The Potential Genetic Effect for Yield and Foliar Disease Resistance in Faba Bean (Vicia faba L.) Assessed via Morphological and SCoT Markers
Abstract
1. Introduction
2. Results
2.1. The Performance of Crosses
2.2. Polymorphism via SCoT Markers
2.3. Genetic Parameters
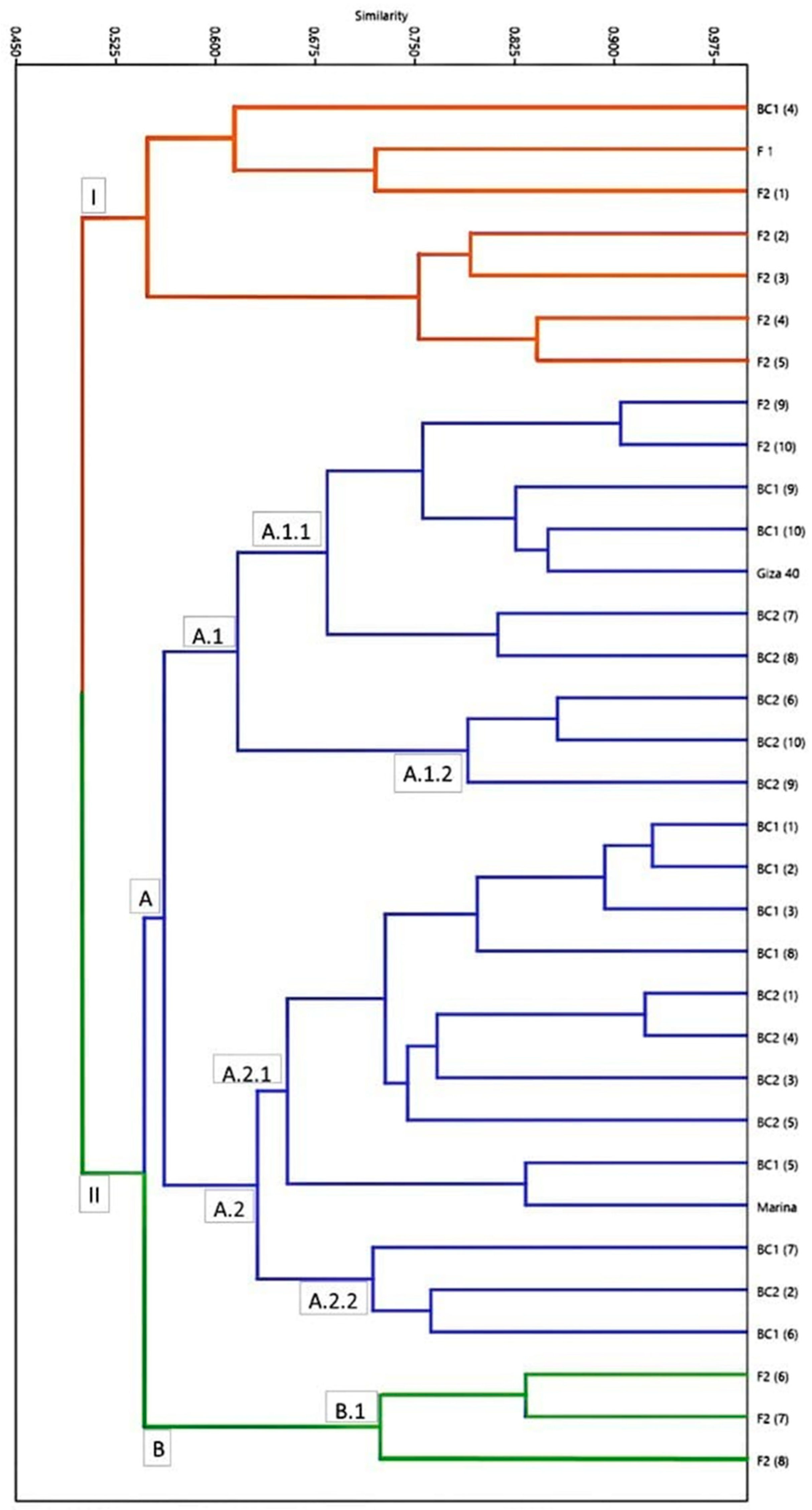
2.4. Genetic Distance and Similarity
2.5. Phylogeny Analysis of ScoT Markers
3. Discussion
4. Materials and Methods
4.1. Field Experiment
4.2. Statistical Analysis
4.3. Types of Gene Effect (Gene Action)
4.4. DNA Extraction and SCoT Amplification Marker
4.5. PCR Scoring and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rubiales, D.; Moral, A.; Flores, F. Agronomic Performance of Broomrape Resistant and Susceptible Faba Bean Accession. Agron. J. 2022, 12, 1421. [Google Scholar] [CrossRef]
- Li, L.; Tao, Y.; Rong, L.; Redden, B.; Maalouf, F.; Zong, X. Food legume production in China. Crop J. 2017, 5, 115–126. [Google Scholar] [CrossRef]
- Suso, M.; Moreno, M.; Melchinger, A. Variation in outcrossing rate and genetic structure on six cultivars of Vicia faba L. as affected by geographic location and year. Plant Breed. 1999, 118, 347–350. [Google Scholar] [CrossRef]
- Suso, M.; Pierre, J.; Moreno, M.; Esnault, R.; Le Guen, J. Variation in outcrossing levels in faba bean cultivars: Role of ecological factors. J. Agric. Sci. 2001, 136, 399–405. [Google Scholar] [CrossRef]
- Suso, M.J.; Maalouf, F. Direct and correlated responses to upward and downward selection for outcrossing in Vicia faba. Field Crops Res. 2010, 116, 116–126. [Google Scholar] [CrossRef][Green Version]
- Kaur, S.; Cogan, N.O.I.; Forster, J.W.; Paull, J.G. Assessment of Genetic Diversity in Faba Bean Based on Single Nucleotide Polymorphism. Diversity 2014, 6, 88–101. [Google Scholar] [CrossRef]
- Oliveira, H.R.; Tomás, D.; Silva, M.; Lopes, S.; Viegas, W.; Veloso, M.M. Genetic Diversity and Population Structure in Vicia faba L. Landraces and Wild Related Species Assessed by Nuclear SSRs. PLoS ONE 2016, 11, e0154801. [Google Scholar] [CrossRef] [PubMed]
- FAO. Online Statistical Database of Food and Agriculture Organization of the United Nations. FAOSTAT, 2020. Available online: http://www.fao.org/faostat/ (accessed on 1 January 2023).
