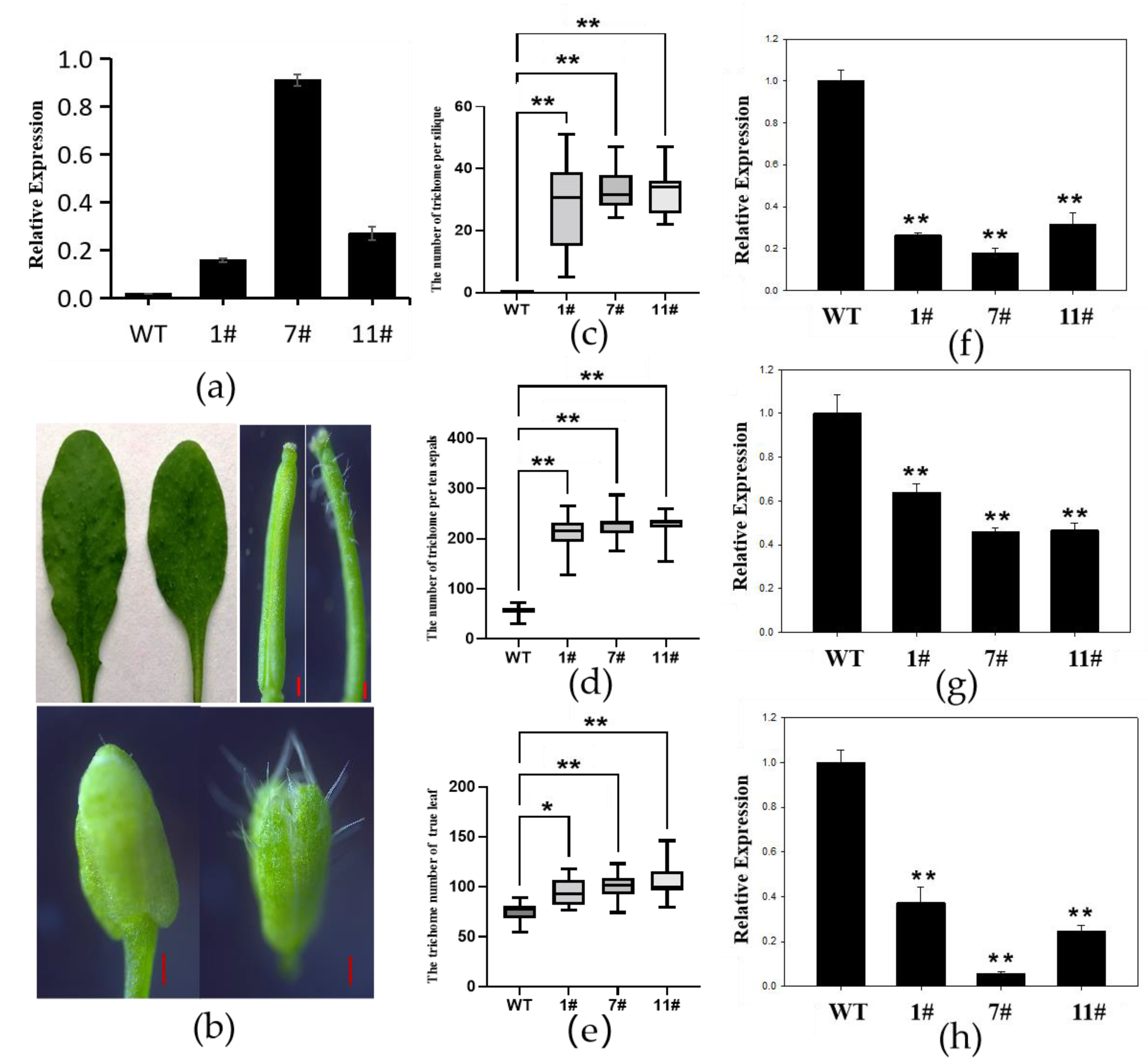
2.4. Overexpression of GhZFP8 Enhanced Formation of Trichomes
2.5. Overexpression of GhZFP8 Inhibited the Growth of Overexpressor Arabidopsis
2.6. Interference of GhZFP8 Regulated the Growth of Cotton
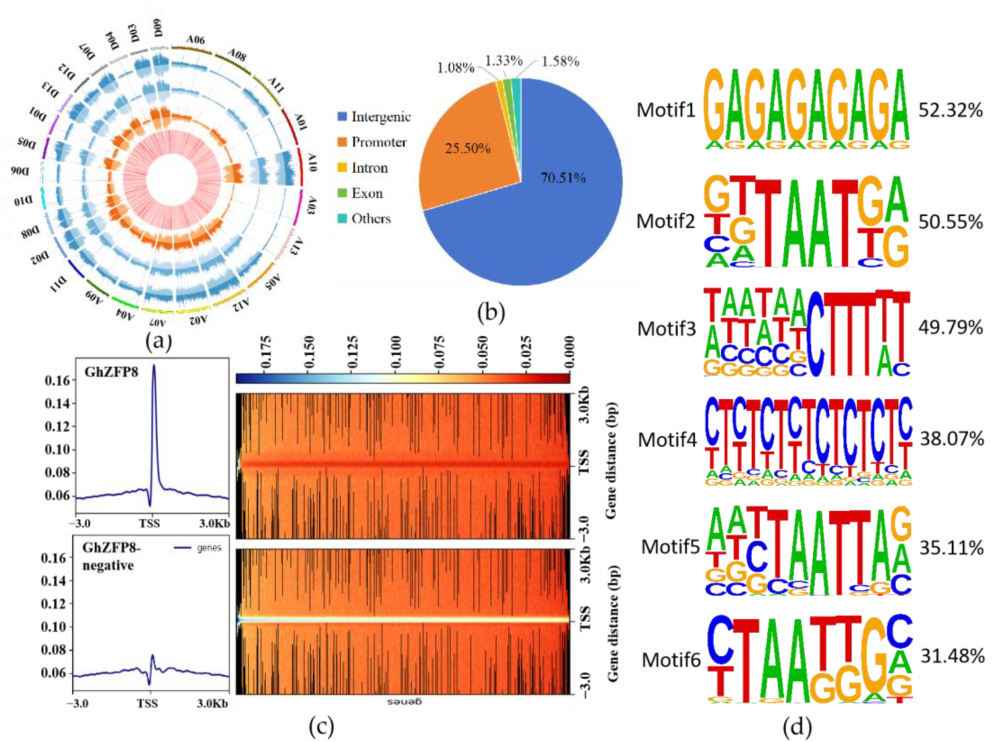
2.7. Identiffcation of GhZFP8-Binding Region by DAP-seq
2.8. GhZFP8 Binds to Genes Associated with Cotton Fiber Development
3. Discussion
3.1. GhZFP8 Might Function as a Suppressor in Arabiodosis
3.2. GhZFP8 Positively Regulates the Elongation of Cotton Fibers
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Construction of RNAi Vector and Generation of RNAi Transgenic Cotton
4.3. RNA Extraction and Quantitative PCR
4.4. Analysis of the Correlation between GhZFP8 and the Development of Cotton Fiber
4.5. Subcellular Localization of GhZFP8
4.6. Measurement of Phytohormone in Arabidopsis
4.7. DNA Afffnity Puriffcation Sequencing Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larkin, J.C.; Brown, M.L.; Schiefelbein, J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 2003, 54, 403–430. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Morohashi, K.; Hatlestad, G.; Grotewold, E.; Lloyd, A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 2008, 135, 1991–1999. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ye, Z. Trichomes as models for studying plant cell differentiation. Cell. Mol. Life Sci. 2013, 70, 1937–1948. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.Y.; Zhou, H.Y.; Xu, L.A.; Liu, X.Y.; Fan, S.X.; Cao, J.S. The zinc-finger transcription factor BcMF20 and its orthologs in Cruciferae which are required for pollen development. Biochem. Biophys. Res. Commun. 2018, 503, 998–1003. [Google Scholar] [CrossRef]
- Prigge, M.J.; Wagner, D.R. The arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 2001, 13, 1263–1279. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Wei, X.; Dong, X.; Wang, C.; Sui, N.; Guo, J.; Yuan, F.; Gong, Z.; Li, X.; Zhang, Y.; et al. Arabidopsis ZINC FINGER PROTEIN1 Acts Downstream of GL2 to Repress Root Hair Initiation and Elongation by Directly Suppressing bHLH Genes. Plant Cell 2020, 32, 206–225. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Li, H.G.; Wang, J.J.; Su, Y.; Wang, H.L.; Feng, C.H.; Yang, Y.; Niu, M.X.; Liu, C.; Yin, W.; et al. PeSTZ1, a C2H2-type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol. J. 2019, 17, 2169–2183. [Google Scholar] [CrossRef]