- Rubiales, D.; Fondevilla, S.; Chen, W.; Gentzbittel, L.; Higgins, T.J.V.; Castillejo, M.Á.; Singh, K.B.; Rispail, N. Achievements and Challenges in Legume Breeding for Pest and Disease Resistance. Crit. Rev. Plant Sci. 2015, 34, 195–236. [Google Scholar] [CrossRef]
- Soliman, A.A.; Ibrahim, M.A.; Mostafa, S.M.; Mohamed, A.M.; Kalboush, S.F. Evaluation of fifteen faba bean (Vicia faba L.) genotypes for Orobanche crenata tolerance and foliar diseases resistance. Egypt. J. Agric. Res. 2023, 101, 997–1006. Available online: https://ejar.journals.ekb.eg/article_314289.html (accessed on 21 September 2023).
- Alowafeir, M. Fungicidal management of chocolate spot of faba bean and assessment of yield losses due to the disease. Ann. Agric. Sci. 2011, 56, 27–35. [Google Scholar] [CrossRef][Green Version]
- Beyene, A.T.; Derera, J.; Sibiya, J.; Fikre, A. Gene action determining grain yield and chocolate spot (Botrytis fabae) resistance in faba bean. Euphytica 2015, 207, 293–304. [Google Scholar] [CrossRef]
- Deneke, S. Review on Epidemiology and Management of Faba Bean (Vicia fabae) Chocolate Spot (Botrytis fabae), Root Rot (Fusarium solani) and Rust (Uromyces vicia fabae) in Ethiopia. Int. J. Sci. Res. Publ. 2018, 8, 105–111. [Google Scholar] [CrossRef]
- Tivoli, B.; Baranger, A.; Ávila, C.M.; Banniza, S.; Barbetti, M.J.; Chen, W.; Davidson, J.A.; Lindeck, K.; Kharrat, M.; Rubiales, D.; et al. Screening techniques and sources of resistance to foliar diseases caused by major necrotrophic fungi in grain legumes. Euphytica 2006, 147, 223–253. [Google Scholar] [CrossRef]
- Emeran, A.A.; Sillero, J.C.; Fernández-Aparicio, M.; Rubiales, D. Chemical control of faba bean rust (Uromyces viciae-fabae). Crop Prot. 2011, 30, 907–912. [Google Scholar] [CrossRef]
- Shifa, H.; Hussien, T.; Sakhuja, P.K. Association of Faba Bean Rust (Uromyces viciae-fabae) with Environmental Factors and Cultural Practices in the Hararghe Highlands, Eastern Ethiopia. East. Afr. J. Sci. 2011, 5, 58–65. [Google Scholar]
- Sillero, J.C.; Villegas-Fernández, Á.M.; Thomas, J.; Rojas-Molina, M.M.; Emeran, A.A.; Fernández-Aparicio, M.; Rubiales, D. Faba bean breeding for disease resistance. Field Crops Res. 2010, 115, 297–307. [Google Scholar] [CrossRef]
- Maalouf, F.; Ahmed, S.; Shaaban, K.; Bassam, B.; Nawar, F.; Singh, M.; Amri, A. New faba bean germplasm with multiple resistances to Ascochyta blight, chocolate spot and rust diseases. Euphytica 2016, 211, 157–167. [Google Scholar] [CrossRef]
- Villegas-Fernández, Á.M.; Sillero, J.C.; Emeran, A.A.; Winkler, J.; Raffiot, B.; Tay, J.U.; Flores, F.; Rubiales, D. Identification and multi-environment validation of resistance to Botrytis fabae in Vicia faba. Field Crops Res. 2009, 114, 84–90. [Google Scholar] [CrossRef]
- Stoddard, F.L.; Nicholas, A.; Rubiales, D.; Thomas, J.; Villegas-Fernández, Á.M. Integrated pest management in faba bean. Field Crops Res. 2010, 115, 308–318. [Google Scholar] [CrossRef]
- Abd El-zaher, I.N. Inheritance of Seed Yield and Some Yield Components of Faba Bean Using Six Populations. Assiut J. Agric. Sci. 2016, 47, 32–40. [Google Scholar] [CrossRef]
- Ashrei, A.A.M.; Ghareeb, Z.E.; Yamani, K.M.M.; Rehab, A.M.; Garhy, A.M.A. Statistical and Genetical Analysis of Yield and Some Related Components in Faba Bean. Egypt. J. Plant Breed. 2013, 17, 57–67. [Google Scholar] [CrossRef]
- Gamble, E.E. Gene effects in corn (Zea mays L.): I. Separation And Relative Importance of Gene Effects For Yield. Can. J. Plant Sci. 1962, 42, 339–348. [Google Scholar] [CrossRef]
- Bishnoi, S.K.; Hooda, J.S.; Yadav, I.S.; Panchta, R. Advances On Heterosis And Hybrid Breeding In Faba Bean (Vicia faba L.). Forage Res. 2012, 38, 65–73. [Google Scholar]
- Bishnoi, S.K.; Hooda, J.S.; Sharma, P.; Kumar, P. Analysis of combining ability and inheritance of breeding parameters in yield component traits in faba bean (Vicia faba L.). J. Pharmacogn. Phytochem. 2018, 7, 1085–1090. [Google Scholar]
- Abd El-Aty, M.S.; El-Hity, M.A.; Abo Sen, T.M.; Abd El-Rahaman, I.F.; Ibrahim, O.M.; Al-Farga, A.; El-Tahan, A.M. Generation Mean Analysis, Heterosis, and Genetic Diversity in Five Egyptian Faba Beans and Their Hybrids. Sustainability 2023, 15, 12313. [Google Scholar] [CrossRef]
- Abdalla, M.M.F.; Saleh, D.D.; El-Hady, M.M.; EEl-Harty, H. Investigations on faba beans, (Vicia faba L.) 16. F1 and F2 diallel hybrids with reciprocals among five parents. Egypt. J. Plant Breed. 2001, 5, 155–179. [Google Scholar]
- Poulsen, M.H. Performance of inbred populations analysis of(Vicia faba L). spp. minor. In Some Current Research on Vicia faba in Western Europe; Bond, A.D., Scarascia-Mugnozza, G.T., Poulsen, M.H., Eds.; Commission of the European Communities: Luxenbourg, 1979; pp. 342–354. [Google Scholar]
- Narayanan, P.S.S.S. Biometrical Techniques in Blant Breeding; Kalyani Publ.: New Delhi, India, 1993; pp. 74–84. [Google Scholar]
- Abid, G.; Mingeot, D.; Udupa, S.M.; Muhovski, Y.; Watillon, B.; Sassi, K.; M’hamdi, M.; Souissi, F.; Mannai, K.; Barhoumi, F.; et al. Genetic Relationship and Diversity Analysis of Faba Bean (Vicia faba L. var. Minor) Genetic Resources Using Morphological and Microsatellite Molecular Markers. Plant Mol. Biol. Report. 2015, 33, 1755–1767. [Google Scholar] [CrossRef]
- Al-Khayri, J.M.; Mahdy, E.M.B.; Taha, H.; Eldomiaty, A.S.; Abd-Elfattah, M.A.; Abdel Latef, A.A.H.; Rezk, A.A.-S.; Shehata, W.F.; Almaghasla, M.I.; Shalaby, T.A.; et al. Genetic and Morphological Diversity Assessment of Five Kalanchoe Genotypes by SCoT, ISSR and RAPD-PCR Markers. Plants 2022, 11, 1722. [Google Scholar] [CrossRef]
- Varshney, R.K.; Thudi, M.; Roorkiwal, M.; He, W.; Upadhyaya, H.D.; Yang, W.; Bajaj, P.; Cubry, P.; Rathore, A.; Jian, J.; et al. Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 2019, 51, 857–864. [Google Scholar] [CrossRef]
- Al-Hadeithi, Z.S.M.; Jasim, S.A. Study of Plant Genetic Variation through Molecular Markers: An Overview. J. Pharm. Res. Int. 2021, 33, 464–473. [Google Scholar] [CrossRef]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Mahjbi, A.; Baraket, G.; Oueslati, A.; Salhi-Hannachi, A. Start Codon Targeted (SCoT) markers provide new insights into the genetic diversity analysis and characterization of Tunisian Citrus species. Biochem. Syst. Ecol. 2015, 61, 390–398. [Google Scholar] [CrossRef]
- Pakseresht, F.; Talebi, R.; Karami, E. Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of landrace chickpea (Cicer arietinum L.) genotypes collected from north-west of Iran. Physiol. Mol. Biol. Plants 2013, 19, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Gowayed, S.M.; Abd El-Moneim, D. Detection of genetic divergence among some wheat (Triticum aestivum L.) genotypes using molecular and biochemical indicators under salinity stress. PLoS ONE 2021, 16, 248890. [Google Scholar] [CrossRef]