- Gan, Y.; Kumimoto, R.; Liu, C.; Ratcliffe, O.; Yu, H.; Broun, P. GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 2006, 18, 1383–1395. [Google Scholar] [CrossRef]
- Gan, Y.; Liu, C.; Yu, H.; Broun, P. Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 2007, 134, 2073–2081. [Google Scholar] [CrossRef]
- Zhou, Z.; An, L.; Sun, L.; Zhu, S.; Xi, W.; Broun, P.; Yu, H.; Gan, Y. Zinc finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol. 2011, 157, 673–682. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, A.; Zhou, Z.; Zhao, Y.; Yan, A.; Bao, S.; Yu, H.; Gan, Y. GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 2015, 206, 220–230. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Lin, S.D.; Zhang, T.; Pei, L.L.; Tang, Q.; Liu, F.; Liu, Y.C. Cloning and expression analysis of GhSIZ1, encoding a C2H2 zinc finger protein in cotton (Gossypium hirsutum). Cotton Sci. 2015, 27, 189–197. [Google Scholar]
- He, P.; Yang, Y.; Wang, Z.; Zhao, P.; Yuan, Y.; Zhang, L.; Ma, Y.; Pang, C.; Yu, J.; Xiao, G. Comprehensive analyses of ZFP gene family and characterization of expression profiles during plant hormone response in cotton. BMC Plant Biol. 2019, 19, 329. [Google Scholar] [CrossRef]
- Kumar, R.; Das, J.; Raghavendra, K.P.; Nandeshwar, S.B. Identification and expression pattern analysis of two Gossypium hirsutum zinc finger transcription factors during cotton fiber initiation. Natl. Acad. Sci. Lett. 2020, 43, 115–119. [Google Scholar] [CrossRef]
- Yuan, Z.H.; Li, Y.; Zhang, B.; Cheng, W.H.; Liu, Y.C. A C2H2 zinc finger protein GhZFP8 from cotton (Gossypium Hirsutum) is involved in salt stress tolerance in Arabidopsis. Pak. J. Bot. 2024, 56, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Yu, T.; Yang, Q.; Li, C.; Xiong, C.; Gao, S.; Xie, Q.; Zheng, F.; Li, H.; Tian, Z.; et al. Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J. 2018, 96, 90–102. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, D.; Khan, A.R.; Liu, B.; Wu, M.; Huang, L.; Wu, J.; Song, G.; Ni, H.; Ying, H.; et al. NbGIS regulates glandular trichome initiation through GA signaling in tobacco. Plant Mol. Biol. 2018, 98, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Gu, Y.; Dai, T.; Wu, Y.; Wu, P.; Xu, Y.; Chen, F. Regulation of trichome development in tobacco by JcZFP8, a C2H2 zinc finger protein gene from Jatropha curcas L. Gene 2018, 658, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yang, L.; Luo, S.; Wang, X.; Wang, W.; Cheng, Y.; Tian, H.; Zheng, K.; Cai, L.; Wang, S. Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in Arabidopsis. BMC Plant Biol. 2018, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Esch, J.J.; Chen, M.A.; Hillestad, M.; Marks, M.D. Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 2004, 40, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Luo, X.; Li, M.; Joldersma, D.; Plunkert, M.; Liu, Z. Mechanism of fertilizat ion-induced auxin synthesis in the endosperm for seed and fruit development. Nat. Commun. 2022, 13, 3985. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wang, K.; Huang, B.; Mila, I.; Frasse, P.; Maza, E.; Djari, A.; Hernould, M.; Zouine, M.; Li, Z.; et al. The auxin-responsive transcription factor SlDOF9 regulates inflorescence and flower development in tomato. Nat. Plants 2022, 8, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, D.; Hu, R.; Hua, C.; Ali, I.; Zhang, A.; Liu, B.; Wu, M.; Huang, L.; Gan, Y. AtGIS, a C2H2 zinc-finger transcription factor from Arabidopsis regulates glandular trichome development through GA signaling in tobacco. Biochem. Biophys. Res. Commun. 2017, 483, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Levitin, B.; Richter, D.; Markovich, I.; Zik, M. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. Plant J. 2008, 56, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, M.; Geng, L.L.; Zhao, J. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 2010, 64, 482–497. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Garcia, G.; Vielle-Calzada, J.P. A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 2004, 16, 2614–2628. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Suzuki, Y.; Chono, M.; Knox, J.P.; Yamaguchi, I. CsAGP1, a gibberellin-responsive gene from cucumber hypocotyls, encodes a classical arabinogalactan protein and is involved in stem elongation. Plant Physiol. 2003, 131, 1450–1459. [Google Scholar] [CrossRef]
- Coimbra, S.; Costa, M.; Jones, B.; Mendes, M.A.; Pereira, L.G. Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J. Exp. Bot. 2009, 60, 3133–3142. [Google Scholar] [CrossRef]
- Yuan, Y.; Teng, Q.; Zhong, R.; Ye, Z.H. TBL3 and TBL31, Two Arabidopsis DUF231 Domain Proteins, are Required for 3-O-Monoacetylation of Xylan. Plant Cell Physiol. 2016, 57, 35–45. [Google Scholar] [CrossRef]