- Maalouf, F.; Hu, J.; O’Sullivan, D.M.; Zong, X.; Hamwieh, A.; Kumar, S.; Baum, M. Breeding and genomics status in faba bean (Vicia faba ). Plant Breed. 2018, 138, 465–473. [Google Scholar] [CrossRef]
- Yu, H.L.; Yang, F.; Hu, C.-X.; Yang, X.; Zheng, A.; Wang, Y.; Tang, Y.; He, Y.-h.; Lv, M. Production status and research advancement on root rot disease of faba bean (Vicia faba L.) in China. Front. Plant Sci. 2023, 14, 1165658. [Google Scholar] [CrossRef] [PubMed]
- Hendawy, F.A. Heterosis and inheritance of some quantitative characters in two intervarietal crosses of field beans (Vicia faba L.). Menofiya J. Agric. Res. 1994, 1757–1771. [Google Scholar]
- Koumber, R.M.; El-Gammaal, A.A. Inheritance and gene action for yield and its attributes in three bread wheat crosses (Triticum aestivum L.). J. Plant Prod. 2012, 3, 415–425. [Google Scholar] [CrossRef]
- Akhshi, K.C.; Nazarian-Firouzabadi, F.; Ahmadi, H. Generation mean analysis for yield components in common bean. Indian J. Plant Phys. 2014, 3, 1079–1085. [Google Scholar]
- Obiadalla-Ali, H.A.; Mohamed, N.E.M.; Glala, A.A. Heterosis and nature of gene action for yield and its components in faba bean ( Vicia faba L.). J. Plant Breed. Crop Sci. 2013, 5, 34–40. [Google Scholar] [CrossRef]
- Falconer, D.S. Introduction to Quantitative Genetics, 1st ed.; John Wiley & Sons, Inc.: New York, NY, USA, 1984. [Google Scholar]
- El-Refaey, R.A.; AEl-Gammal, A.; El-Moselhy, M.A.; El-Nagar, A.A. Quantative inheritance of some barley agronomic traits under water stress condition. Menaufia J. Plant Prod. 2017, 2, 39–51. [Google Scholar]
- Attia, S.M.; Salem, M.M. Analysis of yield and its components using diallel mating among five parents of faba bean. Egypt. J. Plant Breed. 2006, 10, 1–12. [Google Scholar]
- Mulugeta, A.T.; Hussein, M.A.; Habtamu, Z. Inheritance of Primary Yield Component Traits of Common Beans (Phaseolus vulgaris L.): Number of Seeds per Pod and 1000 Seed Weight in an 8 × 8 Diallel Cross Population. World Acad. Sci. Eng. Technol. Int. J. Biol. Biomol. Agric. Food Biotechnol. Eng. 2013, 7, 84–88. [Google Scholar]
- Salama, S.M.; Mohamed, N.A. Estimates of genetic components for some characters in faba bean (Vicia faba L.). Zagazig J. Agric. Res. 2004, 31, 2621–2634. [Google Scholar]
- Johnson, H.W.; Robinson, H.F.; Comstock, R.E. Estimates of Genetic and Environmental Variability in Soybeans. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Dixit, P.K.; Saxena, P.D.; Bahtia, L.K. Estimation of genotypic variability of some quantitative characters in groundnut. Indian J. Agric. Sci. 1970, 40, 197–201. [Google Scholar]
- Abou-Zaid, G.G.A. Genetical analysis of yield and its components for three faba bean crosses using six parameter models. Menaufia J. Plant Prod. 2018, 3, 119–133. [Google Scholar] [CrossRef]
- Aziz, A.; Osman, A.A.M. Variability, Heritability and Genetic Advance in Faba Bean, Vicia faba L. Int. J. Res. Agric. For. 2015, 2, 42–45. [Google Scholar]
- Nosair, H. SCoT polymorphism reveals genetic diversity in some important Fabaceae species. Curr. Sci. Int. 2016, 5, 592–598. [Google Scholar]
- Essa, S.M.; Wafa, H.A.; Mahgoub, E.-S.I.; Hassanin, A.A.; Al-Khayri, J.M.; Jalal, A.S.; El-Moneim, D.A.; Alshamrani, S.M.; Safhi, F.A.; Eldomiaty, A.S. Assessment of Eight Faba Bean (Vicia faba L.) Cultivars for Drought Stress Tolerance through Molecular, Morphological, and Physiochemical Parameters. Sustainability 2023, 15, 3291. [Google Scholar] [CrossRef]