- Yuan, Y.; Teng, Q.; Zhong, R.; Haghighat, M.; Richardson, E.A.; Ye, Z.H. Mutations of Arabidopsis TBL32 and TBL33 Affect Xylan Acetylation and Secondary Wall Deposition. PLoS ONE 2016, 11, e0146460. [Google Scholar] [CrossRef]
- Guan, X.Y.; Li, Q.J.; Shan, C.M.; Wang, S.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol Plant. 2008, 134, 174–182. [Google Scholar] [CrossRef]
- Qin, Y.M.; Zhu, Y.X. How cotton fibers elongate: A tale of linear cell-growth mode. Curr. Opin. Plant Biol. 2011, 14, 106–111. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.W.; Yu, N.; Li, C.H.; Luo, B.; Gou, J.Y.; Wang, L.J.; Chen, X.Y. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 2004, 16, 2323–2334. [Google Scholar] [CrossRef]
- Humphries, J.A.; Walker, A.R.; Timmis, J.N.; Orford, S.J. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 2005, 57, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Haigler, C.H.; Singh, B.; Zhang, D.; Hwang, S.; Wu, C.; Cai, W.X.; Hozain, M.; Kang, W.; Kiedaisch, B.; Strauss, R.E.; et al. Transgenic cotton over-producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol. Biol. 2007, 63, 815–832. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, Y.; Hu, W.; Wang, S.; Snider, J.L.; Zhou, Z. Carbohydrate metabolism in the subtending leaf cross-acclimates to waterlogging and elevated temperature stress and influences boll biomass in cotton (Gossypium hirsutum). Physiol. Plant. 2017, 161, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Meng, Y.; Lv, F.; Chen, J.; Ma, Y.; Wang, Y.; Chen, B.; Zhang, L.; Zhou, Z. Photosynthetic characteristics of the subtending leaf of cotton boll at different fruiting branch nodes and their relationships with lint yield and fiber quality. Front. Plant Sci. 2015, 6, 747. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, Z.; Ge, X.; Lu, L.; Qin, W.; Qanmber, G.; Liu, L.; Wang, Z.; Li, F. Brassinosteroids regulate cotton fiber elongation by modulating very-long-chain fatty acid biosynthesis. Plant Cell 2023, 35, 2114–2131. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Iqbal, A.; Latif, A.; Din, S.U.; Sarwar, M.B.; Wang, X.; Rao, A.Q.; Husnain, T.; Ali Shahid, A. Overexpression of a Sucrose Synthase Gene Indirectly Improves Cotton Fiber Quality Through Sucrose Cleavage. Front. Plant Sci. 2020, 11, 476251. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zhai, Y.; Zhang, L.; Chen, Y.; Zhu, Z.; Chen, G.; Wang, K.; Zhu, Y. Molecular studies of cellulose synthase supercomplex from cotton fiber reveal its unique biochemical properties. Sci. China Life Sci. 2022, 65, 1776–1793. [Google Scholar] [CrossRef]