- Heiba, H.E.; Mahgoub, E.; Mahmoud, A.M.A.; Ibrahim, M.; Mahdy, E.M.B. Genetic improvement of faba bean (Vicia faba L.) genotypes selected for resistance to chocolate spot disease. Agron. Colomb. 2022, 40, 186–197. [Google Scholar] [CrossRef]
- Bosily, M.A.; Noaman, M.M.; El-Banna, M.N.; Azzam, C.R.; Nassar, M.A. Breeding for barley resistance to leaf rust disease using marker-assisted selection. In Proceedings of the 7th Field CropsResearch Institute Conference, Gaza, Egypt, 18–19 December 2018. [Google Scholar]
- Serrote, C.M.L.; Reiniger, L.R.S.; Silva, K.B.d.; Rabaiolli, S.M.d.S.; Stefanel, C.M. Determining the Polymorphism Information Content of a molecular marker. Gene 2019, 726, 144175. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.; Skolnick, M.H.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Tessier, C.; David, J.; This, P.; Boursiquot, J.M.; Charrier, A. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor. Appl. Genet. 1999, 98, 171–177. [Google Scholar] [CrossRef]
- Albrifcany, M.; Askander, H.S.A. Genetic Diversity Among some Parental Varieties of Vicia faba L. Using ISSR and SCoT Molecular Marker Techniques. J. Agric. Chem. Biotechnol. 2022, 13, 129–137. [Google Scholar] [CrossRef]
- Kumawat, G.; Kumawat, C.K.; Chandra, K.; Pandey, S.P.; Chand, S.; Mishra, U.N.; Lenka, D.; Sharma, R. Insights into Marker Assisted Selection and Its Applications in Plant Breeding. In Plant Breeding Current and Future Views; IntechOpen: London, UK, 2020. [Google Scholar]
- Mahmoud, A.F.; Abd El-Fatah, B.E.S. Genetic Diversity Studies and Identification of Molecular and Biochemical Markers Associated with Fusarium Wilt Resistance in Cultivated Faba Bean (Vicia faba). Plant Pathol. J. 2020, 36, 11–28. [Google Scholar] [CrossRef]
- Loarce, Y.; Gallego, R.B.; Ferrer, E. A comparative analysis of the genetic relationships between rye cultivars using RFLP and RAPD markers. Euphytica 2004, 88, 107–115. [Google Scholar] [CrossRef]
- Bernier, C.C.; Hanounik, S.B.; Hussein, M.M.; Mohamed, H.A. Field Manual of Common Faba Bean Diseases in the Nile Valley; International Center for Agricultural Research in the Dry Areas (ICARDA) Information: Beirut, Lebanon, 1993; p. 3. [Google Scholar]
- Steel, R.G.D.; Torrie, J.H. Principles and Procedures of Statistics: A Biometrical Approach, 2nd ed.; McGraw-Hill Book Co. Inc.: New York, NY, USA, 1980. [Google Scholar]
- Mather, K.; Jinks, J.L. Biometrical genetics. Heredity 1972, 29, 101–102. [Google Scholar] [CrossRef][Green Version]
- Mather, K. Biometrical Genetics, the Study of Continuous Variation; Dover Puplications: London, UK, 1949. [Google Scholar]
- Miller, P.A.; Williams, J.C.; Robinson, H.F.; Comstock, R.E. Estimates of Genotypic and Environmental Variances and Covariances in Upland Cotton and Their Implications in Selection1. Agron. J. 1958, 50, 126–131. [Google Scholar] [CrossRef]
- Lazar, I. Gel Analyzer Freeware Developed by the Author. 2010. Available online: http://www.gelanalyzer.com/ (accessed on 1 April 2023).
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. iMEC: Online Marker Efficiency Calculator. Appl. Plant Sci. 2018, 6, e01159. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistical software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Société Vaudoise Des. Sci. Nat. 1908, 44, 223–270. [Google Scholar]

| Traits | Crosses | Mean and Variance | Parameters | |||||
|---|---|---|---|---|---|---|---|---|
| P1 | P2 | F1 | F2 | BC1 | BC2 | |||
| Plant Height | Cross 1 | Mean | 107.33 | 105.67 | 121.00 | 120.33 | 134.33 | 129.03 |
| Variance | 35.24 | 60.24 | 36.43 | 153.27 | 115.06 | 88.31 | ||
| Cross2 | Mean | 107.33 | 128.43 | 149.00 | 118.65 | 111.50 | 105.45 | |
| Variance | 35.24 | 20.53 | 29.29 | 128.70 | 91.64 | 99.06 | ||
| Crosse 3 | Mean | 141.67 | 128.43 | 140.33 | 122.70 | 132.00 | 112.83 | |
| Variance | 41.67 | 20.53 | 48.10 | 134.04 | 83.79 | 97.73 | ||
| No. of Branches/plant | Cross 1 | Mean | 4.33 | 5.20 | 7.00 | 4.02 | 5.40 | 4.11 |
| Variance | 0.95 | 0.74 | 1.14 | 2.89 | 1.63 | 2.50 | ||
| Cross 2 | Mean | 4.33 | 4.50 | 5.00 | 4.80 | 5.00 | 5.18 | |
| Variance | 0.95 | 0.96 | 1.29 | 3.86 | 3.24 | 2.03 | ||
| Cross 3 | Mean | 1.60 | 4.50 | 5.93 | 4.92 | 4.83 | 4.73 | |
| Variance | 0.54 | 0.96 | 0.21 | 3.29 | 1.87 | 3.24 | ||
| No. of Pods/Plant | Cross 1 | Mean | 37.47 | 42.27 | 49.47 | 29.51 | 40.20 | 46.42 |
| Variance | 64.12 | 39.21 | 39.70 | 241.36 | 56.65 | 247.34 | ||
| Cross 2 | Mean | 37.47 | 41.43 | 47.67 | 41.56 | 34.20 | 39.27 | |
| Variance | 64.12 | 50.24 | 56.10 | 265.01 | 131.82 | 194.31 | ||
| Cross 3 | Mean | 31.27 | 41.43 | 38.07 | 30.36 | 38.33 | 37.20 | |
| Variance | 26.78 | 50.24 | 38.07 | 274.49 | 164.57 | 204.51 | ||
| No. of Seeds/Plant | Cross 1 | Mean | 112.27 | 142.40 | 158.27 | 107.88 | 148.47 | 157.39 |
| Variance | 481.07 | 221.69 | 246.21 | 2860.71 | 1161.57 | 2387.56 | ||
| Cross 2 | Mean | 112.27 | 111.14 | 168.60 | 126.47 | 104.73 | 111.48 | |
| Variance | 481.07 | 214.69 | 445.69 | 2588.01 | 1324.20 | 1724.36 | ||
| Cross 3 | Mean | 67.87 | 111.14 | 123.13 | 95.68 | 113.43 | 96.23 | |
| Variance | 97.98 | 214.69 | 260.12 | 2103.87 | 787.08 | 1971.63 | ||
| Seed Yield/Plant | Cross 1 | Mean | 100.43 | 110.94 | 121.14 | 76.84 | 115.61 | 112.83 |
| Variance | 452.45 | 615.64 | 165.97 | 1393.11 | 702.02 | 1199.84 | ||
| Cross 2 | Mean | 100.43 | 62.85 | 101.96 | 78.41 | 68.23 | 69.92 | |
| Variance | 452.45 | 364.13 | 687.20 | 1282.24 | 1086.99 | 883.56 | ||
| Cross 3 | Mean | 32.40 | 62.85 | 70.68 | 50.83 | 58.57 | 51.01 | |
| Variance | 136.50 | 364.13 | 359.69 | 871.34 | 593.13 | 696.41 | ||
| chocolate spot | Cross 1 | Mean | 5.07 | 4.27 | 4.93 | 5.50 | 5.17 | 5.69 |
| Variance | 0.21 | 0.21 | 0.21 | 2.69 | 1.45 | 1.76 | ||
| Cross 2 | Mean | 5.07 | 5.93 | 5.27 | 5.40 | 4.00 | 3.76 | |
| Variance | 0.21 | 0.35 | 0.44 | 1.63 | 0.90 | 1.18 | ||
| Cross 3 | Mean | 3.07 | 5.93 | 4.33 | 4.10 | 3.70 | 3.67 | |
| Variance | 0.21 | 0.35 | 0.38 | 1.67 | 1.04 | 1.13 | ||
| Rust | Cross 1 | Mean | 6.20 | 6.07 | 5.00 | 5.73 | 5.47 | 6.25 |
| Variance | 0.17 | 0.07 | 0.01 | 0.95 | 0.88 | 0.25 | ||
| Cross 2 | Mean | 6.20 | 6.43 | 7.13 | 6.68 | 6.00 | 4.87 | |
| Variance | 0.17 | 0.24 | 0.55 | 1.96 | 1.31 | 1.16 | ||
| Cross 3 | Mean | 4.80 | 6.43 | 5.73 | 5.83 | 4.23 | 4.70 | |
| Variance | 0.31 | 0.24 | 0.35 | 2.08 | 0.94 | 1.67 | ||
| Traits | Crosses | Heterosis | Inbreeding Depression | Potence Ratio | |
|---|---|---|---|---|---|
| BP | MP | ||||
| Plant Height | Cross 1 | 12.74 ** | 13.62 ** | 0.55 | 17.47 |
| Cross 2 | 38.82 ** | 26.40 ** | 20.37 | −2.95 | |
| Cross 3 | −0.95 | 3.91 * | 12.56 | 0.80 | |
| No. of Branches/plant | Cross 1 | 34.62 ** | 46.90 ** | 42.57 ** | −5.14 |
| Cross 2 | 11.11 ** | 13.25 ** | 4.00 | −6.88 | |
| Cross 3 | 31.78 ** | 94.43 ** | 17.03 ** | −1.99 | |