- Guo, K.; Tu, L.; He, Y.; Deng, J.; Wang, M.; Huang, H.; Li, Z.; Zhang, X. Interaction between calcium and potassium modulates elongation rate in cotton fiber cells. J. Exp. Bot. 2017, 68, 5161–5175. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Cheng, F.; Zhang, S.P.; Zheng, Y.; Li, Y.; Li, X.B. Comparative phosphoproteomic analysis reveals that phosphorylation of sucrose synthase GhSUS2 by Ca2+-dependent protein kinases GhCPK84/93 affects cotton fiber development. J. Exp. Bot. 2023, 74, 1836–1852. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; He, S.; Dia, S.; Sun, G.; Liu, X.; Wang, X.; Pan, Z.; Jia, Y.; Liru, W.; Baoyin, P.; et al. Alien genomic introgressions enhanced fiber strength in upland cotton (Gossypium hirsutum L.). Ind. Crop. Prod. 2021, 159, 113028. [Google Scholar] [CrossRef]
- He, S.; Sun, G.; Geng, X.; Gong, W.; Dai, P.; Jia, Y.; Shi, W.; Pan, Z.; Wang, J.; Wang, L.; et al. The genomic basis of geographic differentiation and fiber improvement in cultivated cotton. Nat. Genet. 2021, 53, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, P.; You, C.; Yu, J.; Zhang, X.; Yan, F.; Ye, Z.; Shen, C.; Li, B.; Guo, K.; et al. Combined GWAS and eQTL analysis uncovers a genetic regulatory network orchestrating the initiation of secondary cell wall development in cotton. New Phytol. 2020, 226, 1738–1752. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Ma, X.; Li, Y.; Yang, X.; Cheng, W. Zinc Finger Protein8 (GhZFP8) Regulates the Initiation of Trichomes in Arabidopsis and the Development of Fiber in Cotton. Plants 2024, 13, 492. https://doi.org/10.3390/plants13040492
Liu Y, Ma X, Li Y, Yang X, Cheng W. Zinc Finger Protein8 (GhZFP8) Regulates the Initiation of Trichomes in Arabidopsis and the Development of Fiber in Cotton. Plants. 2024; 13(4):492. https://doi.org/10.3390/plants13040492
Chicago/Turabian StyleLiu, Yongchang, Xiaomei Ma, Ying Li, Xiaoyu Yang, and Wenhan Cheng. 2024. "Zinc Finger Protein8 (GhZFP8) Regulates the Initiation of Trichomes in Arabidopsis and the Development of Fiber in Cotton" Plants 13, no. 4: 492. https://doi.org/10.3390/plants13040492
APA StyleLiu, Y., Ma, X., Li, Y., Yang, X., & Cheng, W. (2024). Zinc Finger Protein8 (GhZFP8) Regulates the Initiation of Trichomes in Arabidopsis and the Development of Fiber in Cotton. Plants, 13(4), 492. https://doi.org/10.3390/plants13040492