| No. of Pods/Plant | Cross 1 | 32.03 ** | 24.08 ** | 40.33 * | −4.00 |
| Cross 2 | 27.22 ** | 20.84 ** | 12.82 | −4.15 | |
| Cross 3 | 21.75 * | 4.73 * | 20.25 | −0.34 | |
| No. of Seeds/Plant | Cross 1 | 40.97 ** | 24.29 ** | 31.84 | −2.05 |
| Cross 2 | 50.17 ** | 50.93 ** | 24.99 | 100.7 | |
| Cross 3 | 81.42 ** | 37.57 ** | 22.29 | −1.55 | |
| Seed Yield/Plant | Cross 1 | 9.19 * | 14.62 ** | 36.57 | −2.94 |
| Cross 2 | 24.89 ** | 24.89 ** | 23.1 | 1.08 | |
| Cross 3 | 12.46 ** | 48.41 ** | 28.08 | −1.51 | |
| Chocolate spot | Cross 1 | −2.76 ** | 5.57 ** | −11.56 ** | 0.65 |
| Cross 2 | −4.18 ** | −4.18 ** | −2.47 | 0.53 | |
| Cross 3 | 41.04 ** | −3.78 ** | 5.31 ** | 0.12 | |
| Rust | Cross 1 | −19.35 ** | −18.50 ** | −14.60 ** | −17.46 |
| Cross 2 | 12.91 ** | 12.91 ** | 6.31 ** | −7.09 | |
| Cross 3 | 19.38 ** | 2.05 ** | −1.75 | −0.14 | |
| Traits | Crosses | Gamble’s Parameters | |||||
|---|---|---|---|---|---|---|---|
| (m) | (a) | (d) | (aa) | (ad) | (dd) | ||
| Plant Height | Cross 1 | 120.33 ** | 5.30 ** | 51.90 ** | 37.40 ** | 4.47 | −109.12 ** |
| Cross 2 | 118.65 ** | 6.05 ** | −9.57 | −40.69 ** | 16.59 ** | 140.54 ** | |
| Cross 3 | 122.70 ** | 19.17 ** | 4.15 | −1.13 | 12.55 ** | 62.23 ** | |
| No. of Branches/plant | Cross 1 | 4.02 ** | 1.29 ** | 5.18 ** | 2.94 ** | 1.73 ** | 1.57 |
| Cross 2 | 4.80 ** | −0.18 | 1.75 | 1.16 | −0.1 | −2.69 | |
| Cross 3 | 4.92 ** | 0.1 | 2.34 | −0.55 | 1.55 ** | −0.62 | |
| No. of Pods/Plant | Cross 1 | 29.51 ** | −6.22 ** | 64.80 ** | 55.20 ** | −3.82 | −49.76 ** |
| Cross 2 | 41.56 ** | −5.07 | −11.08 | −19.29 ** | −3.09 | 46.58 ** | |
| Cross 3 | 30.36 ** | 1.13 | 31.35 ** | 29.63 ** | 6.21 | −31.86 ** | |
| No. of Seeds/Plant | Cross 1 | 107.88 ** | −8.92 | 211.14 ** | 180.20 ** | 6.15 | −220.71 ** |
| Cross 2 | 126.47 ** | −6.75 | −16.55 | −73.44 ** | −7.31 | 201.62 ** | |
| Cross 3 | 95.68 ** | 17.2 | 70.24 ** | 36.61 ** | 38.84 ** | −30.67 ** | |
| Seed Yield/Plant | Cross 1 | 76.84 ** | 2.78 | 164.98 ** | 149.52 ** | 8.04 | −152.75 ** |
| Cross 2 | 78.41 ** | −1.69 | −17.02 | −37.34 | −20.48 ** | 128.26 ** | |
| Cross 3 | 50.83 ** | 7.56 | 38.88 ** | 15.83 | 22.79 ** | 1.61 | |
| Chocolate spot | Cross 1 | 5.50 ** | −0.52 | −0.02 | −0.28 | −0.92 ** | −2.24 |
| Cross 2 | 5.40 ** | 0.24 | −6.32 ** | −6.08 ** | 6.73 ** | 12.10 ** | |
| Cross 3 | 4.10 ** | 0.03 | −1.83 ** | −1.67 ** | 1.46 ** | 4.60 ** | |
| Rust | Cross 1 | 5.73 ** | −0.78 ** | −0.62 | 0.52 | −0.85 ** | −1.69 * |
| Cross 2 | 6.68 ** | 1.33 ** | −4.57 ** | −5.39 ** | 1.45 ** | 10.95 ** | |
| Cross 3 | 5.83 ** | −0.47 | −5.33 ** | −5.45 ** | 0.35 | 10.28 ** | |
| Traits | Crosses | Heritability | Genetic Advance | ||
|---|---|---|---|---|---|
| Broad Sense (h2b) | Narrow Sense (h2n) | GS | GS % | ||
| Plant Height | Cross 1 | 71.31 | 67.31 | 17.17 | 14.03 |
| Cross 2 | 77.79 | 51.83 | 12.11 | 10.21 | |
| Cross 3 | 70.46 | 64.58 | 15.40 | 12.55 | |
| No. of Branches/plant | Cross 1 | 67.28 | 57.20 | 2.00 | 49.85 |
| Cross 2 | 70.92 | 63.47 | 2.57 | 53.52 | |
| Cross 3 | 85.41 | 44.68 | 1.67 | 33.93 | |
| No. of Pods/Plant | Cross 1 | 80.25 | 74.05 | 23.7 | 80.3 |
| Cross 2 | 78.63 | 76.94 | 25.8 | 62.08 | |
| Cross 3 | 86.05 | 65.54 | 22.37 | 73.68 | |
| No. of Seeds/Plant | Cross 1 | 88.94 | 75.94 | 83.67 | 77.55 |
| Cross 2 | 84.67 | 82.20 | 86.15 | 68.12 | |
| Cross 3 | 90.10 | 68.87 | 65.08 | 68.02 | |
| Seed Yield/Plant | Cross 1 | 70.47 | 63.48 | 48.81 | 63.52 |
| Cross 2 | 57.28 | 46.32 | 34.17 | 43.58 | |
| Cross 3 | 65.00 | 52.00 | 31.62 | 62.21 | |
| Chocolate spot | Cross 1 | 92.20 | 80.32 | 2.71 | 49.36 |
| Cross 2 | 77.91 | 72.39 | 1.90 | 35.26 | |
| Cross 3 | 80.24 | 70.06 | 1.87 | 45.49 | |
| Rust | Cross 1 | 91.63 | 81.01 | 1.62 | 28.34 |
| Cross 2 | 80.74 | 73.98 | 2.13 | 31.94 | |
| Cross 3 | 84.98 | 74.52 | 2.21 | 37.98 | |
| Marker Name | Total Bands | MMB | PMB | % Polymorphism | Number of Bands Associated with | |
|---|---|---|---|---|---|---|
| Resistance | Susceptibility | |||||
| SCoT1 | 93 | 66 | 27 | 29 | 1 at 260 bp | --- |
| SCoT2 | 77 | 66 | 11 | 14 | --- | 1 at 327 bp |
| SCoT3 | 47 | 0 | 47 | 100 | --- | 1 at 361 bp |
| SCoT4 | 54 | 0 | 54 | 100 | 1 at 207 bp | --- |
| SCoT5 | 79 | 0 | 79 | 100 | --- | 2 at 270 and 779 bp |
| SCoT6 | 56 | 0 | 56 | 100 | --- | 1 at 392 bp |
| SCoT7 | 116 | 33 | 83 | 72 | 1 at 178 bp | --- |
| Total | 522 | 165 | 357 | --- | 3 | 5 |
| Average | 75 | 24 | 51 | 74 | --- | --- |
| Index | Scored Band | H_0 | PIC_0 | E_0 | H.av_0 | MI_0 | D_0 | R_0 |
|---|---|---|---|---|---|---|---|---|
| SCoT 1 | 93 | 0.416 | 0.330 | 2.818 | 0.0032 | 0.0089 | 0.505 | 0.485 |
| SCoT 2 | 77 | 0.486 | 0.368 | 2.333 | 0.0037 | 0.0086 | 0.662 | 0.667 |
| SCoT 3 | 47 | 0.459 | 0.353 | 1.424 | 0.0035 | 0.0049 | 0.875 | 1.455 |
| SCoT 4 | 54 | 0.483 | 0.367 | 1.636 | 0.0037 | 0.0060 | 0.834 | 2.364 |
| SCoT 5 | 79 | 0.480 | 0.365 | 2.394 | 0.0024 | 0.0058 | 0.842 | 3.152 |
| SCoT 6 | 56 | 0.491 | 0.371 | 1.697 | 0.0050 | 0.0084 | 0.683 | 2.061 |
| SCoT 7 | 116 | 0.418 | 0.330 | 3.515 | 0.0025 | 0.0089 | 0.507 | 2.545 |
| Average | 0.462 | 0.355 | 2.260 | 0.0034 | 0.0074 | 0.701 | 1.818 | |
| Genotype | Pedigree | Origin | Reaction to Foliar Diseases |
|---|---|---|---|
| Nubaria 5 (P1) | landraces of Hamam 10 | * FCRI, ARC, Egypt | Resistant |
| Sakha 1 (P2) | Giza 716 × 620/283/85 | Resistant | |
| Giza 40 (P3) | Selection from Rebaya 40 | Highly susceptible | |
| Marina (P4) | Vicia faba L. | Hungary | Highly resistant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soliman, A.A.; Mousa, M.I.; Mosalam, A.M.; Ghareeb, Z.E.; Ibrahim, S.D.; Rehan, M.; Yu, H.; He, Y. The Potential Genetic Effect for Yield and Foliar Disease Resistance in Faba Bean (Vicia faba L.) Assessed via Morphological and SCoT Markers. Plants 2023, 12, 3645. https://doi.org/10.3390/plants12203645
Soliman AA, Mousa MI, Mosalam AM, Ghareeb ZE, Ibrahim SD, Rehan M, Yu H, He Y. The Potential Genetic Effect for Yield and Foliar Disease Resistance in Faba Bean (Vicia faba L.) Assessed via Morphological and SCoT Markers. Plants. 2023; 12(20):3645. https://doi.org/10.3390/plants12203645
Chicago/Turabian StyleSoliman, Alaa A., Manar I. Mousa, Abeer M. Mosalam, Zeinab E. Ghareeb, Shafik D. Ibrahim, Medhat Rehan, Haitian Yu, and Yuhua He. 2023. "The Potential Genetic Effect for Yield and Foliar Disease Resistance in Faba Bean (Vicia faba L.) Assessed via Morphological and SCoT Markers" Plants 12, no. 20: 3645. https://doi.org/10.3390/plants12203645
APA StyleSoliman, A. A., Mousa, M. I., Mosalam, A. M., Ghareeb, Z. E., Ibrahim, S. D., Rehan, M., Yu, H., & He, Y. (2023). The Potential Genetic Effect for Yield and Foliar Disease Resistance in Faba Bean (Vicia faba L.) Assessed via Morphological and SCoT Markers. Plants, 12(20), 3645. https://doi.org/10.3390/plants12203645








